Abstract
Huntington disease is caused by expansion of a CAG repeat in the huntingtin gene that is translated into an elongated polyglutamine stretch within the N-terminal domain of the huntingtin protein. The mutation is thought to introduce a gain-of-toxic function in the mutant huntingtin protein, and blocking this toxicity by antibody binding could alleviate Huntington disease pathology. Llama single domain antibodies (VHH) directed against mutant huntingtin are interesting candidates as therapeutic agents or research tools in Huntington disease because of their small size, high thermostability, low cost of production, possibility of intracellular expression, and potency of blood-brain barrier passage. We have selected VHH from llama phage display libraries that specifically target the N-terminal domain of the huntingtin protein. Our VHH are capable of binding wild-type and mutant human huntingtin under native and denatured conditions and can be used in Huntington disease studies as a novel antibody that is easy to produce and manipulate.
Electronic supplementary material
The online version of this article (doi:10.1007/s10072-014-1971-6) contains supplementary material, which is available to authorized users.
Keywords: VHH, Huntington disease, PolyQ, N-terminal huntingtin, Huntingtin
Introduction
Huntington disease (HD) is caused by expansion of a CAG repeat within the first exon of the huntingtin gene (4p16.3) [1]. This mutation results in an expanded polyglutamine repeat (polyQ) at the N-terminus of the huntingtin protein (htt), causing HD pathology through a toxic gain-of-function mechanism [2]. Antibody binding could reduce toxicity of the mutant htt protein. Messer et al. showed that a single chain Fv antibody construct, selected against the first 17 N-terminal htt amino acids was capable of reducing HD pathogenesis in various HD models [3, 4]. In our study we make use of llama single domain antibody fragments called VHH [5]. VHH contain four framework regions (FR1–4) for structural integrity and three variable complement determining regions (CDR1–3) that usually determine epitope binding. VHH have distinctive advantages compared with other antibody classes. VHH are thermostable, only ~16 kD in size and their single domain nature simplifies selection and production [6, 7]. VHH have been used for diseases such as oculopharyngeal muscular dystrophy (OPMD), which shares characteristics with HD. OPMD is caused by expansion of a triplet repeat in the PABPN1 gene that encodes for a polyalanine repeat at the N-terminus of the polyA binding nuclear 1-protein (PABN1). VHH binding to an α-helical domain of mutant PABN1 prevented aggregation [8], and alleviated OPMD pathology in a droso philia model [9]. In the current study, we have selected VHH against the N-terminal domain of htt from llama phage display libraries. We show that high resolution melting curve analysis (HRMCA) [10] can successfully identify identical clones prior to sequencing. Our VHH can bind both endogenous and purified human wild-type and mutant htt at an epitope located between amino acids 49–148 and can co- immunoprecipitate htt from human HD brain lysates.
