Abstract
Due to the limited ultrastructural information about the coronavirion, little is known about the interactions acting at the interface between nucleocapsid and viral envelope. Knowing that subtle mutations in the carboxy-terminal endodomain of the M protein are already lethal, we have now probed the equivalent domain of the spike (S) protein by extending it terminally with a foreign sequence of 27 kDa: the green fluorescent protein (GFP). When expressed individually in murine cells, the S-GFP chimeric protein induced the formation of fluorescent syncytia, indicating that it was synthesized and folded properly, trimerized, and transported to the plasma membrane, where it exhibited the two key S protein functions, i.e., interaction with virus receptor molecules and membrane fusion. Incorporation into virus-like particles demonstrated the assembly competence of the chimeric spike protein. The wild-type S gene of mouse hepatitis coronavirus (MHV) was subsequently replaced by the chimeric construct through targeted recombination. A viable MHV-SGFP was obtained, infection by which could be visualized by the fluorescence induced. The efficiency of incorporation of the chimeric protein into particles was, however, reduced relative to that in wild-type particles which may explain, at least in part, the reduced infectivity produced by MHV-SGFP infection. We conclude that the incorporation of spikes carrying the large GFP moiety is apparently impaired by geometrical constraints and selected against during the assembly of virions. Probably due to this disadvantage, deletion mutants, having lost the foreign sequences, rapidly evolved and outcompeted the chimeric viruses during virus propagation. The fluorescent MHV-SGFP will now be a convenient tool to study coronaviral cell entry.
Enveloped viruses acquire their lipid envelope by budding from cellular membranes. This budding process involves the specific association of viral membrane proteins and capsid protein, together with the viral genomic RNA/DNA, into a virus particle. The selective inclusion of viral proteins generally results in the exclusion of the abundantly present cellular proteins from the virus particle in such a way that the protein content of the virion is essentially of viral origin. This is established by affinity interactions among the viral proteins in addition to the concentration of the viral proteins at the site of budding at membrane microdomains. Alphaviruses do not allow the incorporation of host proteins, probably because of the extensive and specific interactions between the spikes themselves and with the nucleocapsid during the budding process. However, retroviruses, dependent for budding only on the Gag protein, seem to incorporate many cellular membrane proteins, which are present at the site of budding (reviewed in reference 18). The inclusion of host proteins apparently depends strongly on the structural organization of enveloped viruses.
Coronaviruses are among the largest enveloped RNA viruses, measuring 80 to 120 nm in diameter. They contain a helical nucleocapsid, which consists of an ∼30-kb positive-stranded RNA genome encapsidated by the nucleocapsid protein N (∼60 kDa). The nucleocapsid is incorporated into a viral particle by budding into the early compartments of the exocytic pathway (23, 24, 44), thereby acquiring the viral envelope. The viral membrane accommodates at least three different membrane proteins: a type III, triple-membrane-spanning membrane protein M (∼25 kDa); a small hydrophobic envelope protein E (∼10 kDa); and the large type I spike protein S (∼200 kDa). Some coronaviruses also contain the hemagglutinin-esterase protein HE (∼65 kDa) (39). The M and E proteins are essential for virus budding. Coexpression of these proteins leads to nucleocapsid independent budding of coronavirus-like particles (VLPs) (45). Incorporation of spikes into virions and VLPs is mediated by interaction of the S protein with the M protein (7, 32, 45). The M protein also facilitates the nucleocapsid packaging into virions by interaction with the nucleocapsid protein (26) and with the genomic RNA (17, 30).
The mouse hepatitis coronavirus A59 (MHV-A59) spike protein is a class I membrane fusion protein (2) and is solely responsible both for virus entry and for cell-cell fusion. The spikes appear on the virion as large characteristic surface projections ca. 20 nm in length. The S protein is synthesized in the rough endoplasmic reticulum, where it folds and oligomerizes slowly (31, 46) into dimers (27, 48) or trimers (9). A fraction of the protein is transported to the plasma membrane where it induces cell-cell fusion. In the virion the 1,324-amino-acid (aa) membrane protein consists of a large (1,263-aa), extensively N-glycosylated amino-terminal ectodomain; a hydrophobic (29-aa) transmembrane domain (TM); and a short (32-aa) hydrophilic carboxy-terminal endodomain. Upon passage through the Golgi compartment, the 180-kDa spike may be cleaved by furin-like enzymes (unpublished results) into two subunits (S1 and S2) of approximately the same size, which remain noncovalently linked. The S1 subunit is responsible for receptor binding (1, 20, 42), whereas the membrane bound S2 subunit is involved in membrane fusion (49). The TM and/or endodomain of S is responsible for interaction with the M protein and incorporation into the virion (19).
In the present study, we investigated the structural requirements for coronavirus assembly. Little is known about the structural organization of coronaviruses. Ultrastructural research on coronaviruses based on crystallography and electron microscopy has been hampered by the pleiomorphic nature of these viruses. The structure of the nucleocapsid is still unresolved, and it is unknown how it fits to the viral membrane and to the luminal parts of the membrane proteins E, M, and S. Mutagenesis studies on the MHV M protein have shown that subtle changes in the carboxy terminus were already fatal and impaired the assembly of viral particles (5). Just like those of the M and E proteins, the membrane-exposed carboxy-terminal part of S is rather short, and the function of this domain is currently unknown. The cytoplasmic domain of S might be involved in interaction with the M protein and/or the nucleocapsid. To get more insight into this matter, we made use of our recently established targeted recombination system in order to test whether modification of the carboxy terminus of S is tolerated. Rather than starting by making subtle mutations, we decided to extend the domain substantially by a foreign sequence. Thus, we tested whether appending the sequence of the green fluorescent protein (GFP) would be tolerated. If so, this would provide us at the same time with a recombinant virus suitable for future studies, for instance on viral entry.
In the S-GFP fusion protein constructed, the cytoplasmic domain was extended by 282 residues. This protein was found to be fusion active and to be incorporated into VLPs. Using targeted RNA recombination, a viable recombinant virus (MHV-SGFP) was obtained in which the spike gene was replaced by the S-GFP gene. This virus indeed contained the fusion protein and was fluorescent. Genetic stability experiments revealed that the GFP gene was not maintained resulting in the gradual loss of GFP sequences during passage of the virus.
MATERIALS AND METHODS
Virus, cells, and antibodies.
The preparation of recombinant wild-type MHV-A59 (MHV-WT) has been described previously (6). Recombinant vaccinia virus encoding the T7 RNA polymerase (vTF7.3) was obtained from B. Moss. OST7-1 (12) (obtained from B. Moss) and LR7 cells (25) were maintained as monolayer cultures in Dulbecco modified Eagle medium (DMEM) containing 10% fetal calf serum (FCS), 100 IU of penicillin/ml, 100 μg of streptomycin/ml (DMEM-FCS) (all from Life Technologies). The monoclonal antibody (MAb) J1.3 against the amino terminus of MHV M (anti-M) (43) and the MAb WA3.10 directed against the MHV S protein (anti-S) (16) were kindly provided by J. Fleming (University of Wisconsin, Madison). The production of polyclonal antiserum K134 to MHV-A59 (anti-MHV) has been described previously (36). The goat anti-rabbit red fluorescent antibody Cy5 was purchased from Jackson Immunoresearch Laboratories, and the polyclonal antiserum against GFP (anti-GFP) was purchased from Clontech.
Plasmid constructs.
