Abstract
Despite remarkable advances in the genomic characterization of adult melanoma, the molecular pathogenesis of pediatric melanoma remains largely unknown. We analyzed 15 conventional melanomas (CMs), 3 melanomas arising in congenital nevi (CNMs), and 5 spitzoid melanomas (SMs), using various platforms, including whole genome or exome sequencing, the molecular inversion probe assay, and/or targeted sequencing. CMs demonstrated a high burden of somatic single-nucleotide variations (SNVs), with each case containing a TERT promoter (TERT-p) mutation, 13/15 containing an activating BRAF V600 mutation, and >80% of the identified SNVs consistent with UV damage. In contrast, the three CNMs contained an activating NRAS Q61 mutation and no TERT-p mutations. SMs were characterized by chromosomal rearrangements resulting in activated kinase signaling in 40%, and an absence of TERT-p mutations, except for the one SM that succumbed to hematogenous metastasis. We conclude that pediatric CM has a very similar UV-induced mutational spectrum to that found in the adult counterpart, emphasizing the need to promote sun protection practices in early life and to improve access to therapeutic agents being explored in adults in young patients. In contrast, the pathogenesis of CNM appears to be distinct. TERT-p mutations may identify the rare subset of spitzoid melanocytic lesions prone to disseminate.
Introduction
Melanoma in children and adolescents is rare (Howlader et al., 2013), but its incidence continues to rise, particularly in the age group of 15–19 years (Wong et al., 2013). Although certain predisposing factors, such as xeroderma pigmentosum, family history of melanoma, and the presence of a large/giant congenital melanocytic nevus, may increase the risk of developing melanoma in early life, most pediatric cases are sporadic (Pappo, 2003). In addition, similar to the adult counterpart, pediatric melanoma is more prevalent among fair-skinned individuals with a propensity to sunburn (Whiteman et al., 1997; Strouse et al., 2005).
Over the past decade, considerable progress has been made in elucidating the molecular pathogenesis of adult melanoma. It has been shown that ∼60% of adult melanoma carry an oncogenic BRAF V600 and ∼20% carry an oncogenic NRAS mutation (Davies et al., 2002; Lee et al., 2011). These discoveries have been rapidly translated into the clinic, as exemplified by promising results using targeted therapy against BRAF (Flaherty et al., 2011).
The development of melanoma is believed to be the result of sequential acquisition of multiple mutations that cooperate to induce overt malignancy. The first event is the acquisition of a driver mutation in the RAS/RAF/mitogen-activated protein kinase pathway that promotes melanocytic proliferation. The oncogenic mutations in BRAF and NRAS or the kinase fusions, however, are insufficient on their own for the development of cancer, as evident by their presence in the nevi of acquired, congenital, or spitzoid types (Pollock et al., 2003; Bauer et al., 2007; Wiesner et al., 2014). Additional genetic events, such as disruption of the RB/p16 tumor suppressor pathway, activation of the phosphatidylinositol 3 kinase/AKT pathway, and reactivation of telomere maintenance mechanisms, are necessary for malignant transformation (Gray-Schopfer et al., 2006; Soo et al., 2011). The recently discovered UV-signature mutations in TP53, RAC1, STK19, PPP6C, PREX2, or those in the core promoter of TERT (telomerase reverse transcriptase) are consistent with this model of melanoma tumorigenesis (Berger et al., 2012; Hodis et al., 2012; Horn et al., 2013; Huang et al., 2013).
Although the genomic characterization of adult melanoma has drawn much attention and has been therapeutically exploited, the genomic landscape of pediatric melanoma has not been investigated. Childhood and adolescent melanomas comprise a heterogeneous group of melanocytic neoplasms, in terms of their clinical and pathological characteristics that can be divided into three categories (Barnhill, 2006). The first type is pediatric conventional melanoma (CM) that shares the histological and clinical features of melanoma arising in intermittently sun-exposed skin in adults. CM is exceedingly rare to occur before puberty; hence, the majority of pediatric patients with CM are teenagers (Barnhill et al., 1995; Scalzo et al., 1997). The second type arises in association with a large/giant congenital melanocytic nevus. The lifetime risk of malignant transformation in a giant congenital melanocytic nevus is 5 to 10% that most often occurs in the first decade of life (Bittencourt et al., 2000; Bett, 2005; Krengel et al., 2006). Finally, in the spectrum of spitzoid melanocytic tumors, spitzoid melanoma (SM) and atypical Spitz tumor are distinct histologic variants with a less aggressive clinical course compared with conventional melanoma and rare extranodal metastatic potential (Barnhill et al., 1995; Berk et al., 2010).
To explore the genomic landscape of pediatric melanoma, we studied a well-annotated cohort of 23 children and adolescents with “de novo conventional melanoma” (CM, n=15), “melanoma arising in a congenital melanocytic nevus” (CNM, n=3), and “spitzoid melanoma” (SM, n=5) (Figure 1 and Supplementary Table S1 and Supplementary Figure S1 online).
