Abstract
The mutation frequency of Turnip crinkle virus can increase 12-fold without inducing error catastrophe. Lesions in a hairpin repressor frequently reverted and led to second-site alterations biased for specific mutations. These results suggest that the hairpin may also function as an RNA chaperone to properly fold the RNA-dependent RNA polymerase.
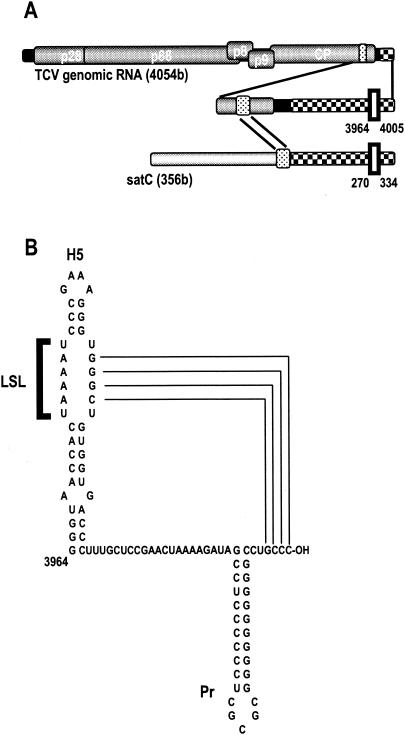
Turnip crinkle virus (TCV), a member of the genus Carmovirus in the family Tombusviridae, has been used as a model for identifying cis-acting elements important for RNA replication. The 4,054-base TCV genome consists of a positive-sense, single-stranded RNA with five overlapping open reading frames (ORF) encoding proteins that function in replication, movement, and packaging (3, 8, 13) (Fig. 1A). p28 and its readthrough product p88 are required for replication in vivo. p88 contains the RNA-dependent RNA polymerase (RdRp) active site and can by itself promote complementary-strand synthesis from cognate templates in an in vitro (cell-free) assay (16).
FIG. 1.
Location and sequence of H5. (A) Genome organization of TCV and satC. p28 and p88 are the virally encoded subunits of the RdRp. p8 and p9 are proteins involved in virus movement. Similar sequences are patterned alike. Open rectangle with solid border indicates the position of H5. (B) Sequence and structure at the 3′ end of TCV. Solid lines indicate base pairing between the H5 LSL and the 3′-terminal bases, which is required for repression of minus-strand synthesis in satC (24).
TCV is also associated with several noncoding satellite RNAs (satRNAs) that range from 194 to 356 bases, with satC (356 bases) sharing its 3′-terminal 166 bases with TCV (19). Based on studies using satC, several elements in the 3′ untranslated region (3′ UTR) of TCV that are likely important for replication have been identified. These include a core promoter hairpin (Pr) for synthesis of minus strands, which is located at the 3′ terminus of plus strands and can function independently as a promoter in in vitro assays (2, 20, 22), and hairpin 5 (H5), a recently identified repressor of minus-strand synthesis (24) (Fig. 1B). Hairpins with similar sequence and/or structural features are located in analogous positions in nearly all carmoviruses (24). H5, which contains a large symmetrical internal loop (LSL), represses minus-strand synthesis in vitro when the 3′ side of the LSL is base paired with the 3′-terminal 4 bases of the RNA (GCCC-OH), thereby sequestering the 3′ terminus from the RdRp (Fig. 1B). Disruption of the interaction between the satC 3′ terminus and the LSL substantially enhances synthesis of both full-length and aberrantly initiated complementary strands in vitro. In addition, localized disruption of H5 (and 3′-proximal sequences) occurred in satC transcripts containing a deletion of 3′-terminal bases. In TCV, compensatory exchanges between the LSL and 3′-terminal bases enhanced replication relative to that of virus containing the individual alterations, strongly suggesting that H5 serves an analogous function in TCV (unpublished data). A repressor of minus-strand synthesis with a large asymmetrical internal loop was also recently discovered in Tomato bushy stunt virus, a virus belonging to the same family as TCV (14). In the present study, we have identified an additional feature of H5. Mutations introduced into the TCV LSL resulted, surprisingly, in an increase of as much as 12-fold in second-site mutations scattered throughout the sequenced region; most of these alterations were uridylate-to-cytidylate or adenylate-to-guanylate substitutions.
