Abstract
Human cytomegalovirus (HCMV) is known to carry host cell-derived proteins and mRNAs whose role in cell infection is not understood. We have identified a phospholipase A2 (PLA2) activity borne by HCMV by using an assay based on the hydrolysis of fluorescent phosphatidylcholine. This activity was found in all virus strains analyzed and in purified strains. It was calcium dependent and was sensitive to inhibitors of cytosolic PLA2 (cPLA2) but not to inhibitors of soluble PLA2 or calcium-independent PLA2. No other phospholipase activity was detected in the virus. Purified virus was found to contain human cellular cPLA2α, as detected by monoclonal antibody. No homology with PLA2 was found in the genome of HCMV, indicating that HCMV does not code for a PLA2. Decreased de novo expression of immediate-early proteins 1 and 2 (IE1 and IE2), tegument phosphoprotein pp65, and virus production was observed when HCMV was treated with inhibitors of cPLA2. Cell entry of HCMV was not altered by those inhibitors, suggesting the action of cPLA2 was postentry. Together, our results indicate a selective sorting of a cell-derived cPLA2 during HCMV maturation, which is further required for infectivity.
Human cytomegalovirus (HCMV) is a β herpesvirus that causes lifelong infection (27). Coevolution of the virus and host under strict immune pressure has directed the virus to develop many subversion mechanisms (3, 29). How HCMV takes advantage of the host's environment begins to be understood (13). These events include activation of NF-κB; extracellular signal-regulated kinases (ERK); transcription of Fos, Jun, and Myc; perturbation of the cell cycle (reviewed in reference 13); and increases of cellular mRNAs (36, 37). One of the mechanisms used by HCMV to exploit cellular metabolism could be the uptake of host proteins and RNA during virus maturation (9, 16, 21, 23, 24, 32, 34). However, a precise role for transport of cell-derived material by HCMV has not been determined up to now.
Phospholipase A2 (PLA2) comprises a family of enzymes that catalyze the hydrolysis of phospholipids at their sn-2 position, leading to the formation of fatty acids and lysophospholipids. Fatty acids such as arachidonic acid are transformed into prostanoids, which are mediators of inflammation, through the action of cyclooxygenases (31). Cytosolic PLA2s (cPLA2s) are members of the expanding superfamily of PLA2s (19, 31). Three isoforms of cPLA2 (α, β, and γ) have been described (31). The three isoforms share homology, but only cPLA2α and cPLA2β have a Ca-dependent lipid-binding domain and thus are calcium dependent. They translocate to the nucleus and endoplasmic reticulum membranes upon activation (19).
Most viruses modify the host cellular environment in order to maximize virus replication. Perturbations of cellular metabolism by HCMV include the induction of cPLA2 and cyclooxygenase 2 (COX-2) mRNAs (36, 37) and increase of cPLA2 activity in cells (1, 2, 25, 30). Inhibition of COX-2 blocks HCMV replication by decreasing immediate-early protein 2 (IE2) synthesis, which clearly emphasizes the role of this pathway in infection (37). Pathogens such as Escherichia coli (12) and Toxoplasma gondii (15) have been reported to depend on their host's cPLA2 activity for infection. Some viruses have been shown to code for their own lipolytic enzymes. For example, the major envelope protein of vaccinia virus, p37, exhibits multiple lipolytic activities (5, 7). It has been recently reported that a parvovirus contains a soluble PLA2 (sPLA2)-type activity encoded by the virus itself (14, 35).
Based on the observations that HCMV carries several cell-derived proteins and that PLA2s are important in infections by several pathogens, we made the hypothesis that HCMV could bear such activity. Our study reveals the presence in HCMV of a cPLA2 that is of cellular origin and is not encoded by the virus itself. This is the first description of a PLA2 selectively taken up from host cells by a virus, which is important to further infect target cells.
(This work was presented as an abstract at the 9th International Cytomegalovirus Workshop, abstr. J02, p. 80, Maastricht, The Netherlands, 20 to 25 May 2003.)
MATERIALS AND METHODS
Reagents.
2-Decanoyl-1-{O-[11-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-propionyl)amino]undecyl}-sn-glycero-3-phosphatidylcholine (BODIPY-PC) was from Molecular Probes, and 1-acyl-2-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl]-sn-glycero-3-phosphatidylcholine (NBD-PC) was from Avanti Polar Lipids. Both were used at 6 μM. Methyl arachidonyl fluorophosphonate (MAFP) irreversible inhibitor of group IV cPLA2 (Ca2+-dependent PLA2) was obtained from Calbiochem. E-6-(Bromomethylene) tetrahydro-3-(1-b naphthalenyl)-2H-pyran-2-one (HELSS [haloenol lactone suicide substrate] ST-340), an irreversible inhibitor of Ca2+-independent PLA2, and Manoalide (EI-177), an irreversible inhibitor of human sPLA2, were purchased from Biomol Research Laboratories. Indoxam and pyrrolidine 1 were kindly supplied, respectively, by Gerard Lambeau, IPMC, UMR 6097, Centre National de la Recherche Scientifique, Sophia-Antipolis, France, and Michael Gelb, Department of Chemistry and Biochemistry, University of Washington, Seattle. All inhibitors were used at 10 μM.
Antibodies.
Anti-cPLA2 monoclonal antibody (MAb) SC-1724 was a generous gift from J.-S. Saulnier-Blache (INSERM U317, Toulouse, France). Anti-pp65 (1C3) was purchased from Argene-Biosoft, Varilhes, France, and anti-IE1 and -2 (E13) were a generous gift from M.-C. Mazeron (Lariboisière Hospital, Paris, France). Anti-β-actin (AC-15) was from Sigma. Secondary antibodies were coupled to either peroxidase (Amersham Biosciences Europe, Orsay, France) or rhodamine (Beckmann Coulter, Roissy, France).
Cell culture.
MRC5 (BioMerieux, France) fibroblasts were grown in Dulbecco's modified Eagle's medium, containing 10% fetal calf serum (FCS) and supplemented with sodium pyruvate (1 mM), penicillin (100 IU/ml), streptomycin (100 μg/ml), and l-glutamine (2 mM) (DMEM-FCS) (Gibco BRL, Life Technologies, Cergy-Pontoise, France) at 37°C in a humidified incubator containing 5% CO2.
Virus.
The HCMV strains used were AD169 (purchased from the American Type Culture Collection), Towne (kindly provided by S. Michelson, Institut Pasteur, Paris, France), Toledo (kindly provided by J. Nelson, University of Portland, Portland, Oreg.), and TB40/E (kindly provided by C. Sinzger, University of Tübingen, Tübingen, Germany).
Preparation of virus stocks.
Subconfluent (60%) MRC5 cells were infected with HCMV laboratory strain AD169, Toledo, Towne, or TB40/E at a multiplicity of infection (MOI) of 0.01. HCMV TB40/E was passaged on endothelial cells, and stocks were prepared on MRC5 cells for no more than 4 rounds. Fourteen days later, virus was collected from cleared medium. Virus titration was performed as described previously (22). Briefly, supernatants were diluted and used to inoculate a monolayer of MRC5 cells in DMEM-FCS. After 24 h, the medium was replaced with 0.8% agarose in DMEM-FCS, and the culture was continued for 7 to 10 days until plaques were clearly observed. The monolayer was then fixed in 10% formaldehyde and stained with 0.05% methylene blue, and the plaques were counted under an inverted microscope.