Figure 1.
The clinical and genomic data for 23 pediatric melanomas analyzed by whole genome, whole exome, and/or the molecular inversion probe assay. The mutation rate plot displays the mutation rates in the coding regions for the three subtypes of pediatric melanoma. The gray horizontal line shows the median coding mutation rate in cutaneous melanoma in adults. The mutation spectrum plot in pediatric melanoma demonstrates a high rate of cytidine to thymidine (C–>T) or guanine to adenine (G–>A) transitions in each conventional melanoma sample. The red horizontal line depicts the median rate of transition mutations in melanoma in adults (adapted from data in Hodis et al., 2012). The MC1R variants shown in red font are associated with complete loss of gene function; the variants shown in black font are associated with partial loss of gene function. * SJMEL001020 denoted with an asterisk is an acral melanoma. CTX, translocation; DEL, deletion; MIP, molecular inversion probe assay; SV, structural variation; WES, whole exome sequencing; WGS, whole genome sequencing.
Results
Whole genome sequencing (WGS) and/or whole exome sequencing (WES) were performed in 20 melanomas and paired nontumor germline samples in parallel from 18 patients (Figure 1). Data were analyzed for single-nucleotide variations (SNVs), small insertion/deletions (indels), structural variations (SVs), including inter- and intra-chromosomal rearrangements, and copy number variations (CNVs). RNA sequencing was successfully carried out in seven melanomas from five patients. The molecular inversion probe (MIP) assay to profile CNVs was performed in 19 melanomas. Targeted gene sequence for BRAF, NRAS, and TERT-p was analyzed in all 23 melanomas. Immunohistochemistry for ALK and fluorescence in situ hybridization (FISH) assays for ROS1, NTRK1, RET, and BRAF were applied to five SMs to explore the presence of selected gene rearrangements. Figure 1 shows major findings in the genomic analysis, including the prevalence of mutations in cancer genes and commonly mutated genes. Supplementary Table S7 online shows the tier 1 variants (SNVs and indels) as detected by WGS/WES in each sample, and Supplementary Tables S9 online shows genes affected by gains and losses in a statistically significant number of subjects. Two patients (SJMEL001003 and SJMEL001004) each contributed two tumor samples (D1 and D2) for WGS. Comparison of the SNVs between D1 and D2 showed a high percentage (>90%) of overlapping mutations in both patients (Supplementary Tables S6 online).
UV-induced mutational spectrum in pediatric CM
Across the 13 CMs analyzed by WGS/WES, the coding mutation rates averaged 14.36 per megabase (range, 3.21 to 52.65) (Supplementary Tables S2 and S3 online), comparable to the mutation rates in the adult counterpart (range, 5–55) (Berger et al., 2012). The ratio of nonsynonymous to silent mutations across the WGS samples ranged from 1.27 to 2.22 (median, 1.82) (Supplementary Table S2 online), consistent with a high load of nonselected passenger mutations. Importantly, an analysis of the type of SNV revealed that >80% of the somatic mutations in CM were cytidine to thymidine (C–>T) or guanine to adenine (G–>A) transitions, and ⩾90% of the mutations occurred 3′ to a pyrimidine base, consistent with UV-induced DNA damage (Supplementary Figure S3 online) (Pfeifer et al., 2005; Daya-Grosjean and Sarasin, 2005; Leiter and Garbe, 2008; Wei et al., 2011). In addition, all of the WGS samples with a high prevalence of C–>T/G–>A mutations contained high numbers of tandem dinucleotide CC–>TT mutations, accounting for >70% of their total dinucleotide mutations (Supplementary Table S2 online). Moreover, when the proportion of C–>T/G–>A mutations was compared in relationship with the strand type, the number of C–>T/G–>A mutations was lower on the transcribed than on the nontranscribed strand (Supplementary Figure S4 online). Taken together, these data show evidence of UV-induced damage in the genomic DNA of pediatric CM. The mutation rates of the 2 CNMs varied by one order of magnitude (1.85 vs. 15.03), likely reflective of their different cumulative sun exposure (Supplementary Table S2 online).