Expression vectors, used in the vTF7.3 expression system, contained the genes under the control of the bacteriophage T7 transcription regulatory elements. The expression constructs pTUMM, pTM5ab, and pTUMS contain the genes encoding the MHV-A59 M, E, and S proteins, respectively, cloned into the pTUG31 plasmid (45, 47). The plasmid pTUGS-GFP, encoding the S-GFP protein, was created in three stages. First, the BamHI-StyI fragment of the pTUMS plasmid (45) corresponding to the ectodomain of the MHV-A59 S protein was blunted by using the Klenow enzyme. The fragment was cloned into the BamHI-linearized and blunted pTUG31 vector (47). The resulting vector was designated pMHVSΔTMCD and contained the sequence encoding the S ectodomain under a T7 promoter with a unique BamHI site at its 5′ end. Second, the pMHVSΔTMCD vector was digested with BamHI and blunted by using Klenow enzyme, followed by heat inactivation of the enzyme. After restriction with the MluI enzyme, the linearized pMHVSΔTMCD vector was isolated from an agarose gel. Primer 974 (5′-GTGGATCCATCGAAGGTCGTAATGCAAATGCTGAAGC-3′) and primer 1508 (5′-AGATCTCTGCAGCGGCCGCGATAGTGGATCCTCATGAGAG-3′) were used to amplify by PCR the ∼1-kb sequence of the 3′ end of the S gene by using the pTUMS plasmid as a template. Primer 1508 lacks the stop codon of spike gene and contains an overhang with a BamHI, a NotI, and a PstI site. The PCR product was digested with MluI, and the 400-bp fragment containing the 3′ end of the S gene was purified from agarose gel and ligated into the linearized and MluI-treated pMHVSΔTMCD vector. The resulting vector contained the entire spike gene except the stop codon under a T7 promoter and was designated pTUGS(stop−). Third, the final pTUGS-GFP vector was created by ligating the BamHI-NotI fragment of pEGFP-N3 (Clontech) into the BamHI-NotI-digested pTUGS(stop−) vector. The final construct was verified by DNA sequencing.
The transcription vector pXHSGFP, needed to produce donor RNA for the construction of recombinant MHV-SGFP via targeted recombination, was created by ligation of the MluI-PstI fragment of pTUGS-GFP into the MluI-Sse8387I-digested pMH54 vector (25).
Construction of recombinant MHV-SGFP.
The chimeric S-GFP gene was transferred into the MHV genome by targeted RNA recombination as described previously (6, 22). Capped, runoff donor RNAs transcribed from the PacI-truncated pXHSGFP vector were electroporated into feline FCWF cells that had been infected 4 h earlier by fMHV (25). These cells were then divided over two T25 flasks containing a monolayer of LR7 cells to obtain two independent recombinant viruses (MHV-SGFP A and B). Progeny viruses released into the medium were harvested, and candidate recombinants were selected by two rounds of plaque purification on LR7 cells.
Confocal laser scanning microscopy (CLSM).
Confocal images of S-GFP transfected OST7-1 cells, MHV-SGFP-infected LR7 cells, or fluorescent MHV-SGFP virions were taken on a Leica inverted fluorescence microscope equipped with an argon and helium ion laser and with a ×40 or ×100 oil immersion objective lens. Enhanced GFP was excited at 488 nm; Cy5 was excited at 568 nm.
Growth curves.
Confluent monolayers of LR7 cells in 35-mm dishes were infected with either MHV-WT or MHV-SGFP at a multiplicity of infection (MOI) of 1, followed by incubation for 1 h at 37°C. Cells were subsequently washed with DMEM and then fed with DMEM-FCS. Samples from the culture medium taken at different times postinfection (p.i.) were titrated by an endpoint dilution assay, and titers expressed as 50% tissue culture infectious doses (TCID50) were calculated.
Passaging of MHV-SGFP and RT-PCR analysis.
The MHV-SGFP virus was passaged for six successive rounds on LR7 cells at an MOI of 0.05. The presence of the inserted GFP gene was checked by reverse transcription-PCR (RT-PCR) on samples taken from the culture fluids. An RT reaction was performed on the MHV-SGFP and MHV-WT genome by using primer 1261 (5′-GCTGCTTACTCCTATCATAC-3′), which is complementary to the open reading frame 5 (ORF5) region. A PCR was performed on the RT product by using primer 935 (5′-GCACAGGTTGTGGCTCATG-3′) and primer 1090 (5′-GATTCAGGTTTGTAACATAATCTAGAGTCTTAGG-3′). These primers map in the ectodomain of the S gene and the ORF4 TRS, respectively. The two plasmid vectors used to construct MHV-WT (pMH54) and MHV-SGFP (pXHSGFP) were taken along as controls. The PCR products obtained for MHV-SGFP A and B passage 6 were cloned into pGEM-T-Easy and subsequently sequenced by using primer 935.
Immunofluorescence.
The percentage of S-GFP expressing viruses of MHV-SGFP A and B at passages 1, 2, 4, and 6 was determined by immunofluorescence. LR7 cells, grown on 12-mm coverslips, were infected in DMEM at an MOI of 0.5. At 1 h p.i., cells were washed with DMEM and overlaid with DMEM-FCS containing 1 μM HR2 (2) to prevent cell-cell fusion. At 8 h p.i., cells were fixed, permeabilized, and labeled as described previously (31) with the anti-MHV serum (1:400) in combination with a red fluorescent Cy5 secondary antibody (1:200). The percentage of green fluorescent cells (GFP positive) over red fluorescent cells (MHV positive) was determined by random counting of 100 cells.
Vaccinia virus infection, DNA transfection, and metabolic labeling of OST7-1 cells.
Subconfluent monolayers of OST7-1 cells in 10-cm2 tissue culture dishes were inoculated with vTF7.3 in DMEM at an MOI of 10. At 1 h p.i., cells were washed with DMEM and overlaid with transfection medium consisting of 0.2 ml of DMEM that contained 10 μl of Lipofectin (Life Technologies), 5 μg of pTUMM, 1 μg of pTM5ab, and 2 μg of either pTUMS or pTUGS-GFP. After 10 min at room temperature, 0.8 ml of DMEM was added, and incubation was continued at 37°C. At 4.5 h p.i., cells were washed with DMEM and starved for 30 min in cysteine- and methionine-free minimal essential medium containing 10 mM HEPES (pH 7.2) without FCS. The medium was subsequently replaced by 600 μl of the same medium containing 100 μCi of 35S in vitro labeling mixture (Amersham). At 6 h p.i., the radioactivity was chased by incubating the cells with 2 ml of DMEM containing 2 mM methionine and cysteine. After 3 h, the dishes were placed on ice, and the culture media were collected and cleared by centrifugation for 15 min at 4,000 × g and 4°C. Cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed with 600 μl of detergent buffer (50 mM Tris-Cl [pH 8.0], 62.5 mM EDTA, 0.5% sodium deoxycholate, 0.5% Nonidet P-40) containing 1 mM phenylmethylsulfonyl fluoride. Lysates were cleared by centrifugation for 10 min at 13,000 × g at 4°C. Viral proteins in the cell lysates and VLPs in the culture medium were subjected to immunoprecipitation in the presence or absence (affinity purification) of detergents, respectively. Cell lysates (100 μl) or cleared culture medium (300 μl) were diluted 2.5 times with detergent buffer or with TEN buffer (10 mM Tris [pH 7.6], 1 mM EDTA, 50 mM NaCl), respectively, and subsequently incubated overnight at 4°C with the anti-MHV serum (2 μl) or the anti-S MAb (20 μl). The immune complexes were adsorbed to Pansorbin cells (Calbiochem) for 30 min at 4°C and collected by low-speed centrifugation. Pellets were washed three times by resuspension and centrifugation with either detergent buffer (cell lysates) or TEN buffer (culture medium). Pellets were resuspended and heated in Laemmli sample buffer at 95°C for 2 min before being analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 15% polyacrylamide gels.