Results
Selection of VHH against N-terminal huntingtin
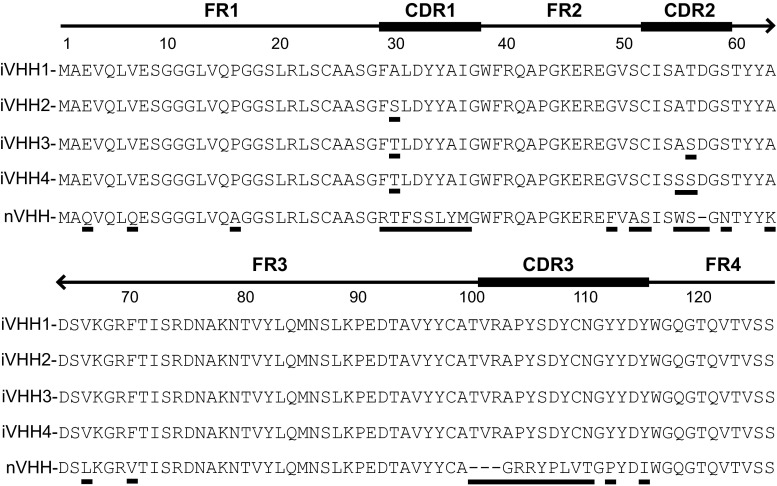
The phage-VHH (P-VHH) display library originated from llamas pre-immunized with an Escherichia coli produced N-terminal htt protein fragment consisting of the first 548 amino acids with 46 polyQs. We performed four different selections; each selection involved two rounds using either a wild-type or mutant N-terminal htt fragment. Enrichment of P-VHH after two rounds of selection was similar for direct coating or pre-capturing of N-terminal htt in the first round, with the optimal concentration being 5 µg N-terminal htt (Online Resource 1a). P-VHH output numbers of up to 3 × 104 were obtained. Screening ELISA revealed that on average, 20 % of selected clones bound htt (Online Resource 1b). Further screening by HRMCA (Online Resource 1c), followed by sequence analysis, revealed four htt specific VHH, (immune)VHH1-4. These differed by one, two, or three amino acid substitutions in the CDR1 and CDR2, while the CDR3 was identical (Fig. 1). The negative control VHH (n-VHH) was selected previously from a naive llama phage display library [10].
Fig. 1.
VHH protein sequences. iVHH 1–4 were selected from an immunized llama phage display library. nVHH was selected from a non-immunized llama phage display library. Underscored amino acid position differs from iVHH1. Amino acid positions, framework (FR, thin line) and complementary determining regions (CDR, thick line) are according to Kabat [25]. −, deletion
Specificity of monoclonal iVHH for N-terminal huntingtin
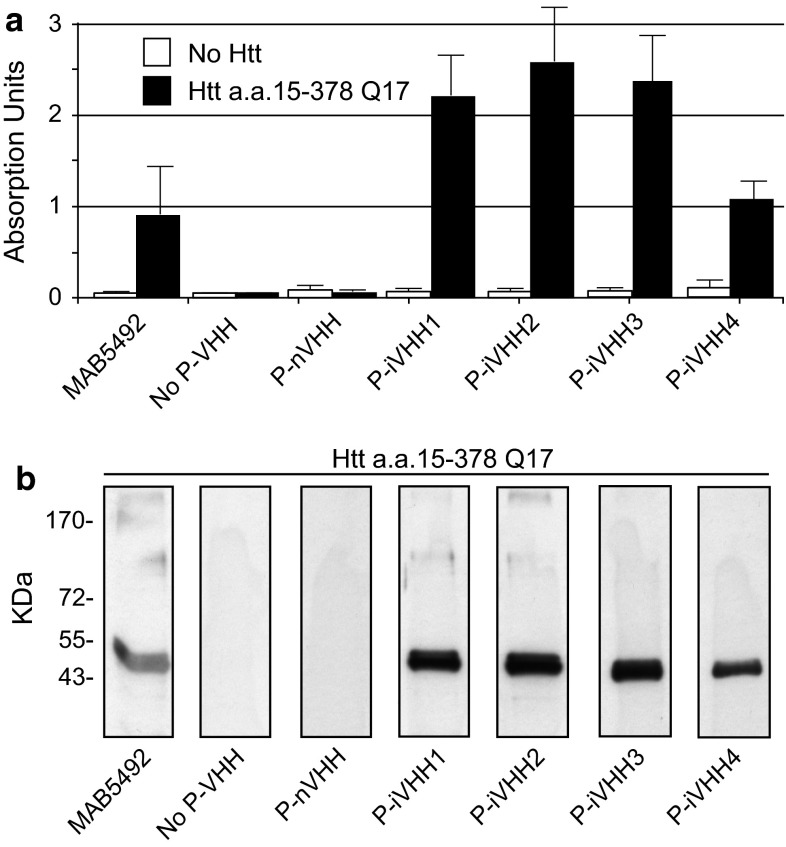
To investigate if the iVHH specifically bind htt, we performed ELISA and western blot analysis on a normal and mutant N-terminal htt fragment using P-iVHH [11]. ELISA resulted in a positive signal for all P-iVHH (Fig. 2a, Online Resource 2a). P-iVHH1, 2 and 3 showed an equally strong ELISA signal, whereas P-iVHH4 gave a weaker signal. On western blot, all P-iVHH showed a band that matched the band obtained with the known htt antibody MAB5492 (Fig. 2b, Online Resource 2b). Western blotting results were in agreement with ELISA results.