Purified laboratory strain AD169 was purchased from ABI (Columbia, Md.). The Towne strain was purified by sorbitol gradient as previously described (21). Briefly, supernatants recovered from infected human fibroblasts were collected when the cytopathic effect was 100% and centrifuged to eliminate the cell debris. After an overnight incubation with 5% (wt/vol) polyethylene glycol 8000 at 4°C and slow agitation, the supernatant was centrifuged and the pellet was washed twice with phosphate-buffered saline (PBS), resuspended in TBS (0.05 M Tris-HCl, pH 7.4, 0.15 M NaCl), and subjected to sorbitol gradient (40 to 70%) purification (58,750 × g at 4°C). The viral “ring” recovered was pelleted, resuspended in TBS, and used or frozen at −80°C.
Western blotting.
HCMV (3 ×108 PFU) was washed three times by ultracentrifugation (100,000 × g, 30 min) in 50 mM Tris (pH 7.3)-2.5 mM CaCl2 (washing buffer). The virus pellet was solubilized in lysis buffer (20 mM Tris, pH 7.5, 1% Triton X-100, 10% glycerol, 10 mM NaCl, 5 mM MgCl2, 1 mM EDTA, protease inhibitor cocktail from Sigma) plus sonication (30 s). Lysates were cleared by centrifugation (15 min at 13,000 rpm at 4°C). MRC5 cells were harvested and lysed with lysis buffer. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide), blotted to nitrocellulose membrane, and incubated with antibodies against cPLA2, pp65, IE1 and IE2, and β-actin. Detection was performed with peroxidase-conjugated secondary antibodies (Amersham Biosciences Europe) and the ECL enhanced chemiluminescence detection kit (Amersham Biosciences Europe).
In some instances, separation of cytosol and membrane fractions with Triton X-114 was performed. Cells and virus were harvested and lysed in 20 mM Tris, pH 8, 10 mM NaCl, 5 mM MgCl2, 1 mM EDTA, with 1 mM dithiothreitol (DTT), 1% Triton X-114, and protease and phosphatase inhibitors (Sigma) as described above. Membrane and cytosolic proteins were separated in two phases—i.e., supernatant (containing cytosolic proteins) and pellet (containing membrane proteins)—by Triton X-114 partitioning at 30°C, as described by Bordier et al. (8), and were separated on an SDS-PAGE (10% polyacrylamide) and blotted to nitrocellulose membranes. Nitrocellulose membranes were then incubated with antibodies against cPLA2. Detection was performed as described above.
Evaluation of phospholipase activity of HCMV.
PLA2 activity was measured with a fluorescence-based assay. We have developed for PLA2 a sensitive method based on that described by Kemken et al. (20) for phospholipase D (PLD) detection, which utilizes a PC substrate containing either a BODIPY or NBD moiety, respectively, at the sn-1 and sn-2 position of the glycerol backbone.
Incubation of virus with lipids and lipid extraction.
When crude preparations of virus were used, HCMV (106 PFU) was washed three times by ultracentrifugation in 50 mM Tris (pH 7.3)-2.5 mM CaCl2 (washing buffer) or calcium- and magnesium-free PBS (PBS CMF) depending on the assay, and resuspended in 1 ml of PBS containing the fluorescent lipid, and incubation was performed for 30 min at 37°C.
Purified virus was used according to the concentration of total proteins. Virus was incubated for 30 min at 37°C with BODIPY-PC and NBD-PC (both 6 μM). Reaction was terminated by adding 1 ml of butanol-1, which extracted fluorescent products as well. Samples were then vortexed and centrifuged (5 min at 16,000 × g). The aqueous fraction was discarded, and the remaining lipids in the butanol phase were collected to be analyzed.
TLC and HPLC analyses of PLA2 activity.
For thin-layer chromatography (TLC), samples were dried under N2, resuspended in 20 μl of 1/1 (vol/vol) chloroform-methanol, and loaded onto TLC plates. TLC was performed either with 45/45/10 (vol/vol) chloroform-methanol-H2O, when BODIPY-PC was used as the substrate, or with 65/35/5 (vol/vol) chloroform-methanol-NH4-OH when NBD-PC was used as the substrate. Fluorescence was revealed under UV light.
When high-performance liquid chromatography (HPLC) separation was performed, each sample was dried under N2 and resuspended in injection solvent, composed of 50/50/1/0.08 (vol/vol) hexane-isopropanol-acetic acid-triethylamine. Lipid separation was accomplished by normal-phase HPLC on a LiChrosphere 100 DIOL (5 μm) column (250 by 3 mm) (Cluzeau Info Labo, Ste. Foy La Grande, France). The analyses were carried out by binary gradient elution with mobile-phase solvents of 82:17:1 (vol/vol) hexane-isopropanol-acetic acid (solvent A) and 85:14:1 (vol/vol) isopropanol-water-acetic acid (solvent B) at a flow rate of 0.4 ml/min. Triethylamine (0.08% [vol/vol]) was added to the solvents. The gradient profile started at 5% for solvent B and was increased to 40% solvent B in 25 min, after which it was increased to 100% of solvent B in 5 min. The HPLC column was regenerated by a switch from 100% solvent B to the initial mobile phase (5% solvent B) within 8 min and maintained at 5% solvent B for another 20 min. Aliquots (20 μl) of lipid samples were injected onto the column, which was kept at 45°C in all runs. The HPLC system was interfaced with a fluorescence detector (SFM 25; Kontron Instruments) set at excitation and emission wavelengths of 475 and 515 nm, respectively. The detector signal was recorded and integrated by a personal computer and a chromatography data software program (KromaSystem 3000; Bio-Tek Instruments Srl). Spontaneous hydrolysis of NBD-PC and BODIPY-PC, when observed, was <1.5% in all experiments and was subtracted from experimental values. Several washes of virus stocks were performed in all experiments in order to eliminate potential contaminating PLA2 originating from serum.
Characterization of PLA2 activity.
PLA2 inhibitors were used at a concentration of 10 μM, which was determined to produce optimal effects in preliminary experiments. Virus was washed three times by ultracentrifugation and resuspended in 1 ml of washing buffer. Washed virus (106 PFU) was incubated in the presence or absence of inhibitor for 15 min at room temperature and then washed again three times. Fluorescent lipids were then added, and incubation was performed for 30 min at 37°C.
Evaluation of Ca2+ dependency.
Virus (106 PFU) was washed in PBS CMF and incubated for 45 min at room temperature in 1 ml of PBS CMF or PBS supplemented only with Mg2+ (10 mM). When indicated, Ca2+ (50 mM) was added 15 min prior to addition of fluorescent lipids and incubation was performed for 30 min at 37°C.
Confocal microscopy.
MRC5 cells were seeded on glass coverslips in six-well plates (Nunc) at a density of 2.5 × 105 cells/well. At the indicated times, cells were washed twice with PBS, fixed in PBS with 3% paraformaldehyde for 20 min, washed, permeabilized with 0.1% Triton X-100 in PBS for 5 min, and then washed again in PBS. pp65 and IE1 and -2 were detected with specific MAbs followed by rhodamine-conjugated antimouse immunoglobulin G (IgG) antibody (Beckmann Coulter, Roissy, France). Fluorescence was analyzed with a Zeiss LSM 510 confocal microscope (Zeiss, Jena, Germany) or a Nikon TE200 inverted fluorescence microscope. Three-dimensional images were reconstructed by using the API Deltavision software system (Applied Precision).
Sequence analysis.