SVs in pediatric melanoma
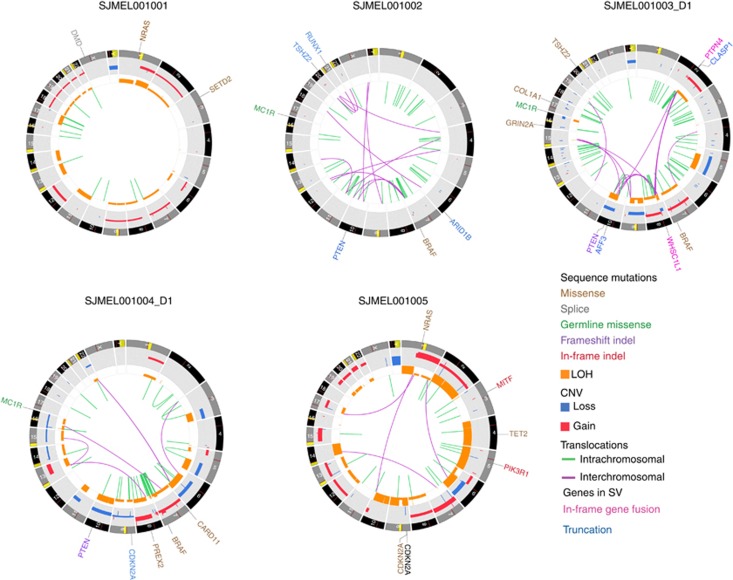
The SVs (deletions, insertions, and translocations) detected by WGS ranged from 11 to 89 events per sample (median, 38) (Supplementary Table S4 online). The 3 CMs harbored a higher number of SVs (33–89) compared with the 2 CNMs (11–19) and exhibited localized clustering of structural rearrangements in the regions of CNV, reminiscent of chromotripsis. For example, we found high clusters of SVs on chromosomes 2, 8, and 10, including a large number of translocations between chromosomes 2 and 8 and high clusters of SVs disrupting CLASP1 (a gene involved in the regulation of microtubule dynamics) in SJMEL001003, clusters of SVs on chromosome 7 (between 135 and 149 Mb) in SJMEL001002, and regional clustering of intrachromosomal translocations in chromosomes 7 and 8 in SJMEL001004 (Figure 2). Such complex SVs were not present in CNM.
Figure 2.
Circos plots showing somatic mutation landscape for the five whole genome sequenced samples. Because of the large number of mutations, only cancer-related genes are labeled in the figure. Inner circle shows interchromosomal (purple) and intrachromosomal (green) translocations. Inner track (orange) denotes area of LOH (loss of heterozygosity). Gray track denotes somatic copy number variations (CNVs) (red=gain; blue=loss). Genes affected by point mutations (single-nucleotide variations (SNVs) and indels) and copy number changes are shown in the outer ring with colors denoting the type of mutation. Truncations and fusion genes caused by translocations are denoted by blue and pink color, respectively. All somatic variants have been validated by targeted deep sequencing.
RNA sequencing with fresh tumor was performed in a subset of CMs (five samples) and CNM (one sample). The detected SVs in each case (Supplementary Table S8 online) matched with the WGS data. RNA sequencing using formalin-fixed, paraffin-embedded (FFPE) material was attempted in 3 SMs, of which only one (SJMEL001032) had high base coverage for data analysis, showing an in-frame GSN/NTRK1 fusion (Supplementary Figure S13 online) that was confirmed by the break-apart NTRK1 FISH assay (Supplementary Figure S5 online). This finding is consistent with the recent observations that identified chromosomal rearrangement-induced fusions involving ROS1, NTRK1, ALK, BRAF, and RET as a common mechanism of oncogene activation in spitzoid neoplasms (Wiesner et al., 2014). A panel of break-apart FISH assays for ROS1, NTRK1, BRAF, and RET, applied to the four other SMs, identified an additional SM (SJMEL001033) with BRAF rearrangement, whereas the three other SMs were negative for gene rearrangements (Figure 1 and Supplementary Figure S5 online). None of the cases expressed ALK protein by immunohistochemistry to suggest ALK being involved.
CNVs in pediatric melanoma
The MIP assay and WGS revealed genomic instability with multiple CNVs in each CM (Supplementary Figures S6 and S11 online). Common regions of alteration in CM were akin to those reported in adult melanoma (Bastian et al., 1998, 2003) (Figure 3). Across the entire cohort of 19 melanomas, MIP analysis found a gain in some portion of chromosome 7q (79%), 7p (63%), 6p (63%), 1q (58%), and 20q (58%), and a loss in some portion of chromosome 9p (63%), 5q (58%), 11q (58%), and 10q (47%) (Supplementary Table S5 online). Supplementary Figure S12 online shows the prevalence of gains and losses across the genome for CMs, CNMs, and SMs separately.
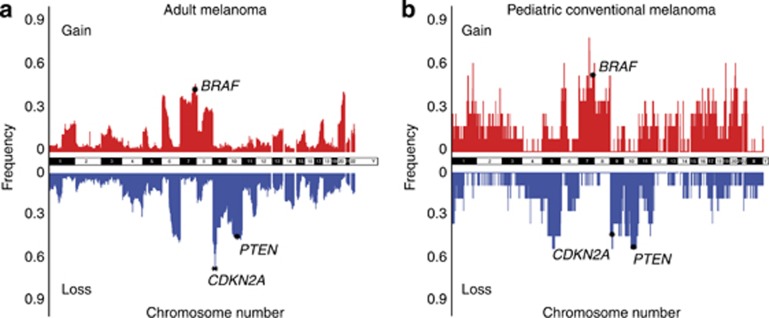
Figure 3.