Infection and metabolic labeling of LR7 cells.
Subconfluent monolayers of LR7 cells in 10-cm2 dishes were inoculated with either MHV-WT or MHV-SGFP A or B (passage 2) in DMEM at an MOI of 1 for 1 h. Subsequently, cells were washed with DMEM and incubated under DMEM-FCS. At 5.5 h p.i., the cells were washed with DMEM and starved for 30 min in cysteine- and methionine-free MEM containing 10 mM HEPES (pH 7.2). The culture medium was then replaced by 600 μl of the same medium but containing 100 μCi of 35S in vitro labeling mixture (Amersham). At 7 h p.i., the radioactivity was chased by incubating the cells with DMEM containing 2 mM methionine and 2 mM cysteine. After 2 h, the cells were placed on ice, and the culture media were collected and cleared by centrifugation for 15 min at 4,000 × g and 4°C. Cells were washed with ice-cold PBS and lysed with lysis buffer (20 mM Tris [pH 7.6], 150 mM NaCl, 0.5% sodium deoxycholate, 1% Nonidet P-40, 0.1% SDS). Lysates were cleared by centrifugation for 10 min at 13,000 × g at 4°C. Cell lysates were diluted 2.5 times with lysis buffer. The cleared culture media were supplemented with 1/4 volume of 5× lysis buffer. Viral proteins in the cell lysates and culture media were incubated overnight at 4°C with the anti-MHV serum (2 μl), the anti-S MAb (20 μl) or the anti-GFP serum (1 μl; Clontech). The immune complexes were adsorbed to Pansorbin cells for 30 min at 4°C and were subsequently collected by low-speed centrifugation. Pellets were washed three times with lysis buffer. Pellets were resuspended and heated in Laemmli sample buffer at 95°C for 2 min before being analyzed by SDS-PAGE in 15% polyacrylamide gels.
For quantification of the relative presence of the viral proteins in coronavirus particles released into the culture medium, the above procedure was modified slightly. The cleared culture media were again diluted 2.5 times with TEN buffer, but the particles were affinity purified with either the anti-M MAb (20 μl) or the anti-S MAb (20 μl). Pellets were washed three times with TEN buffer. Quantification of the radioactivities in the protein bands in the dried gels was carried out by phosphorscanning of dried gels by using a Storm 860 (Molecular Dynamics).
Virus purification.
Virus present in the culture medium of infected cells was partially purified as follows. The virus-containing medium was centrifuged at 3,000 rpm for 5 min, and the supernatant was centrifuged again for 5 min at 15,000 rpm to remove any cell debris and nuclei. Virus in the supernatant was subsequently pelleted through a 20% sucrose cushion for 2 h at 25,000 rpm and 4°C by using an SW28 rotor. The virus pellet was carefully resuspended in 100 μl of PBS for 4 h on ice.
RESULTS
Construction and functional analysis of S-GFP fusion protein.
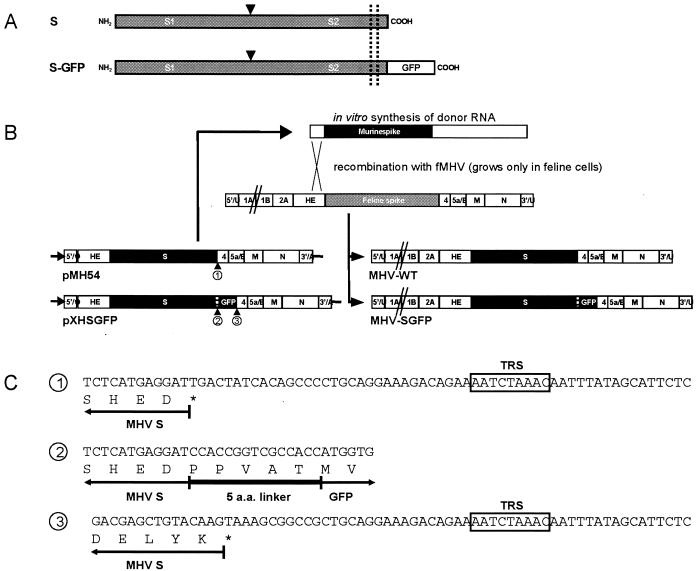
To evaluate whether extension of the relatively short coronavirus spike endodomain by a long polypeptide would affect the protein's biological functions and, in particular, whether this would impair incorporation into particles to yield infectious virus, we appended the (enhanced) GFP gene to the carboxy-terminal end of the S gene in an expression vector via a short linker sequence corresponding to 5 aa residues. The resulting carboxy-terminal 239-residue extension of the endodomain is predicted to increase the S protein's molecular mass by ca. 27 kDa (Fig. 1).
FIG. 1.
(A) Schematic diagram of the MHV S and S-GFP protein. The arrowhead indicates the cleavage site, where posttranslational cleavage of S into two subunits (S1 and S2) takes place. In S-GFP the GFP protein is fused to the carboxy terminus of the S protein. The dashed lines indicate the position of the transmembrane region. (B) Plasmid constructs, targeted recombination, and recombinant viruses. The plasmid pMH54 (described previously [25]) and pXHSGFP (see Materials and Methods) were used to transcribe the defective RNAs in vitro by using T7 polymerase. The arrow at the left end of the vectors indicates the T7 promoter; the solid circle represents the polylinker between the 5′-end segment of the MHV genome (labeled 5′/1) and the HE gene, followed by the structural and group specific genes, the 3′ untranslated region (UTR) and the polyadenylate segment (together labeled as 3′/U). The positions of the numbered sequences shown in panel C correspond to the numbered arrowhead positions. At the top, a scheme is shown for targeted recombination by using the interspecies chimeric fMHV, which grows only in feline cells. Recombinant viruses generated by the indicated crossover event can be selected on the basis of their ability to grow in murine cells. The genomes of these viruses are represented at the right. (C) Sequences of the numbered junctions shown in panel B. The numbers correspond to those under the arrowheads in panel B. The amino acids encoded by the 3′ end of the S and the GFP gene, by the linker sequence and by the 5′ end of the GFP gene are indicated. The asterisk denotes the stop codon. The MHV TRS belonging to gene 4 is boxed.
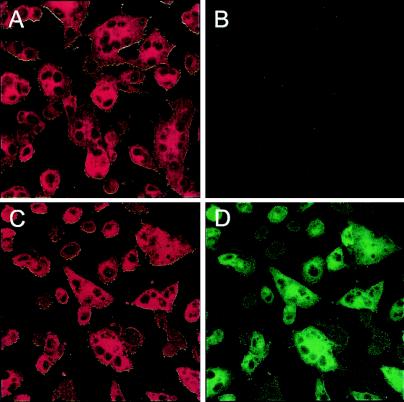
The S protein is responsible for both virus-cell and cell-cell fusion. To examine the functionality of the chimeric protein with respect to membrane fusion, the S-GFP gene and the wild-type S gene were transiently expressed in OST7-1 cells by using the recombinant vaccinia virus-bacteriophage T7 RNA polymerase system. Cells were subjected to immunofluorescence and analyzed by CLSM. In both cases, extensive cell-cell fusion was observed (Fig. 2A and C). When excited at the excitation wavelength of GFP at 488 nm, only cells expressing the S-GFP protein appeared to be green fluorescent (Fig. 2D), in contrast to cells expressing the wild-type S protein (Fig. 2B). Thus, despite its large extension, the spike protein appeared to be properly folded, transported to the plasma membrane, able to bind MHV receptor on neighboring cells, and cause fusion with these cells.