Here, we will briefly sketch the bioinformatic method PRIAM described in more detail elsewhere (11), for the detection of potential enzymatic activities, here PLA2, in a fully sequenced genome, here HCMV. Protein sequences belonging to the cytosolic PLA2 family, as indicated by the ENZYME database (6), were downloaded from SWISSPROT and used to construct profile position-specific scoring matrices in the following manner: a multiple alignment is constructed for the cytosolic PLA2 protein sequences, and the associated profile is a scoring scheme that takes into account the distribution of amino acids at each alignment position (4). These descriptors are subsequently used to search the genome under study, using the PSI-BLAST program (4), for protein sequences sharing sequence similarities with the cPLA2 family. The same procedure was applied to the proteins corresponding to the sPLA2 family. Detection of the sPLA2-type activity in parvovirus was used as a positive control of our analyses.
RESULTS
Detection of PLA2 activity in HCMV.
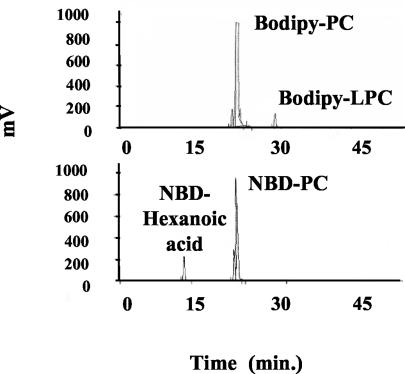
We have used a sensitive assay for PLA2 activity detection that utilizes fluorescent PC substrates (i.e., BODIPY-PC and NBD-PC), with fluorescent moiety labeled fatty acids, respectively, on the sn-1 and sn-2 positions. Cleavage of these substrates by PLA2 liberates fluorescent compounds, BODIPY-LPC and NBD-hexanoic acid, respectively, that can be readily separated from the uncleaved substrate when run on HPLC, and quantified. Figure 1 shows the resulting PLA2-specific metabolites upon incubation with washed AD169 HCMV. HPLC analysis of hydrolysis of BODIPY-PC and NBD-PC by HCMV revealed peaks with the same retention times as standard LPC (32 min) and hexanoic acid (12 min), respectively (Fig. 1). Therefore, a PLA2 activity was detected in HCMV upon incubation with these fluorescent substrates. Interestingly, no other type of phospholipase activity was detected. Fluorescent substrates can also be hydrolyzed by PLC and PLD, but neither diglycerides nor phosphatidic acids were recovered in any experiment (Fig. 1).
FIG. 1.
Hydrolysis of fluorescent substrates by HCMV. Representative HPLC profiles of hydrolysis of BODIPY-PC and NBD-PC by HCMV (AD169, 106 PFU) after 30 min of incubation at 37°C. Lipid extraction and HPLC analysis of fluorescent metabolites were performed as described in Materials and Methods.
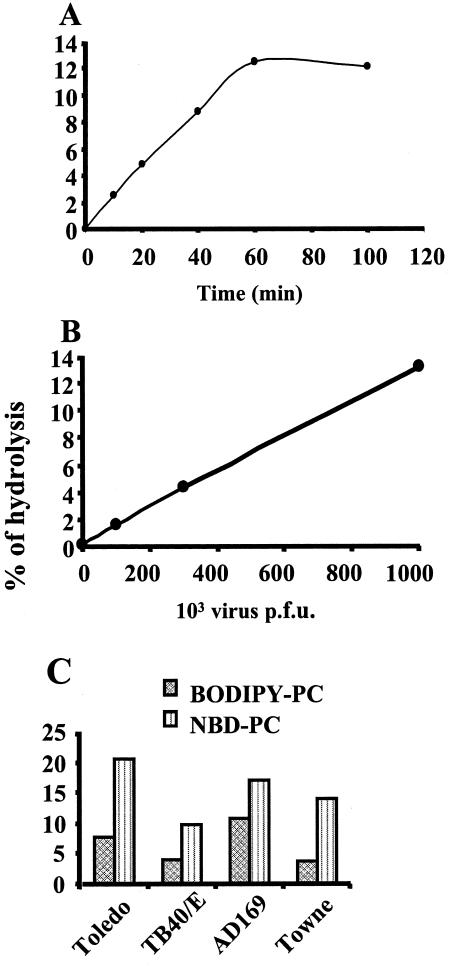
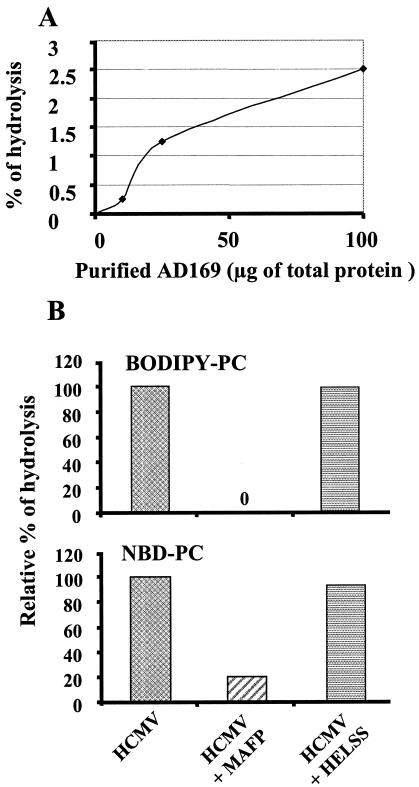
We then tested whether the hydrolysis of fluorescent substrate BODIPY-PC was dependent on the time of incubation and the amount of HCMV added. Figure 2A shows that cleavage of BODIPY-PC was detectable as early as 10 min postincubation and reached a plateau after 60 min of incubation. With regard to the quantity of virus, there was a linear relationship between the number of PFU and the amount of LPC obtained from the hydrolysis of BODIPY-PC as detected by HPLC (Fig. 2B). The specific activity was 135 nmol/h/mg of protein. We then tested whether the PLA2 activity of HCMV could be detected in various strains. Both clinical (Toledo and TB40/E) and laboratory (Towne and AD169) HCMV strains were shown to bear PLA2 activity (Fig. 2C), thus extending our initial observation.
FIG. 2.
Effects of time course, dose of virus, and virus strain on PLA2 activity of HCMV. (A) Effect of time course on hydrolysis of BODIPY-PC by HCMV. HCMV (AD169, 106 PFU) was incubated with BODIPY-PC over a time course of 100 min. (B) Effect of increasing amounts of HCMV on hydrolysis of BODIPY-PC. (C) HCMV was incubated with BODIPY-PC for 30 min at 37°C. HCMV strains Toledo (5.105 PFU), TB40/E (2.105 PFU), AD169 (106 PFU), and Towne (5.105 PFU) were analyzed for hydrolysis of NBD-PC and BODIPY-PC. Background hydrolysis was substracted from experimental values. Lipid extraction and HPLC analysis of fluorescent metabolites were performed as described in Materials and Methods. Data are representative of three similar independent experiments.
Characterization of PLA2 activity. (i) Ca2+ dependency.