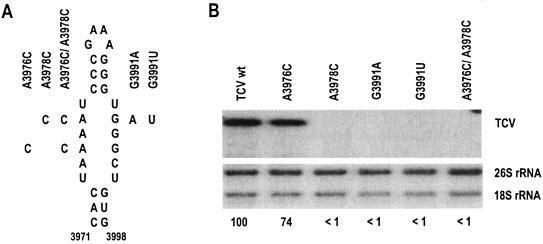
To determine the importance of the TCV H5 for accumulation in vivo, several single and double point mutations were constructed in the LSL (Fig. 2A). Mutations were generated by PCR using pTCV66, a construct containing wild-type (wt) TCV cDNA downstream from a T7 RNA polymerase promoter. Primers were complementary to positions 4035 to 4054 and homologous to positions 3947 to 3993. The latter oligonucleotide contained either an equal mixture of dATP and dCTP at positions 3976 and 3978 or an equal mixture of deoxynucleoside triphosphates at position 3991. PCR products were treated with the Klenow fragment, digested with SpeI, ligated between the SpeI and SmaI sites of pTCV66, and transformed into Escherichia coli. TCV genomic RNA was synthesized by using T7 RNA polymerase following digestion of plasmids with SmaI, which generates transcripts with precise 5′ and 3′ ends. Protoplasts (5 × 106), prepared from callus cultures of Arabidopsis thaliana ecotype Col-0, were inoculated with 20 μg of TCV genomic RNA transcripts by using polyethylene glycol as previously described (11). Northern blots of total RNA were probed with a [γ-32P]ATP-labeled oligonucleotide complementary to positions 3950 to 3970 of TCV genomic RNA.
FIG. 2.
Accumulation of TCV H5 LSL mutants at 40 h after inoculation of protoplasts. (A) Location of the mutations in the LSL. Designations of the mutants are given. (B) (Top) RNA gel blot of TCV genomic RNA. (Bottom) RNA gel blot of rRNA loading control. Levels of TCV (expressed below the gels as percentages of the wt level, taken as 100%) were determined with a scanning laser densitometer and normalized for the level of rRNA.
As shown in Fig. 2B, TCV containing an adenylate-to-cytidylate substitution at position 3976 on the 5′ side of the LSL (A3976C) accumulated to 74% of wt levels at 40 h after inoculation in protoplasts. A3978C, either alone or in combination with A3976C, abolished the detection of TCV, indicating that the 5′ side of the LSL is important for TCV accumulation. TCV containing G3991A or G3991U on the 3′ side of the LSL, which should disrupt the interaction of the LSL with the 3′-terminal bases, also failed to accumulate to detectable levels. These results support an important function for both sides of the H5 LSL in TCV accumulation.
To determine if TCVs containing these mutations accumulate to detectable levels in host plants, 12 μg of mutant or wt TCV genomic RNA transcripts was inoculated onto six turnip seedlings. At 20 days postinoculation, total RNA was extracted from leaves and subjected to reverse transcription-PCR (RT-PCR) amplification to detect any TCV accumulation. A 394-base DNA fragment was amplified by Pyrostase Taq polymerase by using the following PCR parameters: an initial 2-min denaturation step for 1 cycle; 30 s of denaturation, annealing, and elongation for 30 cycles; and a final 5-min elongation step of 1 cycle. TCV primers were homologous to positions 3661 to 3677 (oligonucleotide 3661) and complementary to positions 4035 to 4054 (oligonucleotide KK57). Cloned cDNAs derived from progeny of A3976C (42 clones from six plants), G3991A (11 clones from one plant), A3976C/A3978C (25 clones from five plants), and wt TCV (31 clones from five plants) were sequenced between positions 3800 and 4000 by using oligonucleotide KK57, and progeny of A3978C (26 clones from five plants) were sequenced between positions 3700 and 4000 by using oligonucleotides KK57 and 3661.