FIG. 2.
CLSM of OST7-1 cells expressing the S (A and B) and S-GFP (C and D) proteins (×40 magnification). Cells were fixed at 8 h p.i., permeabilized, and processed for immunofluorescence by using the anti-MHV serum (1:400) and the red fluorescent Cy5 secondary antibody (1:200) and excited at 488 nm (GFP; B and D) or 568 nm (Cy5; A and C).
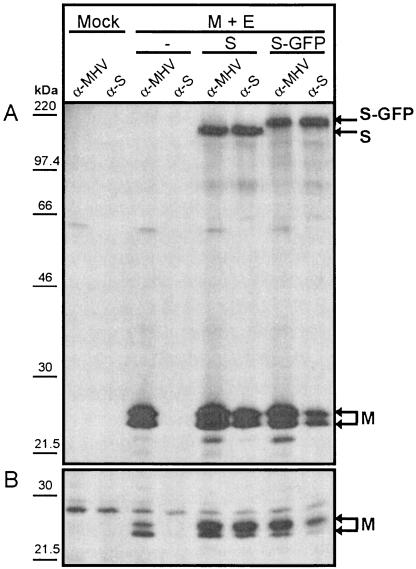
The coronaviral membrane proteins, when coexpressed in cells, assemble into VLPs that are morphologically indistinguishable from normal virions (45). As a first test to study whether the S-GFP protein was able to interact with the M protein and be assembled into VLPs, we coexpressed the M and E protein together or in combination with either the wild-type S or the S-GFP protein by using the vaccinia virus expression system. Cells were labeled with 35S-labeled amino acids from 5 to 6 h p.i., followed by a 2-h chase. Viral proteins were immunoprecipitated from the cell lysates, and VLPs were affinity purified from the culture media with either a polyclonal anti-MHV serum or a monoclonal anti-S antibody and analyzed by SDS-PAGE (Fig. 3).
FIG. 3.
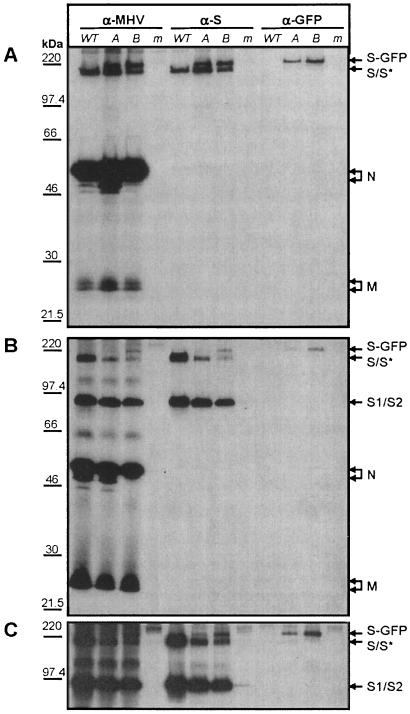
Coimmunoprecipitation of the M protein by S-GFP fusion protein. (A) Coexpression in murine cells of the MHV M and E proteins either in the absence or in combination with the S or S-GFP protein. Cells were labeled with 35S-labeled amino acids from 5 to 6 h p.i., followed by a 2-h chase. Immunoprecipitations were carried out on the cell lysates with the anti-MHV serum (α-MHV) or the anti-S MAb (α-S) and analyzed by SDS-PAGE. (B) SDS-PAGE analysis of VLPs affinity purified from the culture medium with α-MHV and α-S. The molecular mass markers are indicated on the left. Arrows on the right indicate the positions of the expressed proteins.
The results obtained with the anti-MHV serum showed that M, S, and S-GFP were well expressed intracellularly (Fig. 3A). The carboxy-terminal GFP extension of the S protein obviously results in a decreased electrophoretic mobility. The E protein was not detected since it is not recognized by the serum (6). Using the anti-S MAb, we analyzed whether the M and S-GFP protein interact. In the absence of any S protein, no immunoprecipitation of M was seen. However, the M protein was coimmunoprecipitated when either the S protein or, to a lesser extent, when the S-GFP protein was coexpressed (Fig. 3A). To analyze the incorporation of S-GFP into VLPs, an affinity purification was performed with anti-S MAbs by using the appearance of the M protein as a readout parameter. As Fig. 3B shows, the VLPs produced by the coexpression of M and E, detectable by affinity purification from the culture fluid with the anti-MHV serum, could not be isolated by the MAb. They were, however, isolated not only when the wild-type S protein was additionally coexpressed but also in the presence of the S-GFP protein. In conclusion, despite the large endodomain extension of S, the S-GFP is still able to associate with the membrane protein M and to assemble into VLPs.
Construction of recombinant MHV expressing S-GFP.
Thus far, the chimeric S-GFP protein had performed all of the canonical functions of the spike protein. Our next question was whether the cytoplasmic domain extension, which apparently did not prevent its assembly into VLPs, would interfere with the assembly of virions. To this end, we attempted to generate an infectious recombinant virus bearing the S-GFP protein (MHV-SGFP) by targeted RNA recombination (25). Thus, we prepared a transcription vector in which the S gene was replaced by the S-GFP gene, yielding the donor plasmid pXHSGFP (Fig. 1B). As a control, we used the parental vector pMH54 to reconstruct recombinant “wild-type” MHV-A59 (MHV-WT) as we showed earlier (6). The pMH54 transcription vector encodes a defective RNA composed of the genomic 5′ 467 nucleotides fused to the 3′ part (∼8.6 kb) of the genome (Fig. 1B). The arrowheads in Fig. 1B indicate the junctions at the 5′ end of either the S (1) or S-GFP gene (3) and the linker region between the S and the GFP gene (2), the sequences of which are shown in Fig. 1C. The MHV-SGFP recombinant virus was generated by targeted RNA recombination with fMHV, an MHV derivative in which the S protein ectodomain is of feline coronavirus origin (25). Briefly, feline cells were infected with fMHV and transfected with a runoff transcript of the pXHSGFP vector. Recombinant viruses containing the murine spike were selected by growth in murine cells, as we described previously (6). Two independently obtained recombinant viruses (MHV-SGFP A and B) were cloned by two rounds of plaque purification on murine cells by picking fluorescent plaques.
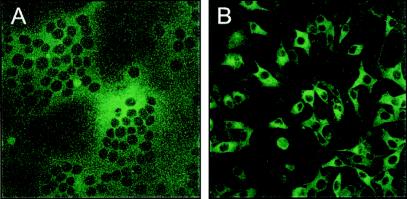
To show the fluorescence induced by the MHV-SGFP virus, cells were infected and viewed 8 h later by fluorescence microscopy (Fig. 4). MHV-SGFP infection was found to induce large syncytia, which were green fluorescent (Fig. 4A). To observe the expression pattern of the S-GFP protein in single cells, we made use of a peptide (HR2) corresponding to the membrane proximal heptad repeat region in the spike protein, which is a potent inhibitor of cell-cell fusion (2). In the presence of this inhibitor (1 μM), the syncytia were no longer induced (Fig. 4B) and the green fluorescent signal was observed predominantly in a typical endoplasmic reticulum pattern throughout the cytoplasm, as seen also after immunofluorescent staining of the wild-type S protein in MHV-A59-infected cells (31).
FIG. 4.
