Abstract
A novel coronavirus (CoV) has been identified as the etiological agent of severe acute respiratory syndrome (SARS). The SARS-CoV genome encodes the characteristic essential CoV replication and structural proteins. Additionally, the genome contains six group-specific open reading frames (ORFs) larger than 50 amino acids, with no known homologues. As with the group-specific genes of the other CoVs, little is known about the SARS-CoV group-specific genes. SARS-CoV ORF7a encodes a putative unique 122-amino-acid protein, designated U122 in this study. The deduced sequence contains a probable cleaved signal sequence and a C-terminal transmembrane helix, indicating that U122 is likely to be a type I membrane protein. The C-terminal tail also contains a typical endoplasmic reticulum (ER) retrieval motif, KRKTE. U122 was expressed in SARS-CoV-infected Vero E6 cells, as it could be detected by Western blot and immunofluorescence analyses. U122 is localized to the perinuclear region of both SARS-CoV-infected and transfected cells and colocalized with ER and intermediate compartment markers. Mutational analyses showed that both the signal peptide sequence and ER retrieval motif were functional.
An outbreak of atypical pneumonia, severe acute respiratory syndrome (SARS) is thought to have originated from Guangdong Province, Republic of China in late 2002. The mortality rate of individuals suffering from SARS can be as high as 15% (1), depending on the age group analyzed. A novel coronavirus (CoV) has recently been shown to fulfill all of Koch's postulates as the primary aetiological agent of SARS, including outcomes of monkey trials (5, 13). SARS-CoV contains a genome of ∼29.7 kbs that encodes the usual CoV replication and structural proteins.
CoVs are positive-sense RNA, enveloped viruses that contain genomes of about 30 kb (9). Traditionally, the CoVs are divided into three groups that include the mammalian viruses in groups 1 and 2 and avian viruses in group 3. The viruses are further classified into species within each group, based on host range, antigenic relationships, and genomic organization (20).
All known CoVs have a common set of essential genes encoding nonstructural proteins involved in replication (replicase gene 1ab) and structural proteins (nucleocapsid, membrane [M], envelope [E], and spike [S]) that are assembled into viral particles (10). Interspersed among these genes are group-specific open reading frames (ORFs), the majority of whose functions have yet to be established. Research on the possible functions of these genes has been limited, but they appear to be nonessential, accessory genes in cell culture (3). Inactivation of group-specific mouse hepatitis virus ORF4 did not affect growth kinetics or cytopathogenicity, indicating that it is not required for growth in cell culture (18). Interestingly, however, deletion of the nonessential genes from the mouse hepatitis virus genome resulted in attenuated viruses when inoculated into their natural hosts (3), indicating a possible in vivo function. On the other hand, Shen et al. (22) showed that continuous passage of infectious bronchitis virus (IBV) in cells resulted in mutations in the 3b gene of IBV. These mutations resulted in a growth advantage in cells and chicken embryos, as well as in an increase in virulence in the embryos.
Most CoVs are fairly host specific, sometimes causing severe upper respiratory or intestinal disease in the host species (9, 14). The human CoVs found in both groups 1 and 2 cause about 30% of colds in humans but rarely cause lower respiratory tract disease (9). Sequence analyses indicate that SARS-CoV is distinct from all known CoVs. Initial reports concluded that SARS-CoV did not belong to any of the three existing CoV groups (16, 20). More recent phylogenetic analysis, however, has identified SARS-CoV as distantly related to members of the group 2 CoVs (23).
This study reports the characterization of the SARS-CoV group-specific gene product encoded by ORF7a (also known as ORFX4 or ORF8) (Fig. 1) (16, 20, 23), which we refer to as U122 (designating a unique protein of 122 amino acids [aa]). To understand the role, if any, that U122 plays in the infectivity of SARS-CoV, characterization of the gene and its product is required. Sequence analysis predicted a 122-aa polypeptide, with a putative signal peptide sequence, C terminus transmembrane domain, and short cytoplasmic tail containing the endoplasmic reticulum (ER) retrieval motif KRKTE (Fig. 2A). Using Western blot and immunofluorescence, we show that U122 was expressed in SARS-CoV-infected cells. The initial characterization of the localization and processing of U122 is presented in this paper. Additionally, mutational analysis was used to characterize the putative signal peptide and ER retrieval sequences. Further work to determine if U122 performs an essential function in viral replication and pathogenesis will be carried out.
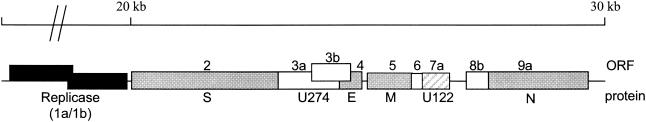
FIG. 1.