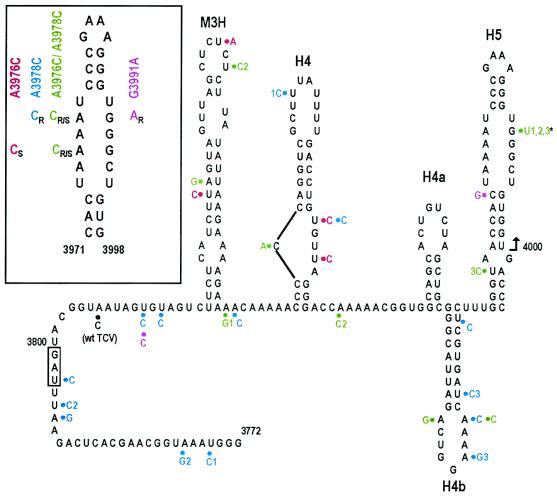
Sequencing of TCV A3976C progeny revealed that the original alteration was stably maintained in all cloned progeny, which supports the finding of only marginally reduced accumulation of A3976C in protoplasts. In addition, 4 of 42 clones contained unique second-site mutations, three of which were uridylate-to-cytidylate transitions (Fig. 3; Table 1). The second-site changes were located in two nearby hairpins (M3H and H4) that have enhancer function in their negative-sense orientations and no known function in their positive-sense orientations (1). There was no discernible relationship between the regions that contained the additional changes and no obvious interactions possible with H5.
FIG. 3.
Locations of second-site mutations in the 3′-terminal region of LSL mutant progeny at 20 days after inoculation of plants. (Inset) The primary mutations in H5 that gave rise to second-site mutations. R, S, and R/S indicate mutations that either reverted to wt, were stable, or were partially stable in progeny virus, respectively. Designations of the mutant TCV constructs are given. The structure of the 3′ UTR of TCV was determined by a combination of solution structure analysis, mFold, version 3.1 (12, 25), computer structural predictions, and phylogenetic comparisons among related carmoviruses. Second-site mutations and their primary-site mutant progenitors are color-coded. The single alteration found in wt TCV progeny is shown in black. Identically colored numbers associated with some second-site mutations indicate their presence in the same clone. The termination codon of the CP is boxed. Asterisk indicates mutation found in five TCV clones isolated from three plants. All other second-site mutations were found only once in independent clones. Arrow indicates location of the first position discernible in the sequencing autoradiographs. A3976C progeny derived from six plants, G3991A progeny derived from one plant, A3976C/A3978C progeny derived from five plants, and wt TCV progeny derived from five plants were sequenced between positions 3800 and 4000. A3978C progeny derived from five plants were sequenced between positions 3700 and 4000.
TABLE 1.
Characterization of second-site mutations in clones derived from wt TCV and TCV mutant constructs
| Construct(s) | No. of substitution types
|
No. of clones with second-site changes/total no. of clones | Total mutations/no. of bases sequenced | Mutation frequency (10−3) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| U to C | A to G | U to A | A to C | C to G | C to A | G to U | ||||
| A3976C | 3 | 0c | 1 | 0c | 0c | 0c | 0c | 4/42 | 4/8,400a | 0.5 |
| A3978C | 9 | 4 | 0c | 1 | 0c | 0c | 0c | 11/25 | 14/7,500b | 1.9 |
| G3991A | 1 | 0c | 0c | 0c | 1 | 0c | 0c | 2/11 | 2/2,200a | 0.9 |
| A3976C/A3978C | 2 | 3 | 0c | 2 | 0c | 1 | 5d | 9/27 | 13/5,400a | 2.4 |
| All mutants | 15 | 7 | 1 | 3 | 1 | 1 | 5 | 26/105 | 33/23,500 | 1.4 |
| wt TCV | 0c | 0c | 0c | 1 | 0c | 0c | 0c | 1/31 | 1/6,200a | 0.2 |
TCV clones sequenced between bases 3800 to 4000 within the 3′UTR.
TCV clones sequenced between bases 3800 to 4000 within the 3′UTR and between bases 3700 to 3800 within the p38 coding region.