CLSM of MHV-SGFP-infected LR7 cells grown in the presence (A) or absence (B) of membrane fusion inhibitor HR2 (×40 magnification).
Viability of MHV-SGFP.
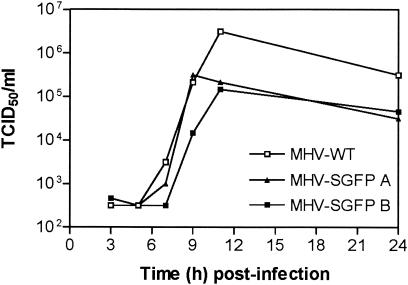
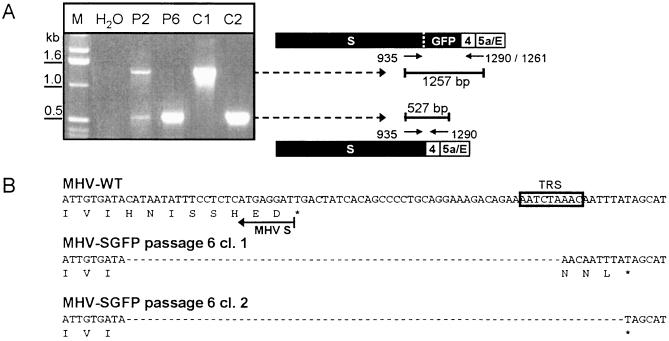
To analyze the effect of the modification of the virus on its multiplication, we compared its growth in culture cells with that of MHV-WT (Fig. 5). Clearly, the replication of MHV-SGFP did not yield the titers of MHV-WT (ca. 1 log lower at t = 11). Since the modification of the S protein apparently affected the growth of the virus, it was of interest to investigate the genetic stability of the mutant virus. To this end, the MHV-SGFP virus was passaged six times on LR7 cells at low MOI (0.05). To determine the presence of the S-GFP gene an RT-PCR was performed at passage 2 (P2) and passage 6 (P6) as shown in Fig. 6A. The RT reaction was performed with primer 1062, which is complementary to the 3′ end of the E gene. The PCR was carried out with the primers 935 and 1090, which localize in the 3′ end of the S gene and the 3′ end of gene 4, respectively (Fig. 6A). As controls, PCR was performed on the pXHSGFP (C1) and pMH54 (C2) plasmid vectors, the vectors used to create MHV-SGFP and MHV-WT, respectively. Two PCR products were obtained with the passage 2 RNA of MHV-SGFP; the larger one of these comigrated with the PCR product obtained for the pXHSGFP vector. The smaller PCR product migrated with a mobility of ∼500 bp, which corresponds to the expected product size of 527 bp for the wild-type situation (C2). The RT-PCR product obtained with passage 6 virus RNA only showed the 500-bp product. Clearly, already after the second passage some of MHV-SGFP recombinant viruses had lost the GFP sequence, which, due to their apparent growth advantage, had completely taken over the population at passage 6. The PCR product of passage 6 MHV-SGFP was cloned and sequenced. The sequence of two clones, both of passage 6 MHV-SGFP A, confirmed that the entire GFP sequence had been deleted (Fig. 6B). In addition, both sequences showed a deletion of 24 nucleotides, corresponding to the carboxy-terminal 8 aa of the cytoplasmic domain of S. This deletion resulted in an extension of the ORF of clone 2 coding for 3 aa. Similar deletions of the GFP gene were observed for MHV-SGFP B (data not shown). The presence of deletion mutant viruses already in passage 2 indicates the strong selection against viruses containing an S protein with an extended cytoplasmic domain. It is noteworthy that the C-terminal 8-aa truncation of the spike protein found in the MHV-SGFP passage 6 recombinants is apparently not required for virus assembly. This is consistent with results from a deletion study showing that a recombinant MHV lacking the C-terminal 12 residues of the spike protein exhibited only a slight decrease (∼1 log) in virus production (data not shown).
FIG. 5.
Growth kinetics of MHV-SGFP and MHV-WT. LR7 cells were infected with either MHV-WT or with the independent MHV-SGFP clones A or B at an MOI of 1. Viral infectivities in the culture media were determined at different times postinfection by a quantal assay on LR7 cells, and the TCID50 values were calculated.
FIG. 6.
(A) PCR analysis of recombinant MHV-SGFP. RT-PCR was performed on purified genomic RNA of MHV-SGFP from passage 2 and 6 (P2 and P6, respectively) to analyze the presence of the GFP sequence. A water control (H2O) is also shown. As additional controls, a PCR was performed on the transcription vectors pXHSGFP (C1) and pMH54 (C2). PCR products were analyzed by electrophoresis in 1% agarose gels and stained with ethidium bromide. Primers used in the experiment, their location on the genome, and the predicted sizes of the PCR products are indicated at the right. In the RT step, primer 1261 was used, whereas primers 935 and 1290 were used in the PCR step. The sizes of relevant DNA fragments of the 1-kb DNA marker ladder (M; Invitrogen) are indicated on the left. (B) Sequence analysis of passage 6 MHV-SGFP. The RT-PCR product obtained from MHV-SGFP clone A passage 6 was cloned into pGEMT-Easy, and plasmid DNA from two colonies was sequenced. The nucleotide sequences of the 3′ region of the spike gene of the two sequenced clones, as well as of MHV-WT, are shown. The translated amino acid sequences corresponding to the S ORF are indicated under the nucleotide sequence. Deletions in the nucleotide sequence are represented by dashed lines. The asterisks mark the stop codons. The TRS sequence of gene 4 is boxed.
In order to investigate the loss of GFP sequences further, we quantified the fraction of viruses expressing fluorescence from MHV-SGFP passages 1, 2, 4, and 6 of clones A and B. LR7 cells were infected with MHV-SGFP at an MOI of 0.5. In order to prevent cell-cell fusion, 1 μM HR2 was added to the culture medium after inoculation. At 8 h p.i., cells were fixed and infected cells were labeled by using the anti-MHV serum in combination with a red fluorescent secondary antibody. The percentage of green fluorescent (GFP positive) over red fluorescent cells (MHV positive) was determined by counting under the microscope (Table 1). Of the cells infected with clone A and B passage 1 (P1) MHV-SGFP, 95 and 98% were positive for GFP, respectively. These numbers had dropped to 50 and 80% in P2 and to 1 and 2% in P4, respectively. By passage 6 no green fluorescent cells could be detected. The rapid loss of GFP expression upon passaging was in agreement with the RT-PCR results. The loss of GFP expression was accompanied by an increase in viral titers (Table 1) obtained with each subsequent virus passage, indicative for a gain in fitness upon deletion of the GFP gene.
TABLE 1.
Loss of GFP expression of MHV-SGFP and increase of viral titers during virus passaginga
| Passageb | MHV-SGFP A
|
MHV-SGFP B
|
||
|---|---|---|---|---|
| % GFP | TCID50 | % GFP | TCID50 | |
| P1 | 95 | 1.0 × 105 | 98 | 1.5 × 105 |
| P2 | 50 | 6.8 × 105 | 80 | 1.0 × 106 |
| P4 | 1 | 2.2 × 106 | 2 | 1.5 × 106 |
| P6 | 0 | 6.8 × 106 | 0 | 1.0 × 107 |
% GFP, infected cells were detected by immunofluorescence microscopy for MHV infection by using an anti-MHV serum and the red fluorescent Cy5 secondary antibody. The percentage of green fluorescent over red fluorescent (% GFP) cells was determined.
LR7 cells were infected with MHV-SGFP clones A or B, passages 1, 2, 4, and 6.