Comparing the prevalence of copy number variations (CNVs) across the entire chromosomes between adult and pediatric conventional melanoma. (a) The frequency of CNVs in 95 nonacral adult cutaneous melanomas using the single-nucleotide polymorphism (SNP) array data from the study by Hodis et al. (2012) and (b) the frequency of CNVs in 11 pediatric conventional melanomas in our study. The 7q34 locus harboring BRAF was a common region of gain, whereas 9p21 and 10q23.3, spanning CDKN2A and PTEN, were common regions of loss in both adult and pediatric melanomas.
Recurrent oncogenes in pediatric melanoma
Of the 15 CMs, 13 contained an activating BRAF mutation (12 BRAF V600E; 1 dinucleotide BRAF V600K) (Figure 1). Of the two CMs with wild-type BRAF, one was an acral melanoma with wild-type KIT (SJMEL001020) and the other was a nodular melanoma with a missense mutation in PIK3R1 (SJMEL001029) (Figure 1). Integrated analysis showed that 55% of all CMs (Figure 3) and 75% of BRAF-mutant melanomas had copy gains of BRAF at 7q34. The three CNMs, in contrast, contained activating NRAS mutations (pQ61K, pQ61R, and pQ61H) (Figure 1). A tandem duplication of MITF (cooperating oncogene) was additionally found in one CNM (SJMEL001005). The five SMs lacked BRAF or NRAS activating point mutations but rather contained kinase fusions in a subset, as described above.
Tumor suppressor genes in pediatric melanoma
The PTEN tumor suppressor gene (the negative regulator of the phosphatidylinositol 3 kinase/AKT pathway) was commonly disrupted in CM. Gene alterations occurred either by gross structural changes (deletion or translocations) or at the nucleotide level (inactivating mutations). Alterations of PTEN, in contrast, were not found in the three CNMs. Considering limitations of various tests, the detection of gene alterations varied across different assays; for example, the FISH and MIP assays revealed that 54% and 50% of CMs, respectively, have a loss in PTEN at 10q23.3, whereas WGS found an alteration in all 3 CMs analyzed, truncating mutations in SJMEL001003 and SJMEL001004, and complex SV in SJMEL001002 (Figures 1 and 2).
Alterations of CDKN2A (the negative regulator of cell cycle G1 progression) were common in all subtypes of pediatric melanoma. Although by immunohistochemistry, 66% of CMs demonstrated complete loss of CDKN2A (p16INK4a) protein, the detection of gene alteration was variable by different assays. For example, by the MIP assay, 45% of CMs had evidence of a copy loss at the locus of CDKN2A (Figure 3), and by FISH, 50% had evidence of CDKN2A gene deletion (mono- or biallelic) (Figure 1 and Supplementary Figure S7 online). Loss of protein without gross gene alterations suggests that the gene might be altered at the nucleotide sequence level (Jonsson et al., 2010). In fact, WGS revealed homozygous CDKN2A microdeletions in SJMEL001004 and putative loss of function SNV in SJMEL001005, whereas FISH in both had shown no alterations.
TP53 mutations were detected in 2 of 18 WGS/WES samples; a germline TP53 missense mutation (R213Q) in a SM (SJMEL001033) and a somatic UV signature TP53 missense mutation (P278S) in a CM (SJMEL001027) (see details in Supplementary Materials online).
TERT promoter (TERT-p) is frequently mutated in pediatric CM
The 23 melanomas were analyzed for SNVs in the TERT-p sequence (Horn et al., 2013; Huang et al., 2013). Of the 15 CMs, 11 tumors had 1 of the 2 known SNVs in mutually exclusive groups (6 SNV 228; 5 SNV 250), 1 tumor had a 242/243 dinucleotide variant, and 2 tumors were marked as indeterminate because of poor sequence quality. Excluding the 2 indeterminate samples, 92% (12 of 13) of CMs had an SNV 228 or an SNV 250 or the 242/243 variant mutation in the TERT-p region. The only CM with wild-type TERT-p was an acral melanoma (Figure 1). Two CMs contained CC->TT mutations: one was the previously described dinucleotide variant (228/229) and the other was a CC–>TT dinucleotide variant (242/243) that generated an ETS1 binding site immediately upstream of the TERT transcription initiation site, similar to the previously described variant (Horn et al., 2013; Huang et al., 2013). Remarkably, only the one SM with hematogenous spread (SJMEL001030), of the total 5 SMs, contained a TERT-p mutation (SNV 228). On the other hand, none of the three CNMs carried such mutations (Figure 1).
Germline mutational analysis of melanoma susceptibility genes
Germline DNA screening for mutations in the high-penetrance melanoma susceptibility genes, CDKN2A and CDK4 (Ward et al., 2012), and for a panel of DNA repair genes (Lange et al., 2011) was negative for a carrier in this cohort. However, 67% (8 of 12) of CMs (excluding the one acral subtype) and 67% (2 of 3) of SMs carried 1 or 2 of the 4 most common germline MC1R variants (V60L, V92M, R151C, and R160W). None of the patients with CNM harbored a germline MC1R variant (Figure 1).