No mutations found for this base change.
All five were at the same position within the LSL (see Fig. 3).
Three of six plants inoculated with A3978C accumulated TCV at or near wt levels, and two additional plants accumulated TCV-specific RNA, as detected by RT-PCR (data not shown). All 25 cloned progeny contained either a reversion to wt (15 of 25) or a primary-site alteration to a uridylate (10 of 25). Unexpectedly, 3 of 25 clones contained two second-site mutations and 8 of 25 clones contained single second-site mutations that were unique for each clone in the 300 bases that were sequenced (Fig. 3; Table 1). One mutation, located at the first position of the coat protein (CP) ORF termination codon, would extend the C terminus of the CP by 10 amino acids. Other mutations in the CP ORF led to conservative amino acid substitutions. The remaining mutations were scattered throughout the UTR, mostly in putative single-stranded regions between hairpins or in internal and terminal loops within hairpins. Of the 14 base changes, 9 were uridylate-to-cytidylate transitions and 4 were adenylate-to-guanylate transitions (Table 1).
Plants inoculated with G3991U failed to accumulate viral RNA detectable by PCR (data not shown). However, one of six plants inoculated with G3991A contained PCR-detectable RNA (data not shown). All 11 clones sequenced contained a reversion to the wt guanylate, and 2 clones had second-site mutations in the 200 bases sequenced (Fig. 3). The second-site alteration in the 5′ side of H5 would expand the LSL, possibly helping to stabilize the interaction of the 3′ LSL with the 3′-terminal bases, which would be weakened by G3991A. The other second-site alteration was near the beginning of the UTR, with no discernible connection between this sequence and H5.
For mutant A3976C/A3978C, which contained the LSL sequence 5′ UACACU, sequencing of 27 clones resulted in the recovery of the following sequences in this location: no change (maintenance of both mutations), 6 of 27; wt (reversion at both positions), 16 of 27; reversion at a single position (5′ UACAAU), 1 of 27; primary-site alteration with a reversion at the other original position (5′ AAAUU), 4 of 27. In addition, within the 200 bases sequenced, six clones contained single second-site mutations, two had double mutations, and one had three mutations (Fig. 3). Five of these alterations, found in clones derived from three plants, were a guanylate-to-uridylate change at position 3991 in the 3′ side of the LSL, directly opposite the original A3978C alteration. Since a uridylate in this location was highly detrimental to TCV accumulation (G3991U; see Fig. 2), this alteration is likely compensating for changes to the 5′ side of the LSL. With this exception, all other second-site changes were scattered throughout the sequenced region and were biased toward uridylate-to-cytidylate (2 of 8) and adenylate-to-guanylate (3 of 8) transitions.
In all, 33 second-site mutations were recovered outside the LSL in 23,500 bases sequenced, which corresponds to a mutation frequency of 1.4 × 10−3 (Table 1). Since most of the region sequenced was in the 3′ UTR, it is possible that a lower mutation frequency exists in coding regions. To determine if the mutation frequency for the TCV LSL constructs, and the mutation bias noted above, were unusual compared with that of wt TCV, wt TCV progeny from identically infected plants were subjected to RT-PCR and 31 clones were sequenced. Only one alteration at position 3806 (adenylate to cytidylate) was found in the 6,200 bases sequenced, corresponding to an approximate mutation frequency of 0.2 × 10−3. This value is consistent with the 0.2 × 10−3 mutation frequency determined for some picornaviruses (5, 10). Alterations in H5 thus increased the mutation frequency 2.5-fold for A3976C to 12-fold for A3976C/A3978C, with an overall increase for all mutants of 7-fold (Table 1).