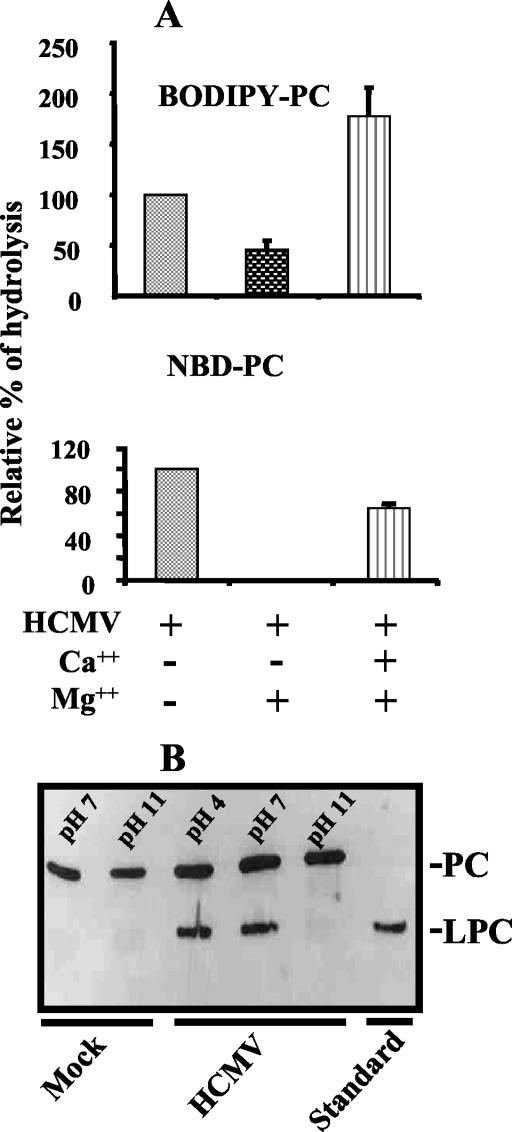
To characterize the PLA2 activity borne by HCMV, Ca2+ dependency was analyzed with high concentrations of Mg2+, which is known to displace Ca2+ from its specific site on cPLA2. Using BODIPY-PC as a substrate, we showed that Mg2+ reproducibly inhibited the hydrolysis of PC (Fig. 3A). When NBD-PC was used, we showed that adding an excess of Mg2+ completely abolished NBD-PC hydrolysis (Fig. 3B). In both cases, an excess of Ca2+ restored the hydrolysis of PC. These experiments further characterized the PLA2 activity as Ca2+ dependent.
FIG. 3.
Calcium and pH dependency of HCMV PLA2 activity. HCMV (AD169, 106 PFU) was incubated for 45 min at room temperature in the presence of MgCl2 (10 mM). Where indicated, CaCl2 (50 mM) was added prior to incubation of HCMV with BODIPY-PC and NBD-PC (A). Error bars represent standard deviations of the mean from three independent experiments. HCMV (106 PFU) was incubated with BODIPY-PC for 30 min at 37°C under different pH conditions. Hydrolysis was tested by TLC (B).
(ii) pH dependency.
The PLA2 activity of HCMV was also observed by TLC to be pH dependent, as shown in Fig. 3B. The absence of activity at basic pH 11 was characteristic of cytosolic PLA2. Further characterization was carried out in subsequent experiments.
(iii) Use of specific inhibitors.
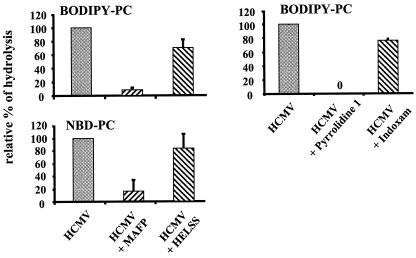
We used specific inhibitors of various classes of PLA2 to more precisely identify the HCMV-associated PLA2 activity. Data using specific inhibitors (Fig. 4) showed that only inhibitors of cPLA2 (MAFP and pyrrolidine 1) but not inhibitors of sPLA2 (indoxam) or calcium-independent PLA2 (iPLA2) (HELSS) reduced hydrolysis of fluorescent PC. This strongly suggested that HCMV was bearing a cPLA2. Similar data were obtained with purified AD169 (Fig. 5). Hydrolysis of BODIPY-PC was tested in the presence of increasing amounts of purified AD169 (Fig. 5A). There was no spontaneous hydrolysis of substrate in this experiment. The relatively low percentage of hydrolysis may be explained by the fact that the purified virus was not infectious and may have lost part of its PLA2 activity during the purification process. The specific activity was evaluated at 2.5 nmol/h/mg of protein. Inhibitors MAFP and HELSS (Fig. 5B) gave results similar to that of washed AD169 on both BODIPY-PC and NBD-PC. Thus, a cPLA2 was identified in purified virus.
FIG. 4.
Effects of inhibitors on PLA2 activity of AD169 HCMV. HCMV (AD169, 106 PFU) was incubated with MAFP, HELSS, pyrrolidine 1, and indoxam. Virus was then washed twice by ultracentrifugation after treatment and incubated with BODIPY-PC and NBD-PC. Error bars represent standard deviations of the mean from three independent experiments. Lipid extraction and HPLC analysis of fluorescent metabolites were performed as described in Materials and Methods. One hundred percent relative hydrolysis was used to represent the hydrolysis of fluorescent substrates observed in the presence of HCMV. The absolute values ranged from 3 to 7% for NBD-PC and 4 to 12% for BODIPY-LPC.
FIG. 5.
Analysis of PLA2 activity of purified HCMV. Various concentrations of purified HCMV AD169 were incubated with BODIPY-PC (A). Purified HCMV (AD169, 50 μg) was incubated with MAFP and HELSS. Virus was then incubated with fluorescent substrates BODIPY-PC and NBD-PC (B). This experiment is representative of two separate experiments (the other one was performed on purified Towne strain; data not shown). Lipid extraction and HPLC analysis of fluorescent metabolites were performed as described in Materials and Methods.
Sequence analysis.
The protein sequences coding for cPLA2 enzymatic activity are synthetically described by a single profile spanning more than 95% of the cPLA2 isoforms (i.e., the multiple alignment used to construct the profile spans more than 95% of the α, β, and γ isoforms). No significant sequences similarities were observed between the predicted proteins of HCMV (AD169 and Toledo strains) and this profile (e-value of >0.1). In addition, an extensive search of the GXSGS motif known to be a consensus lipase motif characteristic of the cPLA2 family (10, 28) was conducted and gave no convincing homology: the only HCMV protein sharing this motif, UL57, shows no sequence similarity with the cPLA2 sequences (e-value of >1). In order to exclude potentially unannotated proteins for this genome, a search was done within the complete HCMV genome, translated in the six possible frames, but produced no significant results. In contrast, the profile characterizing the sPLA2 activity showed significant homology (e-value of <0.05) to the vp1 protein of the parvovirus, with the identified conserved region clearly surrounding the previously described HDXXY conserved motif (35). In conclusion, this sequence analysis excludes a cPLA2 protein encoded by HCMV or a protein homologous to the known cPLA2 proteins.
Identification of a cPLA2 protein in HCMV.