Detection of S-GFP in infected cells and virus particles.
We analyzed the protein synthesis of S-GFP in MHV-SGFP-infected cells and its appearance in virions. LR7 cells were mock infected or infected with either MHV-WT or the two independent clones of MHV-SGFP, A and B. Radiolabeled proteins were immunoprecipitated by using an anti-MHV serum, the anti-S MAb or the anti-GFP serum from the cell lysate or the medium, and analyzed by SDS-PAGE (Fig. 7). Immunoprecipitates performed with the anti-MHV antiserum on MHV-WT-infected cell lysates showed the expression of the MHV structural proteins M, N, and S as indicated in Fig. 7A. Similar precipitates prepared with the MHV-SGFP A- and B-infected cell lysates revealed the presence of an additional band, which was also immunoprecipitated by using the anti-S MAb and the anti-GFP serum. This band, interpreted as the S-GFP fusion protein, migrated at ∼220 kDa, in agreement with the expected size of S-GFP. The gel analysis additionally showed a band (S*) with the size of wild-type S, which is precipitated by the anti-S MAb but not by the anti-GFP serum. This indicates that the passage 2 virus stocks already contained GFP deletion viruses, a finding consistent with the RT-PCR results. The S-GFP band was also immunoprecipitated from the culture media by using the anti-MHV serum, the anti-S MAb, and specifically the anti-GFP serum (Fig. 7B; more clear after a longer exposure in Fig. 7C). This indicates the incorporation of the S-GFP fusion protein into virions. Although the extracellular levels of M and N of MHV-WT and MHV-SGFP viruses were comparable, notably less S*/S-GFP was present in the MHV-SGFP viruses than S in MHV-WT viruses.
FIG. 7.
Viral proteins in the cell lysates and culture media of MHV-SGFP-infected cells. LR7 cells were either mock infected (lanes m), infected with MHV-WT (lanes WT), or infected with MHV-SGFP clones A and B (lanes A and B). (A) At 5 h p.i., cells were labeled for 1 h with 35S-labeled amino acids and lysed immediately. Some cultures were further incubated with chase medium for 2 h, after which the culture supernatant was harvested and diluted with lysis buffer. Immunoprecipitations were performed with the anti-MHV serum (α-MHV), the anti-S MAb (α-S), or the anti-GFP serum (α-GFP) from either the cell lysate (A) or the culture medium (B and C [panel C is a longer exposure of a section of panel B]) and analyzed by SDS-PAGE. The molecular mass marker is indicated on the left. Arrows on the right indicate the positions of the expressed proteins.
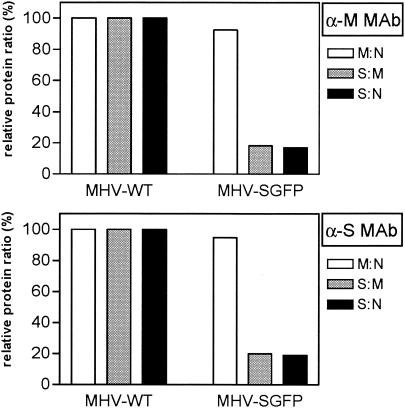
To examine whether the GFP extension lowers the efficiency with which the S protein is incorporated into virions, we quantified the S protein in MHV-SGFP virions relative to that in MHV-WT. Radiolabeled virus particles were affinity purified from the culture medium of infected cells by using an anti-M or an anti-S MAb. The samples were analyzed by SDS-PAGE, and the radioactivities in the viral proteins M, N, and S (i.e., S alone for MHV-WT or S*+S-GFP for MHV-SGFP) were quantitated by phosphorimager scanning of the dried gels. The relative amounts of the viral proteins were determined and normalized to those of the wild-type virus (Fig. 8). Similar results were obtained with the two MAbs. No significant difference was found in the relative amounts of M and N proteins in the two viruses; the calculated ratio of M to N in MHV-SGFP was close to 100% of the MHV-WT ratio. In contrast, the S/M and S/N ratios were dramatically decreased to ca. 20% of the values obtained for MHV-WT. This indicates an approximately fivefold reduction of mutant S protein into MHV-SGFP virions. It is of note that the intracellular S/M and S/N ratios were similar for both viruses (data not shown), ruling out the possibility that the results were caused by a reduced production of mutant S protein in MHV-SGFP-infected cells.
FIG. 8.
Protein quantification of affinity-purified coronavirus particles. LR7 cells were either mock infected, infected with MHV-WT, or infected with MHV-SGFP clone B. At 5 h p.i. cells were labeled for 1 h with 35S-labeled amino acids and chased for another 2 h. Virus particles were affinity purified from the cleared culture medium with either the anti-M MAb (α-M MAb) or the anti-S MAb (α-S MAb) and analyzed by SDS-PAGE. The amounts of radioactivity in the M, S (total, i.e., S+S-GFP), and N proteins in the dried gels were determined by phosphorimager scanning. The ratios of the amounts of M to N, S to M, and S to N in affinity-purified MHV-SGFP relative to those in MHV-WT were calculated.
Incorporation of S-GFP into coronavirus particles produces fluorescent virions.
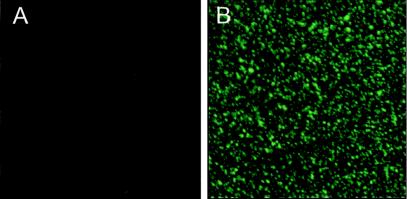
We finally analyzed whether the incorporation of S-GFP molecules into virions gave rise to particles that are fluorescent. MHV-SGFP and MHV-WT were produced in LR7 cells and concentrated by pelleting them through a sucrose cushion. The resuspended particles were examined by CLSM. As Fig. 9B illustrates, numerous fluorescent MHV-SGFP particles were visible. No such particles were seen with MHV-WT.
FIG. 9.
CLSM on MHV-SGFP and MHV-WT. Virus obtained from ∼107 infected LR7 cells was pelleted through a 20% sucrose cushion and resuspended in 100 μl of PBS. A 10-μl aliquot of either MHV-WT (A) or MHV-SGFP (B) was examined by CLSM (×100 magnification).
DISCUSSION
Coronaviruses appear to exhibit an exceptional flexibility regarding the incorporation of their spikes. On the one hand, particle assembly can occur perfectly well without any spikes being assembled, as was demonstrated both by blocking the proper folding of the S proteins in infected cells (21, 35, 37) and by the coexpression of the M and E proteins (45), which resulted in the formation of spike-less virions and VLPs, respectively. On the other hand, it appears that coronavirus assembly is tolerant to dramatic modifications of its spike protein. This was shown earlier by the incorporation into VLPs (19) and virions (25) of chimeric spikes in which the S protein ectodomain was replaced by that of an unrelated coronavirus, a feature now forming the basis of the targeted recombination technology for the genetic engineering of the coronaviruses (25). The present study demonstrates the high degree of tolerance in the endodomain part of the protein, showing that infectious coronavirus was still being efficiently assembled after the S protein's interior domain had been extended by some 27 kDa of foreign protein sequence. Although this virus was somewhat less infectious and genetically unstable, the observations provide new insights into the structural requirements for coronavirus assembly.