Fig. 2.
VHH specificity for N-terminal htt. Assays were performed on a recombinant N-terminal htt fragment consisting of amino acids 15–378 with a polyQ length of 17 (htt a.a. 15–378 Q17). Positive control: MAB5492. Negative control: No P-VHH or P-nVHH. a ELISA on wells with (black bars), or without (white bars) N-terminal htt. Bars represent mean ELISA signal from two independent ELISA assays with standard deviation. Each assay was performed in triplicate. ELISA absorption units are measured at λ = 490 nm. b Western blotting on N-terminal htt. Blots were performed twice. kDa Molecular weight (kilodalton)
Epitope determination of iVHH on N-terminal huntingtin
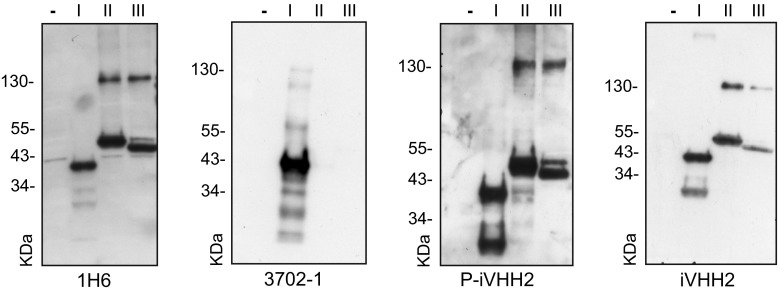
To determine the epitope of our iVHH, we performed western blotting using three different N-terminal htt fragments. Each fragment had a partially overlapping sequence with the next fragment (Fig. 3). All fragments were recognized by the 1H6 antibody [12] while 3702–1, that binds htt at the N-terminal 13 amino acids (Online Resource 3a), only recognized fragment I. All P-iVHH recognized all fragments (Fig. 3, Online Resource 3b) indicating that their epitope is located within the overlapping region of fragments I, II and III that consists of amino acids 49–148. Finally, western blotting with monomeric iVHH was performed. VHH were produced with an average concentration of 1.1 µg/µl. Western blotting using iVHH instead of P-iVHH recognized htt fragments I, II, and III but with less back ground staining.
Fig. 3.
VHH epitope determination. Western blots performed on no htt (−), htt a.a. 1–148 Q46 (I), htt a.a. 15–378 Q17 (II), and htt a.a. 49–415 (III). Primary antibody indicated below each blot. Blots were performed twice. kDa Molecular weight (kilodalton)
iVHH detection of endogenous huntingtin in human HD brain lysates
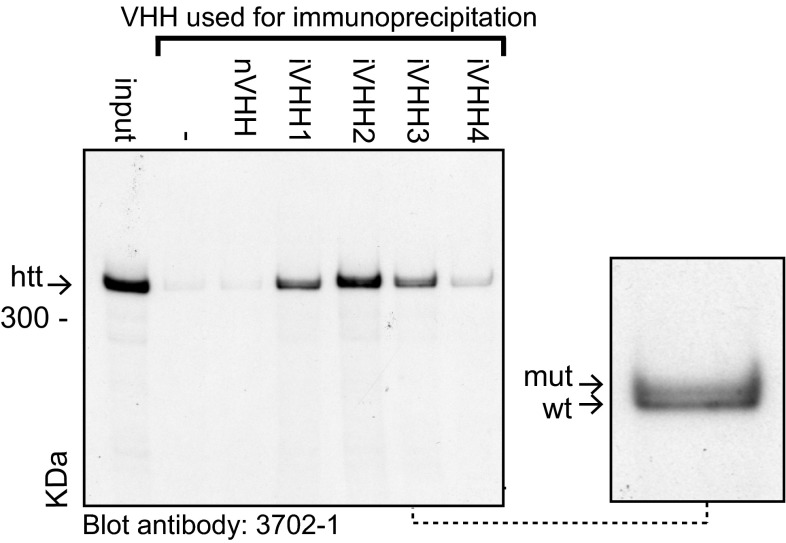
Next, we analysed if our iVHH were capable of binding endogenous human htt. We performed immunoprecipitation experiments using monomeric iVHH and post-mortem human HD brain lysates. Non-specific binding of htt to the protA-beads without VHH (Fig. 4, –lane) was low. Immunoprecipitation in the presence of iVHH resulted in wild-type and mutant full-length htt band on western blot, while the negative control VHH (nVHH) did not show a full-length htt band (Fig. 4, Online Resource 4). This shows that, in concordance with previous ELISA and western blot results on N-terminal wild-type and mutant htt fragments, our iVHH recognize both wild-type and mutant endogenous full-length htt. Additionally, immunoprecipitation with iVHH4 was less efficient compared with the other iVHH.
Fig. 4.