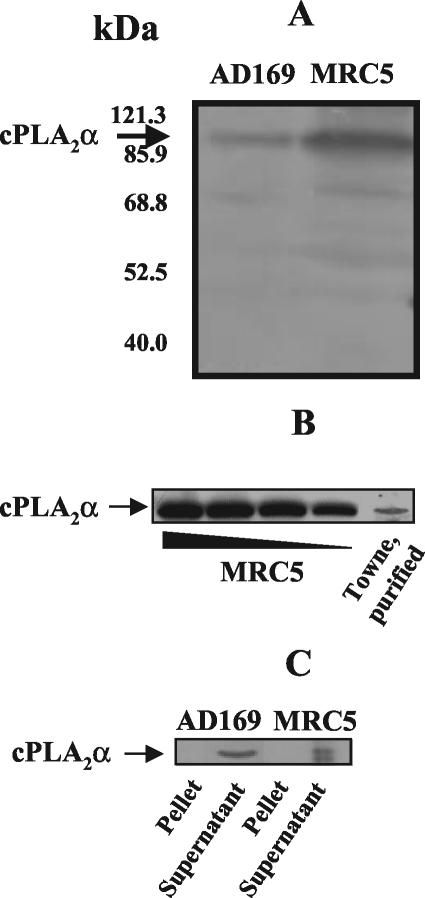
We then tested the hypothesis that HCMV was carrying a cPLA2 that would be detected with a MAb specific for cellular cPLA2. As shown in Fig. 6, Western blots using an antibody specific for human cPLA2α detected a band that comigrated with the cPLA2 observed in the MRC5 cells, whose infection was used for the production of HCMV stocks (Fig. 6A). Similar data were obtained by using gradient-purified Towne HCMV (Fig. 6B). Therefore, HCMV bears a cPLA2 that is not encoded by its genome and is detected by a MAb specific for cellular cPLA2. Partition of membrane-associated and soluble proteins from both HCMV and MRC5 cells showed that cPLA2 was contained in the soluble fraction of Triton X-114-treated cell lysates (Fig. 6C). This observation demonstrated that the cPLA2 was not associated with virus membranes, indicating that the enzyme is likely to be located inside the virus particle. In that respect, no activity could be detected when intact virus was incubated with liposomal substrates (not shown). Instead, the use of permeant fluorescent analogs of PC made the PLA2 activity available to substrates, suggesting an intraviral PLA2 activity.
FIG. 6.
Western blot analyses of cPLA2 in MRC5 cells and HCMV. Samples (cells, 50 μg; and AD169, 100 μg) were analyzed by Western blotting with a cPLA2α-specific MAb (A). The presence of cPLA2α in purified virus (Towne strain; protein amount, 300 μg) was tested, using a cPLA2α-specific MAb, in comparison with various amounts of total proteins from MRC5 cells (125, 100, 75, and 50 μg) (B). Proteins (150 μg/sample) from cells and HCMV (AD169) were separated in two phases with Triton X-144. Thirty micrograms of proteins from supernatant (containing cytosolic proteins) and pellet (containing membrane proteins), respectively, was analyzed in a Western blot with cPLA2α-specific MAb (C).
Role of cPLA2 activity in virus entry.
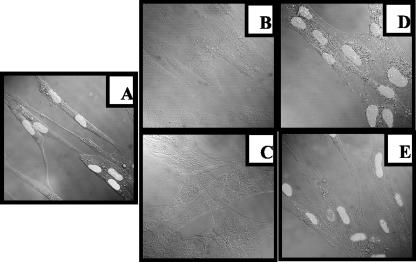
We next explored the possibility that HCMV cPLA2 might be involved in infection of cells by monitoring the fate of tegument protein pp65 inside the cell. pp65 protein is a viral structural protein that is rapidly translocated to the nucleus of infected cells. We first ascertained that soluble pp65 was not contained in the inoculum. Several washes routinely performed before and after treatment with cPLA2 inhibitors (see Materials and Methods) completely abolished detection of soluble pp65 in the inoculum (data not shown), as was previously described for IE1 (22). Figure 7A depicts the intranuclear immunofluorescence staining of pp65 after 12 h of infection (MOI = 3). Preincubation of HCMV with the irreversible cPLA2 inhibitors MAFP (Fig. 7B) and pyrrolidine 1 (Fig. 7C) dramatically reduced the intensity and the number of cells expressing pp65, while incubation with HELSS (Fig. 7D) and indoxam (Fig. 7E) had no effect. Similar results were observed up to 96 h postinfection (data not shown). As a control, electron microscopy experiments showed that treatments with the previously mentioned inhibitors had no effect on virus morphology (data not shown).
FIG. 7.
Infection of cells by HCMV is dependent on cPLA2. MRC5 cells were seeded on glass coverslips in six-well plates (2.5 × 105/well), deprived of FCS for 16 h, and infected for 12 h with HCMV (AD169, MOI = 1) in the absence of inhibitors (A). Preincubations of HCMV with PLA2 inhibitors MAFP (B), pyrrolidine 1 (C), HELSS (D), and indoxam (E) were used to assess for PLA2 dependency. Cells were then stained with specific anti-pp65 MAb. Detection of nuclear pp65 expression was observed with a confocal microscope (original magnification, ×60).
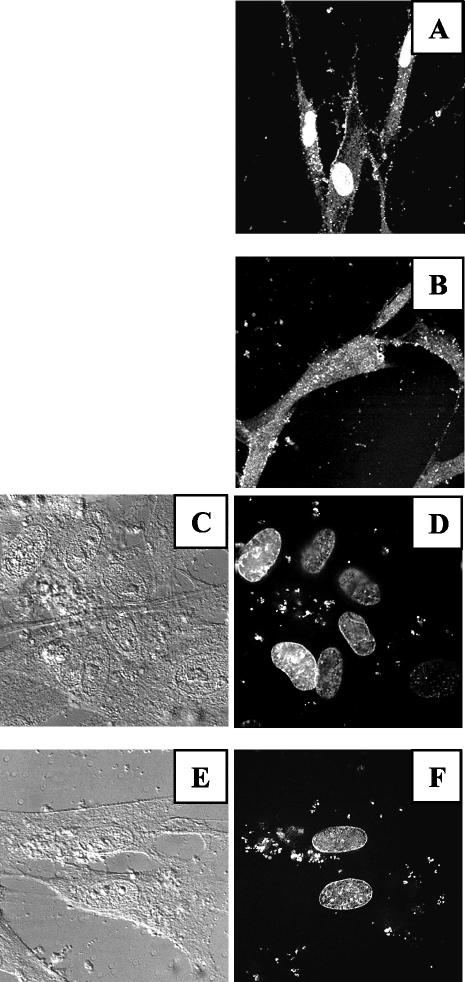
To determine whether cPLA2 was involved in virus entry, we monitored tegument pp65 protein as a marker of virus entry. We used a higher MOI (100 PFU/cell) in order to be able to visualize pp65 in the cytoplasm shortly after virus entry (Fig. 8A). Pretreatment of HCMV with MAFP did prevent translocation of pp65 into the nucleus but not its entry into the cytoplasm, as observed by confocal microscopy (Fig. 8B). It must be stressed that controls were performed to exclude the presence of soluble pp65 in our virus preparations, as already published for IE1 (22). The presence of pp65 in cells was therefore likely to be due to virus entry. To further control that the detection of pp65 was indeed reflecting the entry of virions, the presence of viral DNA in the cytoplasm was verified by using 4′,6′-diamidino-2-phenylindole (DAPI)-labeled HCMV. Figure 8D shows intense intracellular DAPI staining, as observed with a deconvolution microscope, in comparison with a phase-contrast image of the same field (Fig. 8C). It is noteworthy that when using DAPI-labeled HCMV, staining showed leakage of free DAPI to the nucleus as well. This nuclear staining was not due to labeling of viral DNA, but rather of cell DNA, since control experiments performed at 4°C (thus not allowing for virus entry) showed the same pattern (data not shown). Staining of both cellular and viral DNA was thus conveniently used to control that the detection of viral DNA was indeed in the same focal plan as the nucleus (i.e., intracellular). The amount of cytoplasmic viral DNA underwent no modification when HCMV was treated with MAFP (Fig. 8F versus phase-contrast microscopy in Fig. 8E). Altogether, these experiments demonstrated that the role of HCMV-borne cPLA2 was at a postentry step.
FIG. 8.