Expression of GFP by coronaviruses has been reported before. Unlike in the present study, in these earlier studies the GFP gene was inserted as an additional, separate transcription unit rather than as part of a structural protein gene. In the first demonstration that coronaviruses can serve as vectors for foreign gene expression, the GFP gene was inserted in the MHV genome in place of the nonessential gene 4. The recombinant virus grew to high titers and was phenotypically indistinguishable from the wild-type virus with respect to growth in tissue culture (15). Although the GFP protein could be detected in infected cells by Western blot analysis, the expression level was too low to be visualized directly by fluorescence microscopy. Recently, a similar recombinant MHV was prepared containing the enhanced GFP gene; now, the fluorescence expressed in infected cells could readily be detected (38). The expression of the GFP gene appeared to be stable for over six passages. Two similar recombinant transmissible gastroenteritis viruses expressing the GFP protein were reported independently (3, 41). In both, the nonessential gene 3A had been replaced by the foreign gene. Both viruses displayed wild-type growth kinetics and exhibited stable GFP expression for at least 10 passages.
The chimeric S-GFP protein that we expressed exhibited all of the biological functions of the spike protein. The development of fluorescent syncytia upon its independent expression in mouse cells demonstrated that both its receptor binding and its membrane fusion function were operational. These observations implied that the carboxy-terminal extension blocked neither the protein's folding and oligomerization nor the transport of these complexes to the plasma membrane. The chimeric protein was also able to functionally interact with the M protein, as was clearly seen from the production of S-GFP containing VLPs during coexpression with the M and E proteins. When the S-GFP gene was finally incorporated into the MHV genome, replacing the wild-type S gene, the endodomain extension did not prevent the assembly of the nucleocapsid into particles and the formation of infectious virus.
The fitness of the recombinant MHV-SGFP appeared to be affected by the foreign sequences. The virus grew to lower infectious titers than its equivalent wild-type, MHV-WT, whereas propagation of the virus quite rapidly led to loss of foreign sequences. This latter observation is not all that surprising in view of the mutability of RNA viruses in general and of coronaviruses in particular. The determinants for stability of foreign sequences engineered into coronaviruses are currently unknown, but it is clear that the stable maintenance of a foreign gene is, among others, dependent on properties of that particular gene. Thus, whereas the stability of a firefly luciferase gene varied strongly dependent on its location in the MHV genome, the Renilla luciferase gene appeared to be stably maintained at all of the locations tested (8; C. A. M. de Haan and P. J. M. Rottier, unpublished results). In addition to the nature and the genomic position of the foreign gene, the properties (e.g., toxicity) of the translated protein may also have an effect. This may explain, at least in part, why the GFP gene was genetically stable and did not affect viral growth when inserted as a separate transcription unit (13, 15, 38, 40), whereas it was rapidly lost when engineered as part of the S protein.
There are many reasons why carboxy-terminal extension of the S protein could lead to a decrease in fitness. The GFP moiety might interfere with the biogenesis of the chimeric protein, with the formation of trimers, with incorporation of these trimers into particles, or with virion assembly. Furthermore, impaired transport of the S-GFP protein to the plasma membrane might delay the spread of the infection due to reduced syncytium formation. The lower infectious titers, obtained for the virus with the S-GFP compared to its wild-type counterpart, indicate either that fewer particles were produced or that these particles were less infectious. Quantification of the relative presence of the structural proteins in radiolabeled virus particles revealed that the extension indeed affects the incorporation of the S protein rather than particle formation. Because the N/M protein ratio in S-GFP virions was not detectably different from that of MHV-WT, incorporation of the nucleocapsid did not seem to be affected by the extended spike. Also, the amount of virions produced was not significantly changed since the ratio of intracellular versus extracellular nucleocapsid and membrane protein was not altered compared to MHV-WT (data not shown). This is in agreement with previous studies, which show that incorporation of the nucleocapsid seems to involve interaction with the M protein (14, 26, 28-30) but does not require the S protein, as demonstrated by the appearance of spikeless virions after tunicamycin treatment of infected cells (21, 35, 37). Nucleocapsid assembly is apparently not dependent on the cytoplasmic tail of the S protein. All of these observations are consistent with impairment in the incorporation of the S-GFP protein. As a consequence, the virions produced had a relative shortage of spikes, as was confirmed by the reduced S/N and S/M protein ratios, leading to a reduced average infectivity per particle. We can only speculate why the incorporation of the S-GFP was impaired. Inclusion of spikes into virions is driven by heterotypic interactions with the M protein, which may be affected by the extension of the cytoplasmic domain of the S protein. Indeed, interaction with the M protein seemed to be affected as judged from the coimmunoprecipitation analysis (Fig. 3). The large GFP moiety presenting itself on the inner face of the assembling virion may also be actively selected against. Steric hindrance of the proper interactions between the nucleocapsid and the envelope components may thus lead to the partly exclusion of the chimeric spike. In this latter case, it would mean that the fit between the nucleocapsid and its surrounding envelope is not rigid but instead quite pliable, since this exclusion is not complete.
A few other examples have been reported where recombinant viruses were produced expressing a GFP-tagged structural protein that was incorporated into the virion (4, 10, 11, 33, 34). In the case of vesicular stomatitis virus (VSV) a gene construct encoding a VSV G protein cytoplasmically extended by GFP was engineered into the genome of this negative-stranded rhabdovirus as an additional genetic element (4). The protein was found to be incorporated into the virus in the form of mixed trimers, i.e., as complexes composed of both wild-type G and G/GFP fusion proteins. A recombinant VSV in which the G/GFP gene was replacing the G gene was unable to independently replicate but could be propagated by complementing it with the wild-type G protein. The majority of the recombinant viruses grown in this way had, however, lost their GFP expression already after one round of infection by the introduction of a stop codon in the carboxy-terminal domain of the G protein (4). Interestingly, the loss of expression of the GFP moiety from our MHV-SGFP was due to the deletion of the GFP gene rather than to the introduction of a stop codon. This might be a reflection of the different replication strategies that these viruses use. As for the VSV recombinant, the stability of the MHV-SGFP could perhaps be increased by inserting the S-GFP gene into the coronavirus genome in addition to the wild-type S gene.
The expression of foreign proteins as an extension of a structural virion protein rather than through an independent transgene can have its advantages for a number of purposes. Thus, expression of a chimeric structural protein can be useful for studying virus assembly requirements as shown here. However, a foreign protein can also be appended to a structural protein to load the resulting virus particles with a specific biological activity, such as an enzymatic activity to be delivered into cells, or with a specific function, such as a targeting function to direct the recombinant virus to selected cells. The S-GFP protein that we constructed in the present study for our ongoing analysis of coronavirus structure and assembly provides us now with a virus, the fluorescent properties of which may allow us to investigate cell entry of coronaviruses by using time-lapsed imaging fluorescence microscopy.
Acknowledgments
We thank Jack Valentijn for technical assistance with the confocal laser scanning microscope.
This study was supported by The Netherlands Foundation for Chemical Research and The Netherlands Organization for Scientific Research.