Genome organization of SARS-CoV. ORFs encoding the nonstructural proteins (black boxes), as well as ORFs encoding the structural polypeptides (gray boxes) are indicated Also, selected ORFs encoding for putative accessory genes (unshaded boxes) are shown. ORF7a (also called ORFX4 and ORF8) encoding peptide U122 is represented by the striated box. The ORFs shown are labeled according to Snijder et al. (23). S, spike; N, nucleocapsid.
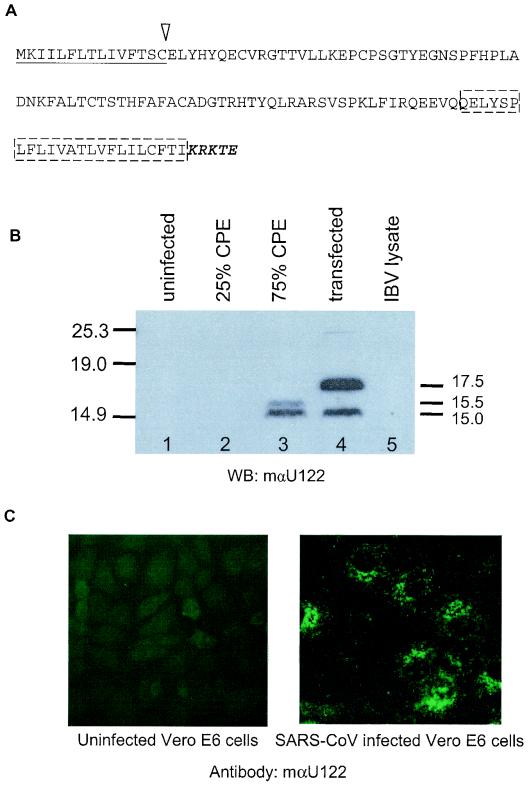
FIG. 2.
U122 is expressed in SARS CoV-infected Vero E6 cells. (A) Analysis of the U122 putative sequence predicts a signal peptide sequence (underlined) at the N terminus, the cleavage site of which is indicated with an arrow. A putative membrane-spanning domain (boxed) and an ER retrieval motif (bold italics) are found at the C terminus. (B) Blots of Vero E6 cells probed with mouse anti-U122 antiserum. Lane 1, 20 μg of protein from uninfected cells; lanes 2 and 3, 20-μg samples of proteins from SARS-CoV-infected cells harvested at about 25% CPE and 50 to 75% CPE, respectively; lane 4, 15 μg of total protein from cells transfected with pXJU122 plasmid; lane 5, 20 μg of total lysate from an IBV-infected cell culture. (C) Uninfected and virus-infected cells at 25% CPE were fixed and stained with mouse anti-U122 antibody. WB, Western blot; mαU122, mouse anti-U122 antibody.
MATERIALS AND METHODS
Viruses and cells lines.
African green monkey kidney fibroblast (Vero E6) cells (American Type Culture Collection, Manassas, Va.) were maintained in complete Dulbecco's modified Eagle medium (Gibco) containing 10% fetal calf serum (HyClone Laboratories), 100 U of penicillin per ml, and 100 μg of streptomycin (Sigma) per ml. SARS-CoV strain SIN2774 (21) was used to infect subconfluent Vero E6 plates at a multiplicity of infection of 0.1. Subsequently, cells were harvested at the desired cytopathic effect (CPE), and total proteins were extracted (24). IBV-infected Vero E6 cell lysates were used as negative controls as indicated.
Raising antibodies to U122.
The cDNA encoding aa 16 to 111 was cloned into pGEX-4T-1 and transformed into Escherichia coli BL21(DE3) cells. These cells were induced to express U122 (aa 16 to 111) with IPTG (isopropyl-β-d-thiogalactopyranoside) and allowed to grow for 4 h at 37°C. Glutathione transferase-fusion proteins were purified, and the preparation was injected into mice for raising polyclonal antibodies (25). After four injections, the mice were bled, and the sera were tested for reactivity to U122. The antibodies showed specific reactivity to U122 expressed in Vero E6 cells infected with the SARS virus or transfected with a U122 expression construct (Fig. 2B and C).
Construction of plasmids and mutations.