The role of cPLA2 in infection is at a postentry step. MRC5 cells were seeded on glass coverslips in six-well plates (2.5 × 105/well), deprived of FCS for 16 h, and incubated for 1 h at 4°C with HCMV (AD169, MOI = 100) untreated (A) or treated (B) with MAFP. Unadsorbed virus was removed with PBS, and cells were further incubated at 37°C for 4 h. Staining was performed with anti-pp65 MAb and observed by confocal microscopy (original magnification, ×60). For panels C to F, MRC5 cells were treated as described for panels A and B, except that DAPI was used to visualize viral DNA. Shown are the phase and DAPI fluorescence pictures for cells infected with untreated HCMV (C and D) or MAFP-treated virus (E and F). Cells were visualized by confocal microscopy (original magnification, ×60).
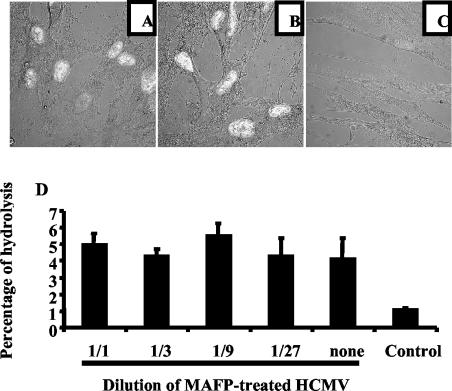
Those experiments suggested that cPLA2 was involved in infection of cells. Extensive washes of virus following treatment with MAFP and pyrrolidine 1 were used to avoid carryover of inhibitor, which could interfere with the host cell cPLA2. Nevertheless, we performed control experiments to exclude this possibility. First, HCMV was treated with MAFP and mixed with untreated HCMV. Figure 9A shows that under such conditions, infection was similar to what was observed with untreated HCMV (Fig. 9B) as measured by IE1 and IE2 expression. As expected, MAFP-treated HCMV did not infect MRC5 cells (Fig. 9C). Therefore, inhibition of infection by MAFP was strictly limited to HCMV placed in the presence of inhibitor, and there was no carryover of inhibitor. To totally exclude that residual MAFP might interfere with cellular PLA2, we incubated MRC5 cells with MAFP-treated HCMV and measured cellular PLA2 activity. Figure 9D shows that this treatment had no effect on the cellular PLA2 activity. As a control, incubation of MRC5 cells with MAFP inhibited cellular PLA2 activity by 76%. This demonstrated that the inhibitory effect of MAFP was not at the cell level but rather at the level of HCMV.
FIG. 9.
Inhibition of infection by MAFP is due to specific inhibition of HCMV-borne, but not cell-borne, cPLA2. MRC5 cells were seeded on glass coverslips in six-well plates (2.5 × 105/well), deprived of FCS for 16 h, and incubated with a mixture of HCMV plus MAFP-treated HCMV (A), untreated HCMV alone (B), and MAFP-treated HCMV alone (C) (AD169, MOI = 0.1) for 1 h at 37°C. Unadsorbed virus was removed with PBS, and cells were further cultured for 5 days. Expression of IE1 and IE2 was analyzed by confocal microscopy (original magnification, ×60). (D) MRC5 cells were seeded in 24-well plates, deprived of FCS for 16 h, and then incubated for 2 h with dilutions of HCMV (original MOI = 0.1) treated with MAFP or not treated. The control represents cells incubated with MAFP only. Cells were washed, harvested, and lysed by sonication. Lysates were incubated for 1 h with BODIPY-PC, and lipid extraction and HPLC analysis of fluorescent metabolites were performed as described in Materials and Methods.
Role of HCMV PLA2 activity in the production of viral proteins and of virus progeny.
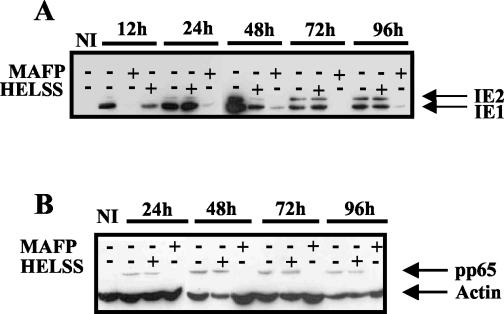
We further looked at the expression of viral proteins IE1, IE2, and pp65 in Western blots as markers of progression of infection. As seen in Fig. 10, incubation of HCMV with MAFP induced a dramatic decrease of IE1, IE2, and pp65 altogether. This effect was observed during the whole duration of the assay (96 h). Expression of IE1 and IE2 was not affected by HELSS, except at 12 and 48 h postinfection. This may be due to a different amount of loaded proteins, as suggested by the actin control. However, pp65 expression was not affected by HELSS, and the slight decrease of IE1 and IE2 did not have any influence on virus production whatsoever (see below). The inhibition of virus entry by inhibitors of cPLA2 correlated well with inhibition of HCMV protein synthesis. Thus, HCMV cPLA2 is crucial in the steps of infection required for synthesis and expression of viral proteins.
FIG. 10.
Western blot analysis of viral proteins production after treatment of HCMV with PLA2 inhibitors. MRC5 cells (5 × 105) were seeded in 25-cm2 flasks, deprived of FCS for 16 h, and infected with HCMV previously treated with MAFP or HELSS. Cell lysates obtained at the indicated time points were analyzed by Western blot using anti-IE1 and -IE2 (A) and anti-pp65 (B) MAbs. Antiactin MAb was used for both panels as a control of the total amount of protein.
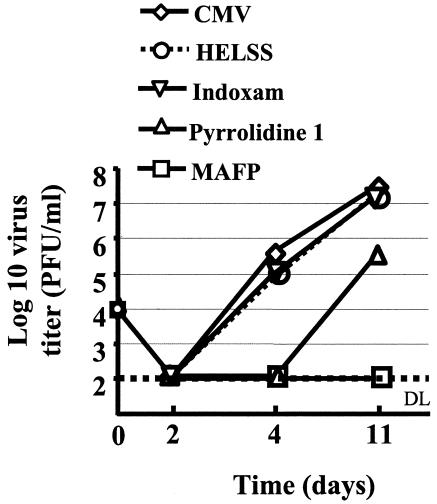
We then tested whether inhibition of infection was reducing the production of virus progeny. HCMV was thus treated with MAFP, pyrrolidine 1, HELSS, or indoxam; washed; and used to infect MRC5 cells. The production of HCMV was then measured in a plaque assay. We observed that MAFP totally abolished virus production while inhibitors of iPLA2 and sPLA2 had no effect (Fig. 11). Pyrrolidine 1 had the same effect as MAFP at day 4 postinfection and inhibited virus production by 2 logs at day 11 postinfection.
FIG. 11.
HCMV production is dependent on cPLA2. MRC5 cells (5 × 105) were seeded in 25-cm2 flasks, deprived of FCS for 16 h, and infected with HCMV (MOI = 0.1) previously treated with MAFP, HELSS, pyrrolidine 1, or indoxam. Supernatants were collected at the indicated times and were tested in duplicate in a virus titration assay. DL, detection limit. Standard deviations were lower than 25% and were omitted for clarity.
DISCUSSION
In this study, we have identified the presence of a cell-derived cPLA2 in HCMV. The enzymatic activity of this cPLA2 was required for infection of cells, since its inhibition resulted in the decrease in viral protein synthesis and virus titers. The dependency of infection on cPLA2 was localized at a post-cell-entry step. Our observation represents an important subversion mechanism of cell metabolism used by HCMV to infect cells.