REFERENCES
- 1.Bonavia, A., B. D. Zelus, D. E. Wentworth, P. J. Talbot, and K. V. Holmes. 2003. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 77:2530-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch, B. J., R. van der Zee, C. A. de Haan, and P. J. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77:8801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curtis, K. M., B. Yount, and R. S. Baric. 2002. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 76:1422-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalton, K. P., and J. K. Rose. 2001. Vesicular stomatitis virus glycoprotein containing the entire green fluorescent protein on its cytoplasmic domain is incorporated efficiently into virus particles. Virology 279:414-421. [DOI] [PubMed] [Google Scholar]
- 5.de Haan, C. A., L. Kuo, P. S. Masters, H. Vennema, and P. J. Rottier. 1998. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 72:6838-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Haan, C. A., P. S. Masters, X. Shen, S. Weiss, and P. J. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Haan, C. A., M. Smeets, F. Vernooij, H. Vennema, and P. J. Rottier. 1999. Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein. J. Virol. 73:7441-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Haan, C. A., L. van Genne, J. N. Stoop, H. Volders, and P. J. Rottier. 2003. Coronaviruses as vectors: position dependence of foreign gene expression. J. Virol. 77:11312-11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delmas, B., and H. Laude. 1990. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 64:5367-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnelly, M., and G. Elliott. 2001. Fluorescent tagging of herpes simplex virus tegument protein VP13/14 in virus infection. J. Virol. 75:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elroy-Stein, O., and B. Moss. 1990. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proc. Natl. Acad. Sci. USA 87:6743-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enjuanes, L., I. Sola, F. Almazan, J. Ortego, A. Izeta, J. M. Gonzalez, S. Alonso, J. M. Sanchez, D. Escors, E. Calvo, C. Riquelme, and C. Sanchez. 2001. Coronavirus derived expression systems. J. Biotechnol. 88:183-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escors, D., J. Ortego, H. Laude, and L. Enjuanes. 2001. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 75:1312-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer, F., C. F. Stegen, C. A. Koetzner, and P. S. Masters. 1997. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J. Virol. 71:5148-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleming, J. O., R. A. Shubin, M. A. Sussman, N. Casteel, and S. A. Stohlman. 1989. Monoclonal antibodies to the matrix (E1) glycoprotein of mouse hepatitis virus protect mice from encephalitis. Virology 168:162-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fosmire, J. A., K. Hwang, and S. Makino. 1992. Identification and characterization of a coronavirus packaging signal. J. Virol. 66:3522-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garoff, H., R. Hewson, and D. J. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godeke, G. J., C. A. de Haan, J. W. Rossen, H. Vennema, and P. J. Rottier. 2000. Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. J. Virol. 74:1566-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes, K. V., J. F. Boyle, D. G. Weismiller, S. R. Compton, R. K. Williams, C. B. Stephensen, and M. F. Frana. 1987. Identification of a receptor for mouse hepatitis virus. Adv. Exp. Med. Biol. 218:197-202. [DOI] [PubMed] [Google Scholar]
- 21.Holmes, K. V., E. W. Doller, and L. S. Sturman. 1981. Tunicamycin resistant glycosylation of coronavirus glycoprotein: demonstration of a novel type of viral glycoprotein. Virology 115:334-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsue, B., T. Hartshorne, and P. S. Masters. 2000. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 74:6911-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klumperman, J., J. K. Locker, A. Meijer, M. C. Horzinek, H. J. Geuze, and P. J. Rottier. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krijnse-Locker, J., M. Ericsson, P. J. Rottier, and G. Griffiths. 1994. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 124:55-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo, L., and P. S. Masters. 2002. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 76:4987-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewicki, D. N., and T. M. Gallagher. 2002. Quaternary structure of coronavirus spikes in complex with carcinoembryonic antigen-related cell adhesion molecule cellular receptors. J. Biol. Chem. 277:19727-19734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narayanan, K., C. J. Chen, J. Maeda, and S. Makino. 2003. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 77:2922-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narayanan, K., A. Maeda, J. Maeda, and S. Makino. 2000. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 74:8127-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narayanan, K., and S. Makino. 2001. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J. Virol. 75:9059-9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Opstelten, D. J., P. de Groote, M. C. Horzinek, H. Vennema, and P. J. Rottier. 1993. Disulfide bonds in folding and transport of mouse hepatitis coronavirus glycoproteins. J. Virol. 67:7394-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Opstelten, D. J., M. J. Raamsman, K. Wolfs, M. C. Horzinek, and P. J. Rottier. 1995. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 131:339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potel, C., K. Kaelin, I. Gautier, P. Lebon, J. Coppey, and F. Rozenberg. 2002. Incorporation of green fluorescent protein into the essential envelope glycoprotein B of herpes simplex virus type 1. J. Virol. Methods 105:13-23. [DOI] [PubMed] [Google Scholar]
- 34.Rietdorf, J., A. Ploubidou, I. Reckmann, A. Holmstrom, F. Frischknecht, M. Zettl, T. Zimmermann, and M. Way. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 3:992-1000. [DOI] [PubMed] [Google Scholar]
- 35.Rossen, J. W., R. de Beer, G. J. Godeke, M. J. Raamsman, M. C. Horzinek, H. Vennema, and P. J. Rottier. 1998. The viral spike protein is not involved in the polarized sorting of coronaviruses in epithelial cells. J. Virol. 72:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rottier, P., J. Armstrong, and D. I. Meyer. 1985. Signal recognition particle-dependent insertion of coronavirus E1, an intracellular membrane glycoprotein. J. Biol. Chem. 260:4648-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rottier, P. J., M. C. Horzinek, and B. A. van der Zeijst. 1981. Viral protein synthesis in mouse hepatitis virus strain A59-infected cells: effect of tunicamycin. J. Virol. 40:350-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarma, J. D., E. Scheen, S. H. Seo, M. Koval, and S. R. Weiss. 2002. Enhanced green fluorescent protein expression may be used to monitor murine coronavirus spread in vitro and in the mouse central nervous system. J. Neurovirol. 8:381-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siddell, S. G. 1995. The Coronaviridae: an introduction. Plenum Press, Inc., New York, N.Y.
- 40.Sola, I., S. Alonso, C. Sanchez, J. M. Sanchez-Morgado, and L. Enjuanes. 2001. Expression of transcriptional units using transmissible gastroenteritis coronavirus derived minigenomes and full-length cDNA clones. Adv. Exp. Med. Biol. 494:447-451. [DOI] [PubMed] [Google Scholar]
- 41.Sola, I., S. Alonso, S. Zuniga, M. Balasch, J. Plana-Duran, and L. Enjuanes. 2003. Engineering the transmissible gastroenteritis virus genome as an expression vector inducing lactogenic immunity. J. Virol. 77:4357-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taguchi, F. 1995. The S2 subunit of the murine coronavirus spike protein is not involved in receptor binding. J. Virol. 69:7260-7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taguchi, F., and J. O. Fleming. 1989. Comparison of six different murine coronavirus JHM variants by monoclonal antibodies against the E2 glycoprotein. Virology 169:233-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tooze, S. A., J. Tooze, and G. Warren. 1988. Site of addition of N-acetyl-galactosamine to the E1 glycoprotein of mouse hepatitis virus-A59. J. Cell Biol. 106:1475-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by coexpression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vennema, H., L. Heijnen, A. Zijderveld, M. C. Horzinek, and W. J. Spaan. 1990. Intracellular transport of recombinant coronavirus spike proteins: implications for virus assembly. J. Virol. 64:339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vennema, H., R. Rijnbrand, L. Heijnen, M. C. Horzinek, and W. J. Spaan. 1991. Enhancement of the vaccinia virus/phage T7 RNA polymerase expression system using encephalomyocarditis virus 5′-untranslated region sequences. Gene 108:201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vennema, H., P. J. Rottier, L. Heijnen, G. J. Godeke, M. C. Horzinek, and W. J. Spaan. 1990. Biosynthesis and function of the coronavirus spike protein. Adv. Exp. Med. Biol. 276:9-19. [DOI] [PubMed] [Google Scholar]
- 49.Yoo, D. W., M. D. Parker, and L. A. Babiuk. 1991. The S2 subunit of the spike glycoprotein of bovine coronavirus mediates membrane fusion in insect cells. Virology 180:395-399. [DOI] [PMC free article] [PubMed] [Google Scholar]