cDNAs were cloned into pXJ40-3′HA (GLAXO Group, Institute of Molecular and Cell Biology, Singapore, Republic of Singapore) for the expression of untagged proteins in mammalian cells; all constructs were untagged. All forward primers used were designed to incorporate a Kozak sequence. To create untagged proteins, all reverse primers (with the exception of mutL14-18R) contained the translation stop codon. The full-length 366-bp SARS-CoV ORF7a was amplified by PCR by using forward primer U122F1 (5′-CGGGATCCACCATGGGAATGAAAAT-3′) and reverse primer U122R2 (5′-CCGCTCGAGTCATTCTGTCTT-3′) incorporating BamHI and XhoI endonuclease restriction sites (underlined), respectively. The amplicon was purified, digested, and cloned into the compatible restriction sites of the expression vector to form pXJU122. To mutate the U122 signal peptide cleavage site, the amino acids SCELY located at positions 14 to 18 were mutated to leucines LLLLL by a two-step PCR-directed mutagenesis approach (27) using plasmid pXJU122 as a template. Briefly, the overlapping primer set consisting of forward primer mutL14-18F (5′-GTATTTACATTGTTGCTGCTACTTCACTATCAGGAG-3′) and reverse primer mutL14-18R (5′-CCTGATAGTGAAGTAGCAGCAACAATGTAAATACAATC-3′) containing the incorporated mutations (underlined), were used in combination with primers U122F1 and U122R2 to create amplicon U122-L. The amplicon was purified, digested, and cloned in the vector to create pXJU122L. Plasmid pXJmatU122 consisting of residues 16 to 122 was constructed by using PCR with forward primer U122F2, 5′-CGGGATCCATGGAGCTATATCACT-3′, and reverse primer U122R2; the BamHI restriction site is underlined and the incorporated ATG is indicated in bold. To study the signal retrieval motif the lysine residues in the 3′-terminal amino acids KRKTE at positions 118 and 120 were mutated to glutamic acid residues. This was done by using PCR with forward primer U122F1 and reverse primer U122RK>E (5′-CCGCTCGAGTCATTCTGTCTCTCTCAAT-3′; the XhoI restriction site is underlined). This construct (pXJU122K>E) expresses the untagged retrieval mutant. All sequences were confirmed by DNA sequencing.
In vitro transcription and translation.
A total of 0.5 μg of plasmid pXJU122 was transcribed and translated by using the TNT T7 coupled reticulocyte lysate system (Promega) in a 10-μl reaction mixture for 1.5 h at 30°C. [35S]cysteine (>1,000Ci/mmol; NEN) was used to label U122, and samples were immunoprecipitated by using U122-specific antibodies with protein A-Sepharose. Proteins were resolved on sodium dodecyl sulfate (SDS)-15% polyacrylamide gel electrophoresis (PAGE) gels and visualized by radioautography with Amplify reagent (Amersham).
Transfection, pulse-chase radiolabeling, and immunoprecipitation.
The transfection of recombinant plasmids was accomplished by using liposomes (Lipofectamine 2000 reagent; Invitrogen). Generally, for a 6-cm plate, 0.5 μg of plasmid DNA was used for transfection according to the manufacturer's protocol.
For pulse-chase experiments, 2.0 μg of plasmid DNA was used for transfection per 6-cm plate. At 6 h posttransfection, confluent Vero E6 cells were starved for 30 min in prewarmed depletion medium lacking methionine and cysteine. The depletion medium was replaced with medium containing 100 μCi of [35S]methionine-cysteine mix per ml (EXPRE35S35S protein labeling mix; NEN) for 10 or 30 min. [35S]cysteine (100 μCi; NEN) was used to supplement the [35S]methionine-cysteine mix to enhance the labeling efficiency. Subsequently, the cells were washed and chased with complete Dulbecco's modified Eagle medium containing a 5 mM concentration of unlabeled methionine and cysteine. Radioimmunoprecipitation assay (RIPA; 1×) buffer was used to extract proteins, and immunoprecipitation was done with protein A-Sepharose-coupled antibodies as described by Nguyen and Hogue (17). Proteins were resolved by SDS-15% PAGE, and gels were fixed and treated with Amplify fluorographic reagent (Amersham). Subsequently, gels were dried and visualized by radioautography. The amount of labeled U122 was quantified by using a Bio-Rad model GS-700 imaging densitometer with Bio-Rad Multi-Analyst version 1.02/Mac software (number of radioautographs quantified for each experiment, 2).
Immunofluorescence of SARS-CoV-infected and transfected cells.
SARS-CoV-infected Vero E6 cells were grown on coverslips until they showed a CPE of 25% (24). The coverslips were fixed in acetone for 20 to 30 min on ice and then air dried before being stored at −20°C. Before use, the coverslips were fixed again in methanol at −20°C and air dried. For transfected proteins, Vero E6 cells were grown on coverslips and transfected as described above. An immunofluorescence assay was performed at about 16 h posttransfection as described by Goh et al. (7). Briefly, the medium was removed, and the coverslips were fixed in methanol for 5 min at −20°C, after which the coverslips were lifted out and completely air dried. To decrease background staining, mouse anti-U122 sera were adsorbed against fixed Vero E6 cells. Uninfected cells showed no background staining with absorbed mouse anti-U122 (Fig. 2C). Mouse anti-U122 was used at a dilution of 1:200, and all other antibodies were used at dilutions of 1:100. Fixed cells were incubated with the appropriate primary antibody combination of mouse anti-U122 and rabbit anti-GS28 (Golgi marker; BD, Singapore, Republic of Singapore) or rabbit anti-Sec31 (intermediate compartment marker Sec31; HWJ Group, Institute of Molecular and Cell Biology, Singapore, Republic of Singapore). Following washing, cells were incubated with the secondary antibody combination of fluorescein isothicyanate (FITC)-conjugated goat antimouse and rhodamine (Rh)-conjugated antirabbit antibodies (Santa Cruz Biochemicals). When cells were double-labeled with mouse anti-U122 and rat anti-GRP94, fixed cells were sequentially incubated with rat anti-GRP94 (ER marker; ITS Science and Medical, Singapore, Republic of Singapore) and FITC-conjugated anti-rat secondary antibody, followed by incubation with mouse anti-U122 and subsequently in Rh-conjugated antimouse (Santa Cruz Biochemicals). This was done to minimize the cross-reaction of secondary antibodies with both the primary antibodies.