Discussion
To our knowledge, this study is the most comprehensive genomic analysis of pediatric melanoma reported to date. Using a combination of sequencing platforms, we found unique genomic features for the three subtypes of pediatric melanoma evaluated in this study. Our data showed that pediatric CM contained a high burden of somatic SNV, far greater than any other pediatric tumor sequenced in the Pediatric Cancer Genome Project to date (Supplementary Figure S9 online). Although compelling epidemiological evidence suggests that UV exposure in early childhood causes melanoma later in life, the genotoxic effect of UV light has not been previously explored at the genomic level in pediatric melanoma. Our study revealed that the vast majority of SNVs in pediatric CM are C–>T/G–>A transitions, reflective of UV-induced DNA damage. In addition, with the exception of the one acral melanoma, all CMs contained a TERT-p C–>T mutation, providing a molecular basis for the contribution of UV light in the pathogenesis of the disease in this young population. These mutations are pathogenically relevant and have been shown to increase the transcriptional activity of TERT and to confer oncogenic advantage by enabling melanocytes to sustain their telomere length and become immortalized (Horn et al., 2013; Huang et al., 2013).
Our study revealed that 87% of CMs have an activating BRAF V600 mutation and commonly an alteration in PTEN. In contrast, CNMs were characterized by NRAS Q61 mutations and no detectable defect in PTEN. The varied frequency in PTEN aberration between NRAS- and BRAF-driven melanomas reflects their difference in activating intracellular signaling pathways. NRAS activates both the mitogen-activated protein kinase and phosphatidylinositol 3 kinase/AKT pathways, whereas BRAF solely stimulates the mitogen-activated protein kinase pathway and requires cooperating genetic modifiers, such as loss of PTEN, to facilitate activation of the phosphatidylinositol 3 kinase/AKT pathway (Tsao et al., 2004).
Although all NRAS-mutant melanomas in our series arose within a congenital nevus, a distinct histopathological subtype has not been linked to this melanoma genotype in adults (Curtin et al., 2005; Broekaert et al., 2010). Larger studies are needed to determine whether or not NRAS-mutant melanomas in the pediatric population are exclusive to those arising in association with congenital nevi. Moreover, the oncogenic role of UV light in CNM will have to be further explored. Although one CNM (SJMEL001001), a multiply recurrent scalp melanoma in an infant (Supplementary Figure S1 online), contained a high burden of UV-signature mutations, the role of UV radiation in the genesis or progression of melanoma in this case must be viewed with caution. Malignant melanocytes are prone to acquire additional UV-induced passenger mutations without contributing to tumor initiation. Regrettably, the primary diagnostic material was not available to explore the genomic DNA of this tumor in its earlier phase of development. Notably, neither this nor any other patient with CNM in our series carried germline DNA mismatch repair genes or MC1R variants (see below). Importantly, none of them carried a UV-signature TERT-p mutation, suggesting that these tumors are under a different set of genetic constraints to maintain their telomere integrity relative to the other subtypes of melanomas. The clinical and genomic data together suggest that CNM is a biologically distinct melanoma subtype (Kinsler et al., 2013; Krengel et al., 2006).
A subset of SMs in our cohort had kinase fusions, consistent with the oncogenic mechanism recently described in spitzoid tumors (Botton et al., 2013; Wiesner et al., 2014). SMs and atypical Spitz tumors belong to a histologic spectrum of melanocytic tumors with a morphologic resemblance to Spitz nevus, affecting more commonly younger individuals. Despite frequent regional nodal involvement, hematogenous metastasis is rare in these tumors, particularly in children (<10 years of age). Disease dissemination, however, can occur on rare occasions but cannot be predicted with certainty using morphological parameters (Barnhill et al., 1995; Paradela et al., 2009). The homozygous CDKN2A (9p21) deletion, recently suggested as a marker of high-risk spitzoid melanocytic tumors, was not a reliable predictor for extranodal spread in our cohort (Gerami et al., 2013). For example, two SMs in our study had biallelic loss of CDKN2A, but neither one developed extranodal metastasis; in contrast, the metastatic lung nodule in the one disseminating SM retained both of its CDKN2A alleles (Supplementary Figure S8 online). Although the most commonly altered gene in melanoma (Curtin et al., 2005), CDKN2A is retained in a subset (Bartkova et al., 1996), as demonstrated at the gene and protein levels in 13% of CMs in our study that overtly expressed p16 protein (Figure 1). A marker to more accurately predict the prognosis is desperately needed to assist in the decision-making process in the management of these lesions.
The one disseminating SM in our study contained a TERT-p mutation that was identical to the variant seen in CM, whereas no other SM carried these mutations, suggesting that a common mechanism of telomere maintenance may be implicated in SM with extranodal metastatic potential and CM. The prognostic relevance of TERT-p mutations as a potential marker of aggressive behavior in spitzoid lesions will have to be further explored in larger cohorts with long-term follow-up data.