To ascertain whether mutations were spuriously introduced during in vitro transcription by T7 RNA polymerase prior to inoculation of turnip seedlings (which, if inoculated, would be more evident in the poorly replicating mutants than in wt TCV), a construct containing satC downstream of a T7 promoter was digested with SmaI and in vitro transcribed. The RNA transcripts were directly subjected to RT-PCR by using an oligonucleotide homologous to satC positions 1 through 19 and complementary to positions 336 to 356, and cDNA was subsequently cloned. No mutations were found in 7,750 bases sequenced from 25 clones. Interestingly, when similar lesions were introduced into either the 5′ or the 3′ side of the satC LSL and mutant satC was coinoculated onto Arabidopsis with wt TCV, no significant increase in the mutation frequency of satC was found (J. Zhang et al., unpublished data). However, it is possible that the limited size of satC (356 bases) imposes sequence constraints on the molecule that are more rigid than the constraints on the 3′ UTR of TCV.
While the effects of individual second-site mutations have not been evaluated for the TCV H5 LSL constructs, several observations lead to the possibility that the vast majority of second-site changes are not compensatory. First, these mutations were found in 26 different positions scattered throughout the sequenced region (Fig. 3). Second, the composition of the changes was strongly biased towards uridylate-to-cytidylate (15 of 33) and adenylate-to-guanylate (7 of 33) transitions (Table 1), suggesting a specific replication defect. Third, the second-site changes were nearly all coupled with reversion of the original alterations, suggesting that the second-site changes were not compensating for the original defects in the H5 LSL. Finally, although A3976C accumulated to 74% of wt TCV levels in protoplasts (Fig. 2B), the progeny still exhibited an increased mutation frequency, suggesting that this phenomenon is more complex than a simple association between poorly replicating constructs and enhanced mutation frequency.
One explanation for the biased second-site alterations may be that poorly replicating mutant viruses multiply undetectably until a mutation occurs in the RdRp that allows for reversion of the original LSL alteration while reducing RdRp fidelity. A second explanation is that H5, in addition to its role as a repressor of minus-strand synthesis, functions as an RNA chaperone that nucleates the formation of an active RdRp complex. An incorrectly assembled or misfolded RdRp could have lower fidelity and more readily mispair templated uridylates with guanylates during transcription of plus and minus strands. RNA elements in the 3′ UTR of Alfalfa mosaic virus (23) and the 3′ UTR and intercistronic regions in RNA 3 of Brome mosaic virus (BMV) in yeast (15) are known to be required for formation of an active RdRp. In addition, studies using BMV replicase suggest that RdRp's are inherently flexible and able to adjust their structure based on contact with a few key nucleotides (21).
Since RNA templates are not substrates for postpolymerization mismatch repair, RNA replication leads to a dynamic quasispecies population consisting of a dominant “master” sequence and master sequence variants that differ in abundance and level according to the intrinsic properties of the virus and perturbations in the host microenvironment (6, 9, 17). Increases of as little as 1.2-fold in the mutation frequency of poliovirus following treatment with nucleoside analogues resulted in steep losses in virus viability and rapid population extinction (error catastrophe) (4, 18). These observations suggest that RNA viruses exist precariously close to the error threshold in order to maximize the genetic diversity of their viral population (quasispecies cloud size) and their potential for adaptation to fluctuating environmental conditions (4, 7). A mechanism that could transiently increase the mutation frequency in excess of the error threshold while avoiding extinction of the population by high-frequency reversion of the original mutation would increase the rate of evolution and quasispecies cloud size of the virus, thereby enhancing adaptive potential without permanently altering the RdRp. Observations presented in this study suggest that this novel attribute may exist for TCV.
Acknowledgments
This work was supported by grants from the NIH (RO1 GM61515-01) and NSF (MCB-0096274) to A.E.S. J.C.M. was supported by NIH training grant T32AI51967.
We thank Robert Stuntz and Kari Linstrom for technical assistance.