RESULTS AND DISCUSSION
U122 expressed in SARS-CoV-infected cells.
Sequence analysis of the SARS-CoV protein translated from ORF7a is predicted to be a 122-aa polypeptide containing a signal peptide at the N terminus, a transmembrane domain, and a retrieval signal at the C terminus (16, 20) (Fig. 2A).
To determine if U122 was expressed in SARS-CoV-infected Vero E6 cells, cells were infected with SARS-CoV (strain SIN2774) as described in Materials and Methods. Total proteins were harvested from Vero E6 cells showing 25 and 50 to 75% CPE and subjected to Western blotting. No signal was detected in mock-infected cells or IBV-infected cells (Fig. 2B, lanes 1 and 5), indicating the specificity of the mouse antibody against U122. By using our mouse anti-U122 antiserum, the protein was only detected in SARS-CoV-infected cells at a CPE of 50 to 75% (Fig. 2B, lane 3) and in Vero E6 cells transfected with a U122 DNA construct (Fig. 2B, lane 4). Two bands of about 15.5 and 15.0 kDa were detected in SARS-CoV late-infected cells (lane 3, 75% CPE), but a larger band of ∼17.5 kDa and a band of ∼15.0 kDa were detected in transient transfected cells expressing untagged U122 (lane 4). In SARS-CoV-infected cells, the immature U122 may have been processed efficiently, so that the immature form was not observed in infected cells. There appeared to be an additional band slightly larger than the mature form, indicating an intermediate form only present in virus-infected cells. U122 was not detected in the early phase of infection (lane 2, 25% CPE), even when the total protein of the sample with a 25% CPE that was used for Western blotting and immunodetection was double that of the sample with a 75% CPE (data not shown).
Immunofluorescence was used to determine the cellular localization of U122 in SARS-CoV-infected cells. By using mouse anti-U122 antibody, the protein was detected in SARS-CoV-infected Vero E6 cells (Fig. 2C). U122 was detected in the perinuclear region and associated with ER. This cellular localization is similar to that observed in U122-expressing Vero E6 cells (see Fig. 5). We do not understand why U122 was clearly detected by immunofluorescence in infected cells at 25% CPE but not in Western blots of total proteins from cells at 25% CPE. The antibodies may have stronger affinity to conformational epitopes present in cells than to linear epitopes that are present in denatured proteins on Western blots.
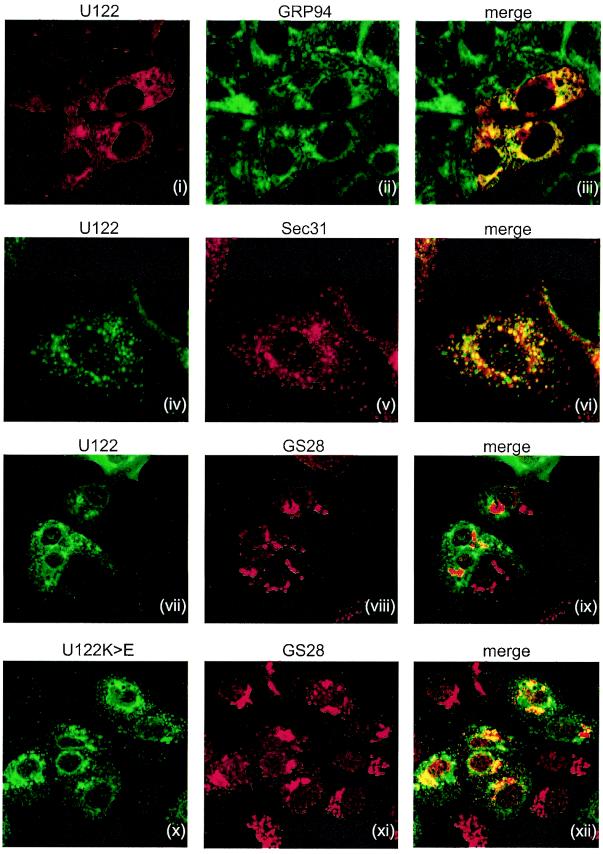
FIG. 5.