Seeking to explain an early onset of melanoma, we searched potential environmental and genetic predisposing factors in our patient cohort. A history of excessive sun exposure or blistering sunburn could not be fully explored in the historical cases in our cohort, but environmental factors were considered to be a contributor in the development of CM in two teenagers engaged in habitual indoor tanning practices. None of our patients carried the known melanoma susceptibility genes CDKN2A or CDK4, but 67% of those with CM and SM carried 1 or 2 germline MC1R variants (Figure 1). MC1R is a highly polymorphic gene in Caucasians, and its variants are associated with certain phenotypic traits, such as red hair and fair skin, and an increased susceptibility to melanoma (Landi et al., 2006; Raimondi et al., 2008; Ward et al., 2012). The discovery of other potential germline polymorphisms implicated in melanoma predisposition in young people needs to be explored through large-scale correlative studies.
In summary, our study has demonstrated that CM in younger patients has many genomic similarities to adult melanoma, including an enrichment of UV-induced mutations, a high prevalence of UV-signature TERT-p mutations, and involvement of similar oncogenes and tumor suppressor genes. The controversies surrounding the diagnosis and prognosis of childhood and adolescent melanoma have led to the almost uniform exclusion of these patients to date from promising trials offered for adults, a strategy that has hampered research efforts and access to treatments in this population. Collectively, the data from our study suggest that therapeutic targets for genotype-specific melanoma in adults might be applicable to pediatric patients. Moreover, our study has revealed molecular evidence for the contribution of UV light in pediatric melanoma development. Fighting melanoma should start with prevention, public awareness, and changing the attitude toward sun exposure by acquiring a habit of sun protection starting in early life.
Materials and methods
Institutional approval of experiments was obtained. Written, informed patient consent was waived because the research involved no more than minimal risk to the subjects. The human investigations were performed after approval by the local institutional review board.
Study population
CM occurred in 15 Caucasian patients aged 11–20 years (median, 16), with the histologic subtypes of nodular (7), superficial spreading (5), acral (1), and unknown primary (2) (Figure 1). At last follow-up (mean, 30 months), 10 patients had died of disease, 3 were alive with disease, and 2 had no evidence of disease. CNM affected 3 children ≤5 years of age who died of disease within an average of 22 months. SM occurred in 5 patients (3 children, 1 adolescent, and 1 borderline age), of whom 1 died of disease and 4 were alive with no evidence of disease at last follow-up (mean, 32 months) (Figure 1 and Supplementary Table S1 online).
Whole genome sequencing
DNA was isolated from seven fresh frozen tumor samples and matched blood from five patients (Figure 1). Sequencing was conducted on a HiSeq 2000 (Illumina, San Diego, CA) using the paired end 2 × 100 cycle protocol, as previously described (Zhang et al., 2012). Validation of variants identified in WGS data was performed as described previously (Chen et al., 2014).
Whole exome sequencing
DNA was isolated from 14 paired FFPE tumor (11 CMs and 3 SMs) and normal tissue/blood samples. WES was completed in 10 samples (8 CMs and 2 SMs) with 87.4 and 90.3% of exonic bases covered at least 20-fold in the tumor and matched normal germline DNA (details in Supplementary Materials online; Supplementary Figure S2 and S10 online).
RNA sequencing
RNA was isolated from frozen tumors (5 CMs and 1 CNM) and FFPE tumors (3 SMs) (details in Supplementary Materials online). The sequencing data were generated as previously described (Parker et al., 2014).
Affymetrix OncoScan MIP assay
Genomic DNA was extracted from FFPE sections of 19 melanomas (Figure 1), guided by the corresponding hematoxylin and eosin–stained slides, to attain >80% tumor purity. DNA was extracted using the QIAamp DNA FFPE tissue kit (Qiagen Sciences, Germantown, MD) according to the manufacturer's instruction (details in Supplementary Materials online). A minimum quantity of 200 ng of genomic DNA was used as input per Affymetrix OncoScan guidelines (Affymetrix, Santa Clara, CA).
Mutation analysis of BRAF and NRAS
Mutational hotspots for BRAF (exon 15) and NRAS (exons 1 and 2) were screened in genomic DNA of 23 melanomas. PCRs were performed using GoTaq Long PCR Master Mix (Promega, Madison, WI). Direct sequencing of PCR products was performed using BigDye version 3.1 and a 3730XL DNA analyzer (Applied Biosystems, Foster City, CA). Results were screened using CLC Main Workbench sequence analysis software version 6.0.2 (CLC bio, Cambridge, MA).
Mutation analysis of TERT-p
A portion of the TERT-p (HG19 coordinates, chr5: 1295151-1295347) was amplified in 23 melanomas by PCR. Briefly, 20–50 ng of sample DNA was added to a 25 μl reaction containing amplitaq gold 360 master mix (Applied Biosystems) with 400 nM each of amplification primers (details in Supplementary Materials online). The sequencing and analysis were performed as described in the previous section.