We have used a sensitive technique taking advantage of soluble fluorescent PC substrate (18, 20). This approach proved to be much more sensitive than the otherwise used liposome technique (30). It appears that the substrate had to penetrate the virus to allow the monitoring of phospholipase activity. This has been possible with the permeant fluorescent PC used in this study. The PLA2 detected in HCMV could be ascribed to the cPLA2 family for several reasons: (i) dependency on Ca2+, (ii) pH dependency, (iii) inhibition by specific inhibitors of cPLA2, and (iv) more precisely, among the cPLA2 family, the fact that identification of cPLA2α relied on its detection by a specific anti-cPLA2α MAb and its apparent molecular weight (19). The absence of homology of open reading frames encoded by HCMV with any motif characteristic of PLA2 strongly argues in favor of a cellular origin of the cPLA2α carried by HCMV. Since no other phospholipase activity was detected in virus preparations, it is likely that the cPLA2 activity detected results from a selective sorting of proteins from the host cell during HCMV maturation. This sorting could be subsequent to the presence of virion proteins engaging in a strong interaction with the host cell cPLA2. Selective sorting also rules out a possible passive contamination of virus preparations by host cell PLA2 proteins.
Inhibitors of cPLA2 did not prevent entry of pp65 and viral DNA into cells. Since (i) no free pp65 was found in washed virus preparations and (ii) entry of pp65 in cells correlated with entry of viral DNA, we concluded that the importance of cPLA2 in infection was at a postentry step. This is in accordance with experiments using Triton X-114, which showed that the cPLA2α is not associated with virion envelope. However, additional experiments are required to test whether the inhibition of infection is related to inhibition of nuclear translocation of virus genetic material.
The involvement of cellular PLA2 activities in infections has been described for various pathogens, including parasites (15) and bacteria (12, 26). In addition, phospholipase activities have been reported to be involved in viral infections. For instance, the parvovirus capsid protein, vp1, which bears an sPLA2 activity, is required for infectivity (35). The envelope protein p37 from vaccinia bears multiple lipase activities, including PLA2 (5). Deletion of the gene coding for p37 results in the loss of cell-to-cell transmission and the inability to produce enveloped extracellular virus (7).
The signaling pathways linked to host cell PLA2 has been shown to be involved in HCMV infection in several ways. (i) HCMV infection stimulates arachidonic acid metabolism associated with activation of PLA2 (1, 2, 25) and a cellular cPLA2 has been shown to be activated in the first 15 min postinfection (30). (ii) Both mRNAs encoding for cPLA2 and COX-2 are increased in infected fibroblasts (36, 37). (iii) Blocking the cellular pathway of PLA2 signaling inhibited infection (32, 33, 37). Finally, (iv) our present paper indicates that a cPLA2 taken up by virus particles from infected cells plays a role in infection. Although all those reports link HCMV infection to PLA2, they seem to point to different levels of the cPLA2 pathways. It will be of interest to determine how these pathways cooperate in the development of HCMV infection. The inhibition, by specific inhibitors, of cPLA2 contained in the virus preparations clearly demonstrated that the cellular cPLA2 activated within the host cell at the moment of infection could not compensate for the acquired cPLA2 activity carried by HCMV. Downstream effectors of the cellular PLA2 signaling pathway such as COX-2 have been previously reported to be involved in HCMV infection (25, 30, 37). However, they seem to be distinct from those observed in our experiments. This is particularly evident from our experiments showing that IE1, as well as IE2 and pp65, was inhibited, whereas the use of COX-2 inhibitors selectively impaired the production of IE2 (37). Thus the inhibition of HCMV-borne cPLA2 had broader consequences on HCMV infection. We also excluded that the treatment of virus with MAFP would interfere with cellular cPLA2 and showed that the inhibition was specific to the MAFP-treated virus (Fig. 9). Therefore the cPLA2 pathways appear to be involved at different steps, in an additive way, in infection by HCMV. Besides the previously reported role of the host cPLA2 (37), we bring new insight related to the key role of the cPLA2 carried by the virion.
It is intriguing why HCMV, contrary to parvovirus and vaccinia virus, has developed a mechanism of “highjacking” cellular cPLA2α rather than coding for its own PLA2. With regard to PP2A, it is noteworthy that, similar to cPLA2α, while HCMV carries a cell-derived PP2A (24), other viruses (vaccinia virus and baculovirus) code for such activity (17). HCMV has been known to carry several host-derived proteins: CD55 and CD59, as regulators of complement may interfere with the complement cascade (32), and serine/threonine phosphatase PP2A may contribute to perturb cell metabolism (24). Other activities found in HCMV particles include a DNase activity (21), a DNA polymerase activity (23), and annexin II (34).
Definitive roles of transported proteins (21, 23, 24, 32, 34) and mRNAs (9, 16) in infection have not yet been determined. It appears that entrapment of cellular constituents, which occurs in virions, is selective. Our data are in accordance with these reports, since PLA2 activities other than cPLA2 were not found in HCMV. Moreover, no PLC or PLD activity was observed in HCMV (Fig. 1), also arguing in favor of sorting of cellular proteins. Better knowledge of the pathways of HCMV maturation will help determine how cell proteins are incorporated into HCMV particles.
We have described a new mechanism through which HCMV subverts cell metabolism. How HCMV has acquired active cPLA2α, the characteristics of interaction of cPLA2 with other proteins of virions, and how this interaction is involved in infection are under investigation. Besides contributing to the understanding of HCMV pathophysiology, our findings may open new ways to anti-HCMV therapy. Combinations of inhibitors of the cPLA2 pathway with other antiviral drugs may be useful in preventing and/or treating HCMV infections.
Acknowledgments
We thank J.-S. Saulnier Blache, M. Gelb, G. Lambeau, M.-C. Mazeron, S. Michelson, J. Nelson, and C. Sinzger for the gift of reagents; Jean-Luc Duteyrat from the Centre de Microscopie Électronique Appliquée à la Biologie, Toulouse, and Sabina Müller for confocal microscopy (IFR 30 Toulouse); and C. Davrinche for discussions.