Intracellular localization of expressed U122 and retrieval signal mutant. Vero E6 cells were transfected with plasmid pXJU122 or pXJU122K>E. At 16 h posttransfection, cells were fixed with methanol and labeled with mouse anti-U122 antibody (left panels) and either antibodies to the ER marker GRP94 (rat anti-GRP94), the intermediate compartment marker Sec31 (rabbit anti-Sec31), or Golgi marker GS28 (rabbit anti-GS28) (middle panels). When anti-U122 was used with anti-GS28 or anti-Sec31, FITC-conjugated goat anti-mouse and Rh-conjugated anti-rabbit antibodies (Santa Cruz Biochemicals) were used as secondary antibodies. For double labeling with anti-U122 and anti-GRP94, FITC-conjugated anti-rat and Rh-conjugated anti-mouse (Santa Cruz Biochemicals) antibodies were used as secondary antibodies. Merged images showed colocalization of U122 and K>E proteins with the marker proteins (right panels).
SARS-CoV U122 protein in transfected cells.
The SARS-CoV genome contains five group-specific ORFs larger than 50 amino acids (16, 20). SARS-CoV proteins, excluding the replication gene products of ORF 1ab, are translated from a set of 5′ nested subgenomic mRNAs (sgmRNAs). Each sgmRNA contains a 5′ end derived from the genomic 5′ leader sequence, subgenomic sequences, and a common 3′ end (8, 20, 26). Between five and eight SARS-CoV sgmRNAs are detected by Northern hybridization analysis from infected cells, ranging from 8.3 to 1.7 kb in size (20, 23). This includes a polycistronic 2.5-kb mRNA that contains a conserved transcription regulation sequence immediately upstream of ORF7a. This indicates that ORF7a is likely to be translated (20) to give U122.
To express untagged SARS-CoV U122 in vitro, full-length ORF7a was cloned into mammalian expression vector pXJ40-3′HA to form pXJU122. Untagged U122 was translated in vitro by using the TNT coupled reticulocyte lysate system (Promega) in the presence of [35S]cysteine. Following immunoprecipitation with antibodies specific for U122, a single ∼17.5-kDa band was observed (Fig. 3A). To detect U122 expressed in Vero E6 cells, total proteins on Western blots were probed with mouse anti-U122. Two bands of about 17.5 and 15.0 kDa were observed (Fig. 3B, lane 3). The smaller protein band could be due to proteolytic cleavage of the signal peptide and was not observed in the in vitro translated product, even with the addition of canine microsomal membranes (data not shown), indicating the possible need for additional host cofactor(s) for efficient processing of U122.
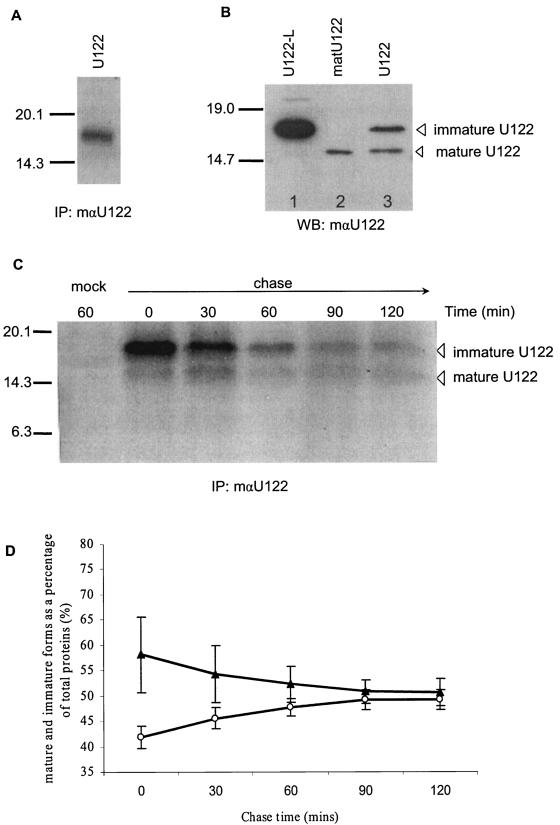
FIG. 3.
Processing of U122 expressed in Vero E6 cells. (A) Untagged U122 was translated in vitro and visualized by autoradiography. A single band of ∼17.5 kDa was detected. (B) Untagged U122-L (lane 1), matU122 (lane 2), and U122 (lane 3) proteins were expressed in Vero E6 cells. Total proteins were extracted by using RIPA buffer, and 20 μg of total protein was used for SDS-15% PAGE. Western blots of these proteins were probed with mouse anti-U122 antibodies. (C) Vero E6 cells were transfected with plasmid pXJU122 for pulse-chase analysis. At 6 h posttranfection, cells were starved for 30 min in cysteine- and methionine-deficient medium and subsequently labeled for 30 min with 35S-labeled amino acids. Cells were either lysed directly (time zero) or chased for 30, 60, 90, or 120 min. Cell lysates were immunoprecipitated with mouse anti-U122 antibodies and separated on an SDS-15% PAGE gel, followed by autoradiography. (D) Quantification of the pulse-chase experiment. The amount of immature (▴) and mature (○) 35S-labeled U122 protein was determined with a densitometer and expressed as a percentage of the total labeled protein at each time point (results are means ± standard error of the means for two experiments). IP, immunoprecipitation; WB, Western blot; mαU122, mouse anti-U122 antibody.