Data analysis and statistical methods
See Supplementary Materials online.
Acknowledgments
This work was supported, in part, by Cancer Center Support (CA21765) from the NCI, grants to MAD from the NIH (EY014867 and EY018599 and CA168875), and the American Lebanese Syrian Associated Charities (ALSAC). MAD is a Howard Hughes Medical Institute Investigator. The whole genome sequencing was supported as part of the St Jude Children's Research Hospital–Washington University Pediatric Cancer Genome Project.
Glossary
- CM
conventional melanoma
- CNM
melanomas arising in a congenital nevus
- CNV
copy number variation
- FFPE
formalin-fixed, paraffin-embedded
- FISH
fluorescence in situ hybridization
- MIP
molecular inversion probe assay
- SM
spitzoid melanoma
- SNV
single-nucleotide variation
- SV
structural variation
- TERT-p
promoter of telomerase reverse transcriptase
- WES
whole exome sequencing
- WGS
whole genome sequencing
The authors state no conflict of interest.
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/jid
Supplementary Material
References
- Barnhill RL, Kerl H.2006Childhood melanoma and melanoma arising in giant congenital naeviIn: Leboit PE, Burg G, Weedon D, Sarasin A (eds)World Health Organization Classification of Tumours: Pathology and Genetics of Skin Tumours Lyon: IARC; 83–85. [Google Scholar]
- Barnhill RL, Flotte TJ, Fleischli M, et al. Cutaneous melanoma and atypical Spitz tumors in childhood. Cancer. 1995;76:1833–1845. doi: 10.1002/1097-0142(19951115)76:10<1833::aid-cncr2820761024>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Lukas J, Guldberg P, et al. The p16-cyclin D/Cdk4-pRb pathway as a functional unit frequently altered in melanoma pathogenesis. Cancer Res. 1996;56:5475–5483. [PubMed] [Google Scholar]
- Bastian BC, LeBoit PE, Hamm H, et al. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res. 1998;58:2170–2175. [PubMed] [Google Scholar]
- Bastian BC, Olshen AB, LeBoit PE, et al. Classifying melanocytic tumors based on DNA copy number changes. Am J Pathol. 2003;163:1765–1770. doi: 10.1016/S0002-9440(10)63536-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J, Curtin JA, Pinkel D, et al. Congenital melanocytic nevi frequently harbor NRAS mutations but no BRAF mutations. J Invest Dermatol. 2007;127:179–182. doi: 10.1038/sj.jid.5700490. [DOI] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk DR, LaBuz E, Dadras SS, et al. Melanoma and melanocytic tumors of uncertain malignant potential in children, adolescents and young adults—the Stanford experience 1995-2008. Pediatr Dermatol. 2010;27:244–254. doi: 10.1111/j.1525-1470.2009.01078.x. [DOI] [PubMed] [Google Scholar]
- Bett BJ. Large or multiple congenital melanocytic nevi: occurrence of cutaneous melanoma in 1008 persons. J Am Acad Dermatol. 2005;52:793–797. doi: 10.1016/j.jaad.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Bittencourt FV, Marghoob AA, Kopf AW, et al. Large congenital melanocytic nevi and the risk for development of malignant melanoma and neurocutaneous melanocytosis. Pediatrics. 2000;106:736–741. doi: 10.1542/peds.106.4.736. [DOI] [PubMed] [Google Scholar]
- Botton T, Yeh I, Nelson T, et al. Recurrent BRAF kinase fusions in melanocytic tumors offer an opportunity for targeted therapy. Pigment Cell Melanoma Res. 2013;26:845–851. doi: 10.1111/pcmr.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekaert SM, Roy R, Okamoto I, et al. Genetic and morphologic features for melanoma classification. Pigment Cell Melanoma Res. 2010;23:763–770. doi: 10.1111/j.1755-148X.2010.00778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Bahrami A, Pappo A, et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014;7:104–112. doi: 10.1016/j.celrep.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Daya-Grosjean L, Sarasin A. The role of UV induced lesions in skin carcinogenesis: an overview of oncogene and tumor suppressor gene modifications in xeroderma pigmentosum skin tumors. Mutat Res. 2005;571:43–56. doi: 10.1016/j.mrfmmm.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Flaherty KT, Yasothan U, Kirkpatrick P. Vemurafenib. Nat Rev Drug Discov. 2011;10:811–812. doi: 10.1038/nrd3579. [DOI] [PubMed] [Google Scholar]
- Gerami P, Cooper C, Bajaj S, et al. Outcomes of atypical spitz tumors with chromosomal copy number aberrations and conventional melanomas in children. Am J Surg Pathol. 2013;37:1387–1394. doi: 10.1097/PAS.0b013e31828fc283. [DOI] [PubMed] [Google Scholar]