C. Allal was supported by a postdoctoral fellowship from ARC (Association pour la Recherche contre le Cancer).
REFERENCES
- 1.AbuBakar, S., I. Boldogh, and T. Albrecht. 1990. Human cytomegalovirus stimulates arachidonic acid metabolism through pathways that are affected by inhibitors of phospholipase A2 and protein kinase C. Biochem. Biophys. Res. Commun. 166:953-959. [DOI] [PubMed] [Google Scholar]
- 2.AbuBakar, S., I. Boldogh, and T. Albrecht. 1990. Human cytomegalovirus. Stimulation of [3H] release from [3H]-arachidonic acid prelabelled cells. Arch. Virol. 113:255-266. [DOI] [PubMed] [Google Scholar]
- 3.Alcami, A., and U. H. Koszinowski. 2000. Viral mechanisms of immune evasion. Trends Microbiol. 8:410-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baek, S. H., J. Y. Kwak, S. H. Lee, T. Lee, S. H. Ryu, D. J. Uhlinger, and J. D. Lambeth. 1997. Lipase activities of p37, the major envelope protein of vaccinia virus. J. Biol. Chem. 272:32042-32049. [DOI] [PubMed] [Google Scholar]
- 6.Bairoch, A. 2000. The ENZYME database in 2000. Nucleic Acids Res. 28:304-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blasco, R., and B. Moss. 1991. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J. Virol. 65:5910-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 9.Bresnahan, W. A., and T. Shenk. 2000. A subset of viral transcripts packaged within human cytomegalovirus particles. Science 288:2373-2376. [DOI] [PubMed] [Google Scholar]
- 10.Brown, W. J., K. Chambers, and A. Doody. 2003. Phospholipase A2 (PLA2) enzymes in membrane trafficking: mediators of membrane shape and function. Traffic 4:214-221. [DOI] [PubMed] [Google Scholar]
- 11.Claudel-Renard, C., C. Chevalet, T. Faraut, and D. Kahn. 2003. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 31:6633-6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das, A., L. Asatryan, M. A. Reddy, C. A. Wass, M. F. Stins, S. Joshi, J. V. Bonventre, and K. S. Kim. 2001. Differential role of cytosolic phospholipase A2 in the invasion of brain microvascular endothelial cells by Escherichia coli and Listeria monocytogenes. J. Infect. Dis. 184:732-737. [DOI] [PubMed] [Google Scholar]
- 13.Fortunato, E. A., A. K. McElroy, I. Sanchez, and D. H. Spector. 2000. Exploitation of cellular signaling and regulatory pathways by human cytomegalovirus. Trends Microbiol. 8:111-119. [DOI] [PubMed] [Google Scholar]
- 14.Girod, A., C. E. Wobus, Z. Zadori, M. Ried, K. Leike, P. Tijssen, J. A. Kleinschmidt, and M. Hallek. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 83:973-978. [DOI] [PubMed] [Google Scholar]
- 15.Gomez-Marin, J. E., H. El'Btaouri, A. Bonhomme, F. Antonicelli, N. Pezzella, H. Burlet, D. Aubert, I. Villena, M. Guenounou, B. Haye, and J. M. Pinon. 2002. Involvement of secretory and cytosolic phospholipases A2 during infection of THP1 human monocytic cells with Toxoplasma gondii. Effect of interferon gamma. Parasitol. Res. 88:208-216. [DOI] [PubMed] [Google Scholar]
- 16.Greijer, A. E., C. A. J. Dekkers, and J. M. Middeldorp. 2000. Human cytomegalovirus virions differentially incorporate viral and host cell RNA during the assembly process. J. Virol. 74:9078-9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakes, D. J., K. J. Martell, W. G. Zhao, R. F. Massung, J. J. Esposito, and J. E. Dixon. 1993. A protein phosphatase related to the vaccinia virus VH1 is encoded in the genomes of several orthopoxviruses and a baculovirus. Proc. Natl. Acad. Sci. USA 90:4017-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendrickson, H. S. 1994. Fluorescence-based assays of lipases, phospholipases, and other lipolytic enzymes. Anal. Biochem. 219:1-8. [DOI] [PubMed] [Google Scholar]
- 19.Hirabayashi, T., and T. Shimizu. 2000. Localization and regulation of cytosolic phospholipase A2. Biochim. Biophys. Acta 1488:124-138. [DOI] [PubMed] [Google Scholar]
- 20.Kemken, D., K. Mier, H. A. Katus, G. Richardt, and T. Kurz. 2000. A HPLC-fluorescence detection method for determination of cardiac phospholipase D activity in vitro. Anal. Biochem. 286:277-281. [DOI] [PubMed] [Google Scholar]
- 21.Landini, M. P., and A. Ripalti. 1982. A DNA-nicking activity associated with the nucleocapsid of human cytomegalovirus. Arch. Virol. 73:351-356. [DOI] [PubMed] [Google Scholar]
- 22.Le Roy, E., M. Baron, W. Faigle, D. Clement, D. M. Lewinsohn, D. N. Streblow, J. A. Nelson, S. Amigorena, and J. L. Davignon. 2002. Infection of APC by human cytomegalovirus controlled through recognition of endogenous nuclear immediate early protein 1 by specific CD4+ T lymphocytes. J. Immunol. 169:1293-1301. [DOI] [PubMed] [Google Scholar]
- 23.Mar, E. C., P. C. Patel, and E. S. Huang. 1981. Human cytomegalovirus-associated DNA polymerase and protein kinase activities. J. Gen. Virol. 57:149-156. [DOI] [PubMed] [Google Scholar]
- 24.Michelson, S., P. Turowski, L. Picard, J. Goris, M. P. Landini, A. Topilko, B. Hemmings, C. Bessia, A. Garcia, and J. L. Virelizier. 1996. Human cytomegalovirus carries serine/threonine protein phosphatases PP1 and a host-cell derived PP2A. J. Virol. 70:1415-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nokta, M. A., M. I. Hassan, K. Loesch, and R. B. Pollard. 1996. Human cytomegalovirus-induced immunosuppression. Relationship to tumor necrosis factor-dependent release of arachidonic acid and prostaglandin E2 in human monocytes. J. Clin. Investig. 97:2635-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pace, J., M. J. Hayman, and J. E. Galan. 1993. Signal transduction and invasion of epithelial cells by S. typhimurium. Cell 72:505-514. [DOI] [PubMed] [Google Scholar]
- 27.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2706. In P. M. Howley and D. M. Knipe (ed.), Fields virology. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 28.Pickard, R. T., B. A. Strifler, R. M. Kramer, and J. D. Sharp. 1999. Molecular cloning of two new human paralogs of 85-kDa cytosolic phospholipase A2. J. Biol. Chem. 274:8823-8831. [DOI] [PubMed] [Google Scholar]
- 29.Reddehase, M. J. 2002. Antigens and immunoevasins: opponents in cytomegalovirus immune surveillance. Nat. Rev. Immunol. 2:831-844. [DOI] [PubMed] [Google Scholar]
- 30.Shibutani, T., T. M. Johnson, Z. X. Yu, V. J. Ferrans, J. Moss, and S. E. Epstein. 1997. Pertussis toxin-sensitive G proteins as mediators of the signal transduction pathways activated by cytomegalovirus infection of smooth muscle cells. J. Clin. Investig. 100:2054-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Six, D. A., and E. A. Dennis. 2000. The expanding superfamily of phospholipase A2 enzymes: classification and characterization. Biochim. Biophys. Acta 1488:1-19. [DOI] [PubMed] [Google Scholar]
- 32.Spear, G. T., N. S. Lurain, C. J. Parker, M. Ghassemi, G. H. Payne, and M. Saifuddin. 1995. Host cell-derived complement control proteins CD55 and CD59 are incorporated into the virions of two unrelated enveloped viruses. Human T cell leukemia/lymphoma virus type I (HTLV-I) and human cytomegalovirus (HCMV). J. Immunol. 155:4376-4381. [PubMed] [Google Scholar]
- 33.Tanaka, J., T. Ogura, H. Iida, H. Sato, and M. Hatano. 1988. Inhibitors of prostaglandin synthesis inhibit growth of human cytomegalovirus and reactivation of latent virus in a productively and latently infected human cell line. Virology 163:205-208. [DOI] [PubMed] [Google Scholar]
- 34.Wright, J. F., A. Kurosky, E. L. G. Pryzdial, and S. Wasi. 1995. Host cellular annexin II is associated with cytomegalovirus particles isolated from cultured human fibroblasts. J. Virol. 69:4784-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zadori, Z., J. Szelei, M. C. Lacoste, Y. Li, S. Gariepy, P. Raymond, M. Allaire, I. R. Nabi, and P. Tijssen. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1:291-302. [DOI] [PubMed] [Google Scholar]
- 36.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu, H., J. P. Cong, D. Yu, W. A. Bresnahan, and T. E. Shenk. 2002. Inhibition of cyclooxygenase 2 blocks human cytomegalovirus replication. Proc. Natl. Acad. Sci. USA 99:3932-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]