The putative SARS-CoV U122 signal sequence.
Since the deduced SARS-CoV U122 amino acid sequence contains a putative signal peptide sequence of 15 residues (16), we speculated that the smaller ∼15.0-kDa product was due to cleavage of the signal peptide from the larger ∼17.5-kDa protein. Signal peptides play a major role in membrane integration and the translocation of secretory and membrane proteins from the ER (4). Many enveloped virus glycoproteins are synthesized as inactive precursors, which are usually unable to mediate membrane fusion and, hence, viral entry (28). Therefore, the endoproteolytic cleavage of viral envelope glycoproteins is crucial for virus maturation, and the availability of cellular enzymes capable of processing the inactive precursors can be major determinants of viral tropism and pathogenicity (15, 28). To determine whether the SARS-CoV signal peptide sequence is active, we used PCR-directed mutagenesis to create two mutations in U122, one in the cleavage site of the putative signal peptide and the other to delete the N-terminal signal peptide up to the cleavage site. Mutation of the residues spanning the predicted cleavage site (residues SCELY at positions 14 to 18) to leucines (U122-L) abolished the cleavage of U122 in Vero E6-transfected cells (Fig. 3B, lane 1). The resultant ∼17.5-kDa product was similar in size to the untagged U122 immature product. Also, transient expression of the deletion mutant matU122 resulted in a product similar in size (∼15.0 kDa) to the mature form of the U122 peptide (Fig. 3B, lane 2). Pulse-chase analysis was done to determine the kinetics of wild-type untagged U122 expression in mammalian cells. U122 was expressed and radiolabeled in Vero E6 cells, and the radiolabeled U122 was immunoprecipitated. During the 2-h chase, the level of immature U122 slowly decreased, while the detectable level of mature U122 remained fairly constant (Fig. 3C). Expressed as a percentage of total U122 proteins, the immature form decreases, while the mature form increases through the 2-h chase (Fig. 3D), indicating that the uncleaved form was processed into the mature form. Both immature and mature forms could be detected from the start of the pulse-chase (Fig. 3C, time zero), indicating that the immature protein was cotranslationally processed to form the smaller mature protein. The total amount of labeled protein decreases, indicating that U122 is degraded in the course of the experiment. Combining these observations, we hypothesized that the constant level of the mature form may be due to the replacement of degraded mature protein by the conversion of immature protein to a mature form. Most of the conversion of immature to mature protein occurred cotranslationally (about 40%), and the remainder of the cleavage was posttranslational (about 10%). At the end of the 2-h pulse-chase, the processing was still not complete and reached a plateau of about 50% by 90 min, indicating that although posttranslational cleavage occurred, it was fairly inefficient.
Mutation of the ER retrieval motif leads to rapid proteolytic processing of U122.
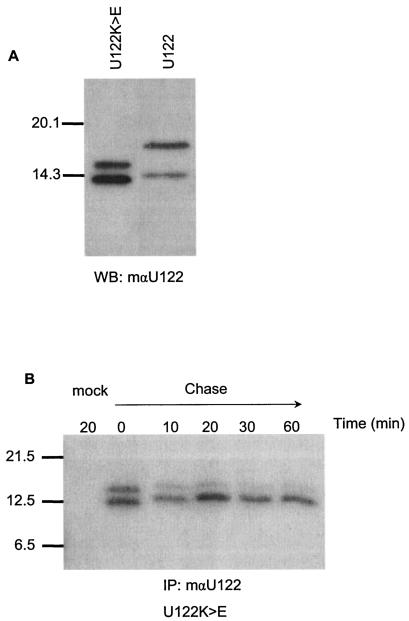
The short C-terminal tail of U122 contains a typical ER retrieval motif, KRKTE. In mammals, plants, and yeasts, this typical cytosolic C-terminal dilysine motif (KKXX or KXKXX, where X is any amino acid) is crucial for ER localization of type I membrane proteins (2, 11). If the C-terminal X is considered the −1 position, the two lysine residues must be in the −3 and −4 or the −3 and −5 positions, respectively. These two lysine residues cannot be replaced by any other basic amino acid, and mutation of these residues results in the abolishment of the ER localization of the reporter proteins of mammalian cells (11). Studies have shown that the carboxyl-terminal sequence of Lys-X-Lys-X-X in integral ER membrane proteins functions either as a retrieval or retention signal, with signals important for the transport of these proteins back to the ER from the intermediate compartment (19). To determine whether this retrieval motif was functional, the lysine residues at positions 118 and 120 were mutated to glutamic acid by using PCR-directed mutagenesis (U122K>E). Transient expression of plasmid pXJU122K>E in Vero E6 cells was done to determine the effect of the mutation on protein expression and localization. Total proteins were harvested at 16 h posttransfection, subjected to Western blotting, and probed with mouse anti-U122 (Fig. 4A). Two bands (∼15.5 and ∼12.5 kDa) were detected in the cells expressing U122K>E (Fig. 4A, lane 1), which were different in size from those observed for U122. The processing of the immature form appears to be very efficient, as