- Gray-Schopfer VC, Cheong SC, Chong H, et al. Cellular senescence in naevi and immortalisation in melanoma: a role for p16. Br J Cancer. 2006;95:496–505. doi: 10.1038/sj.bjc.6603283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- Howlader N, Noone AM, Krapcho M, et al. (eds). (2013SEER Cancer Statistics Review, 1975-2010 National Cancer Institute: Bethesda, MD; http://seer.cancer.gov/archive/csr/1975_2010/http://seercancergov/csr/1975_2010/ based on November 2012 SEER data submission [Google Scholar]
- Huang FW, Hodis E, Xu MJ, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson A, Tuominen R, Grafstrom E, et al. High frequency of p16(INK4A) promoter methylation in NRAS-mutated cutaneous melanoma. J Invest Dermatol. 2010;130:2809–2817. doi: 10.1038/jid.2010.216. [DOI] [PubMed] [Google Scholar]
- Kinsler VA, Thomas AC, Ishida M, et al. Multiple congenital melanocytic nevi and neurocutaneous melanosis are caused by postzygotic mutations in codon 61 of NRAS. J Invest Dermatol. 2013;133:2229–2236. doi: 10.1038/jid.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krengel S, Hauschild A, Schafer T. Melanoma risk in congenital melanocytic naevi: a systematic review. Br J Dermatol. 2006;155:1–8. doi: 10.1111/j.1365-2133.2006.07218.x. [DOI] [PubMed] [Google Scholar]
- Landi MT, Bauer J, Pfeiffer RM, et al. MC1R germline variants confer risk for BRAF-mutant melanoma. Science. 2006;313:521–522. doi: 10.1126/science.1127515. [DOI] [PubMed] [Google Scholar]
- Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Choi JW, Kim YS. Frequencies of BRAF and NRAS mutations are different in histological types and sites of origin of cutaneous melanoma: a meta-analysis. Br J Dermatol. 2011;164:776–784. doi: 10.1111/j.1365-2133.2010.10185.x. [DOI] [PubMed] [Google Scholar]
- Leiter U, Garbe C. Epidemiology of melanoma and nonmelanoma skin cancer—the role of sunlight. Adv Exp Med Biol. 2008;624:89–103. doi: 10.1007/978-0-387-77574-6_8. [DOI] [PubMed] [Google Scholar]
- Pappo AS. Melanoma in children and adolescents. Eur J Cancer. 2003;39:2651–2661. doi: 10.1016/j.ejca.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Paradela S, Fonseca E, Pita S, et al. Spitzoid melanoma in children: clinicopathological study and application of immunohistochemistry as an adjunct diagnostic tool. J Cutan Pathol. 2009;36:740–752. doi: 10.1111/j.1600-0560.2008.01153.x. [DOI] [PubMed] [Google Scholar]
- Parker M, Mohankumar KM, Punchihewa C, et al. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature. 2014;506:451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- Raimondi S, Sera F, Gandini S, et al. MC1R variants, melanoma and red hair color phenotype: a meta-analysis. Int J Cancer. 2008;122:2753–2760. doi: 10.1002/ijc.23396. [DOI] [PubMed] [Google Scholar]
- Scalzo DA, Hida CA, Toth G, et al. Childhood melanoma: a clinicopathological study of 22 cases. Melanoma Res. 1997;7:63–68. doi: 10.1097/00008390-199702000-00010. [DOI] [PubMed] [Google Scholar]
- Soo JK, Mackenzie Ross AD, Kallenberg DM, et al. Malignancy without immortality? Cellular immortalization as a possible late event in melanoma progression. Pigment Cell Melanoma Res. 2011;24:490–503. doi: 10.1111/j.1755-148X.2011.00850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strouse JJ, Fears TR, Tucker MA, et al. Pediatric melanoma: risk factor and survival analysis of the surveillance, epidemiology and end results database. J Clin Oncol. 2005;23:4735–4741. doi: 10.1200/JCO.2005.02.899. [DOI] [PubMed] [Google Scholar]
- Tsao H, Goel V, Wu H, et al. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol. 2004;122:337–341. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward KA, Lazovich D, Hordinsky MK. Germline melanoma susceptibility and prognostic genes: a review of the literature. J Am Acad Dermatol. 2012;67:1055–1067. doi: 10.1016/j.jaad.2012.02.042. [DOI] [PubMed] [Google Scholar]
- Wei X, Walia V, Lin JC, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman DC, Valery P, McWhirter W, et al. Risk factors for childhood melanoma in Queensland, Australia. Int J Cancer. 1997;70:26–31. doi: 10.1002/(sici)1097-0215(19970106)70:1<26::aid-ijc4>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Wiesner T, He J, Yelensky R, et al. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun. 2014;5:3116. doi: 10.1038/ncomms4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JR, Harris JK, Rodriguez-Galindo C, et al. Incidence of childhood and adolescent melanoma in the United States: 1973-2009. Pediatrics. 2013;131:846–854. doi: 10.1542/peds.2012-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Benavente CA, McEvoy J, et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012;481:329–334. doi: 10.1038/nature10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.