REFERENCES
- 1.Carpenter, C. D., J.-W. Oh, C. Zhang, and A. E. Simon. 1995. Involvement of a stem-loop structure in the location of junction sites in viral RNA recombination. J. Mol. Biol. 245:608-622. [DOI] [PubMed] [Google Scholar]
- 2.Carpenter, C. D., and A. E. Simon. 1998. Analysis of sequences and predicted structures required for viral satellite RNA accumulation by in vivo genetic selection. Nucleic Acids Res. 26:2426-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrington, J. C., L. A. Heaton, D. Zuidema, B. I. Hillman, and T. J. Morris. 1989. The genome structure of turnip crinkle virus. Virology 170:219-226. [DOI] [PubMed] [Google Scholar]
- 4.Crotty, S., C. E. Cameron, and R. Andino. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA 98:6895-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Torre, J. C., E. Wimmer, and J. J. Holland. 1990. Very high frequency of reversion to guanidine resistance in clonal pools of guanidine-dependent type 1 poliovirus. J. Virol. 64:664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 7.Drake, J. W., and J. J. Holland. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 96:13910-13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hacker, D. L., I. T. D. Petty, N. Wei, and T. J. Morris. 1992. Turnip crinkle virus genes required for RNA replication and virus movement. Virology 186:1-8. [DOI] [PubMed] [Google Scholar]
- 9.Holland, J., K. Spindler, F. Horodyski, E. Grabau, S. Nicho, and S. van de Pol. 1982. Rapid evolution of RNA genomes. Science 215:1577-1585. [DOI] [PubMed] [Google Scholar]
- 10.Holland, J. J., J. C. de la Torre, and D. A. Steinhauer. 1992. RNA virus populations as quasispecies. Curr. Top. Microbiol. Immunol. 176:1-20. [DOI] [PubMed] [Google Scholar]
- 11.Kong, Q., J.-W. Oh, C. D. Carpenter, and A. E. Simon. 1997. The coat protein of turnip crinkle virus is involved in subviral RNA-mediated symptom modulation and accumulation. Virology 238:478-485. [DOI] [PubMed] [Google Scholar]
- 12.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 13.Oh, J.-W., Q. Kong, C. Song, C. D. Carpenter, and A. E. Simon. 1995. Open reading frames of turnip crinkle virus involved in satellite symptom expression and incompatibility with Arabidopsis thaliana ecotype Dijon. Mol. Plant-Microbe Interact. 8:979-987. [DOI] [PubMed] [Google Scholar]
- 14.Pogany, J., M. R. Fabian, K. A. White, and P. D. Nagy. 2003. A replication silencer element in a plus-strand RNA virus. EMBO J. 22:5602-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quadt, R., M. Ishikawa, M. Janda, and P. Ahlquist. 1995. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc. Natl. Acad. Sci. USA 92:4892-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajendran, K. S., J. Pogany, and P. D. Nagy. 2002. Comparison of turnip crinkle virus RNA-dependent RNA polymerase preparations expressed in Escherichia coli or derived from infected plants. J. Virol. 76:1707-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider, W. L., and M. J. Roossinck. 2001. Genetic diversity in RNA virus quasispecies is controlled by host-virus interactions. J. Virol. 75:6566-6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sierra, S., M. Dávila, P. R. Lowenstein, and E. Domingo. 2000. Response of foot-and-mouth disease virus to increased mutagenesis: influence of viral load and fitness in loss of infectivity. J. Virol. 74:8316-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon, A. E., and S. H. Howell. 1986. The virulent satellite RNA of turnip crinkle virus has a major domain homologous to the 3′ end of the helper virus genome. EMBO J. 5:3423-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song, C., and A. E. Simon. 1995. Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J. Mol. Biol. 254:6-14. [DOI] [PubMed] [Google Scholar]
- 21.Stawicki, S., and C. C. Kao. 1999. Spatial perturbations within an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase (RdRp) reveal that RdRp can adjust its promoter binding sites. J. Virol. 73:198-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stupina, V., and A. E. Simon. 1997. Analysis in vivo of turnip crinkle virus satellite RNA C variants with mutations in the 3′-terminal minus-strand promoter. Virology 238:470-477. [DOI] [PubMed] [Google Scholar]
- 23.Vlot, A. C., L. Neeleman, H. J. M. Linthorst, and J. F. Bol. 2001. Role of the 3′ untranslated regions of alfalfa mosaic virus RNAs in the formation of a transiently expressed replicase in plants and in the assembly of virions. J. Virol. 75:6440-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, G., J. Zhang, and A. E. Simon. 2004. Repression and derepression of minus-strand synthesis in a plus-strand RNA virus replicon. J. Virol. 78:7619-7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]