the larger band (17.5 kDa) is no longer visible. The two bands produced by the mutant untagged protein differed in size from the smaller band (15.0 kDa) of U122. The mutations from K to E may cause the small change in mobility, or mutations in the recycling signal may influence the processing of the U122 at the N terminus so that the alternative cleavage sites are used. Pulse-chase analysis of U122K>E was performed to verify the efficiency of this processing. Following a 10-min pulse-labeling, the immature ∼17.5-kDa product was not detected at the start of the chase, indicating that the immature product has been converted cotranslationally to two smaller products of ∼12.5 kDa (Fig. 4B). Benghezal et al. (2) observed similar results with green fluorescent protein-fusion proteins fused to mutated retrieval signals in plants. They suggested that the mutated retrieval signal caused rapid protein exit from the ER to a distal compartment where the processing activity is located. The larger band of the doublet is eventually converted to the smaller ∼12.5-kDa product after the 60-min chase, suggesting an additional proteolytic cleavage site in the K>E mutant (Fig. 4B).
FIG. 4.
The U122K>E protein is rapidly processed in transfected cells. (A) Untagged U122 and U122K>E proteins were expressed in transfected Vero E6 cells. At 16 h posttransfection total proteins were extracted by using RIPA buffer and 20 μg of protein separated on an SDS-15% PAGE gel. Western blots of these proteins were probed with mouse anti-U122 antibodies. (B) Vero E6 cells were transfected with pXJU122K>E for pulse-chase analysis. At 6 h posttranfection, cells were starved for 30 min in cysteine- and methionine-deficient medium and subsequently labeled for 10 min with 35S-labeled amino acids. Cells were either lysed directly (time zero) or chased for 10, 20, 30, or 60 min. Cell lysates were immunoprecipitated with mouse anti-U122 antibodies, followed by separation on an SDS-15% PAGE gel and autoradiography. IP, immunoprecipitation; WB, Western blot; mαU122, mouse anti-U122 antibody.
SARS-CoV U122 is localized to the ER and ER-Golgi intermediate compartment in transfected cells.
The subcellular localization of U122 in Vero E6 cells was studied. At 16 h posttransfection, cells were fixed with methanol and stained with both mouse anti-U122 antibodies (Fig. 5, left frames) and antibodies to either the ER marker GRP94 (Fig. 5, ii), the ER intermediate compartment marker Sec31 (frame v) or the Golgi marker GS28 (frames viii and xi). U122 was observed to colocalize with GRP94 (Fig. 5, iii), indicating that U122 was present in the ER compartment. In cells showing weaker U122 expression, U122- and Sec31-labeled punctate structures could be detected. These probably corresponded to the ER-Golgi intermediate compartment (Fig. 5, vi). U122 did not colocalize with the cis-Golgi marker GS28 (Fig. 5, ix). This is likely due to the rapid movement of U122 from the Golgi compartment back to the ER, as seen in recycled proteins with retrieval signals (6). On the other hand, mutant U122K>E was predominantly localized to the Golgi (Fig. 5, xii). Following cleavage of the signal sequence, it is rapidly transported to the Golgi but cannot be recycled back to the ER because of the mutated retrieval sequence, resulting in a large fraction of U122K>E remaining in the Golgi apparatus. Collectively, immunofluorescence results indicated that untagged U122 was cycled between the ER and Golgi compartments and that this process was mediated by the retrieval motif at the C terminus.
Interestingly, convalescent-phase human sera could not detect bacterially expressed U122-glutathione transferase fusion protein by Western blot analysis (25). This suggests that U122 is either not a viral structural protein or that it is not exposed or sufficiently immunogenic in vivo. Importantly though, U122 has been shown to interact specifically with another unique group-specific SARS-CoV protein (ORF3a on sgRNA 3), designated U274 (Fig. 1). The latter has been shown to interact with the SARS-CoV structural proteins E, S, and M (24). Also, in this study U122 was shown to localize to the intermediate compartment, where CoVs are known to assemble and bud (12). Taken together, these results indicate that U122 might have some yet undetermined role in virus replication, assembly, or infection at least during in vivo infection. Further work will be done to determine if U122 is a structural protein, whether it plays a role in viral replication in vitro or in vivo, and whether it is a novel SARS-CoV structural protein.
Acknowledgments
We thank the In Vivo Model System Unit, Choi Yook Wah, Eileen Teng, and Tham Puay Yoke for technical support.
This work was supported by grants from the Agency for Science, Technology, and Research (A*STAR), Singapore.
REFERENCES
- 1.Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. [DOI] [PubMed] [Google Scholar]
- 2.Benghezal, M., G. O. Wasteneys, and D. A. Jones. 2000. The C-terminal dilysine motif confers endoplasmic reticulum localization to type I membrane proteins in plants. Plant Cell 12:1179-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Haan, C. A., P. S. Masters, X. Shen, S. Weiss, and P. J. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eichler, R., O. Lenz, T. Strecker, and W. Garten. 2003. Signal peptide of Lassa virus glycoproteins GP-C exhibits an unusual length. FEBS Lett. 538:203-206. [DOI] [PubMed] [Google Scholar]
- 5.Fouchier, R. A., T. Kuiken, M. Schutten, G. van Amerongen, G. J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stohr, and A. D. Osterhaus. 2003. Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaynor, E. C., S. te Heesen, T. R. Graham, M. Aebi, and S. D. Emr. 1994. Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J. Cell Biol. 127:653-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goh, P.-Y., Y.-J. Tan, S. P. Lim, S. G. Lim, Y. H. Tan, and W. J. Hong. 2001. The hepatitis C virus core protein interacts with NS5A and activates its caspase-mediated proteolytic cleavage. Virology 290:224-236. [DOI] [PubMed] [Google Scholar]
- 8.Haijema, B. J., H. Volders, and P. J. Rottier. 2003. Switching species tropism: an effective way to manipulate the feline coronavirus genome. Virology 77:4528-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmes, K. V. 2003. SARS-associated coronavirus. N. Engl. J. Med. 348:1948-1951. [DOI] [PubMed] [Google Scholar]
- 10.Holmes, V. H., and M. C. Lai. 1996. Coronaviridae: the viruses and their replication, p. 541-560. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology, 3rd ed. Lippicott-Raven, Philadelphia, Pa.
- 11.Jackson, M. R., T. Nilsson, and P. A. Peterson. 1990. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 9:3153-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klumperman, J., J. K. Locker, A. Meijer, M. C. Horzinek, H. J. Geuze, and P. J. Rottier. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuiken, T., R. A. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stohr, J. S. Peiris, and A. D. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindemann, D., T. Pietschmann, M. Picard-Maureau, A. Berg, M. Heinkelein, J. Thurow, P. Knaus, H. Zentgraf, and A. Rethwilm A. 2001. A particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 75:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen, V. P., and B. G. Hogue. 1997. Protein interactions during coronavirus assembly. J. Virol. 71:9278-9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ontiveros, E., L. Kuo, P. S. Masters, and S. Perlman. 2001. Inactivation of expression of gene 4 of mouse hepatitis virus strain JHM does not affect virulence in the murine CNS. Virology 289:230-238. [DOI] [PubMed] [Google Scholar]
- 19.Pelham, H. R. B., K. G. Hardwick, and M. J. Lewis. 1988. Sorting of soluble ER proteins in yeast. EMBO J. 7:1757-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, D. A. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 21.Ruan, Y.-J., C. L. Wei, L. A. Ee, V. B. Vega, H. Thoreau, S. Y. S. Thoe, J.-M. Chia, P. Ng, K. P. Chiu, L. Lim, T. Zhang, K. P. Chan, L. E. L. Oon, M. L. Ng, S. Y. Leo, L. F. P. Ng, E. C. Ren, L. W. Stanton, P. M. Long, and E. T. Liu. 2003. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet 361:1779-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen, S., Z. L. Wen, and D. X. Liu. 2003. Emergence of a coronavirus infectious bronchitis virus mutant with a truncated 3b gene: functional characterization of the 3b protein in pathogenesis and replication. Virology 311:16-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan, Y.-J., E. Teng, S. Shen, T. H. P. Tan, P.-Y. Goh, B. C. Fielding, E.-E. Ooi, H.-C. Tan, S. G. Lim, and W. Hong. 2004. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 78:6723-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan, Y.-J., P.-Y. Goh, B. C. Fielding, S. Shen, C.-F. Chou, J.-L. Fu, H. N. Leong, Y. S. Leo, E. E. Ooi, A. E. Ling, S. G. Lim, and W. Hong. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 11:362-371. [DOI] [PMC free article] [PubMed]
- 26.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 27.Wang, S.-H., W.-J. Syu, K.-J. Huang, H.-Y. Lei, C.-W. Yao, C.-C. King, and S.-T. Hu. 2002. Intracellular localization and determination of a nuclear localization signal of the core protein of dengue virus. J. Gen. Virol. 83:3093-3102. [DOI] [PubMed] [Google Scholar]
- 28.Wool-Lewis, R. J., and P. Bates. 1999. Endoproteolytic processing of the ebola virus envelope glycoprotein: cleavage is not required for function. J. Virol. 73:1419-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]