Immunoprecipitation of endogenous human htt with VHH. Western blot analysis of VHH-htt immunoprecipitation complexes. VHH used for htt immunoprecipitation indicated at top, Input, 10 µg of brain lysate; –, No VHH. Arrow indicates full-length htt. kDa Molecular weight (kilodalton). Blot was analysed with 3702-1 anti htt antibody. Right bracket 3× enlargement of the iVHH3 immunoprecipitation result, showing wild-type (wt) and mutant (mut) huntingtin
Discussion
To our knowledge, our study is the first in which VHH have been selected against N-terminal htt protein fragments from an immunized llama phage display library. Monoclonal antibodies are increasingly used as therapeutic agents [13]. Proof of concept for using antibodies in HD has been provided [3, 4]. Whilst this is promising, there are high demands on any therapeutic or imaging antibody. Because HD is a brain disorder, it is important to cross the blood–brain barrier (BBB). Some VHH were able to cross the BBB [14, 15]. Also, VHH diffusion throughout the brain, and cellular uptake were demonstrated [15]. Although VHH selection from non-immune phage-VHH libraries is possible [16], selection from immune llama phage-VHH libraries is expected to yield more high-affinity binders because of affinity maturation. However, diversity will be less as affinity maturation selects for the strongest binder. This is reflected in our results. Our iVHH share a high degree of sequence homology. This indicates that the binding epitope of our iVHH is very similar. Our iVHH bind full-length and N-terminal htt in a native and denatured conformation. Binding characteristics for iVHH1-3 were comparable, whereas iVHH4 showed a lower binding efficiency. This is probably due to a single amino acid change in iVHH4 at position 55 in CDR2 where the a-polar alanine was replaced with a polar serine. Because both alanine and serine have side-chains of comparable sizes, it is conceivable that the lower binding efficiency of iVHH4 is due to a shift from an a-polar to polar amino acid. The epitope of all iVHH was mapped to a region between amino acids 49 and 148 downstream of the polyQ repeat, explaining why our iVHH bind both wild-type and mutant htt. In this region, htt contains several domains associated with HD pathogenesis. There is a proline rich region involved in sequestration of other proteins into htt aggregates [17]. Furthermore, proteolytic cleavage between amino acids 104 and 114 was linked with formation of intranuclear aggregates associated with increased toxicity [12]. Binding of our iVHH in this region could alleviate HD pathogenesis. However, it has to be noted that our huntingtin-specific VHH could also block neuroprotective properties of wild-type htt [18]. Additional research is needed to assess the therapeutic properties of our iVHH or their possible applications in HD research. We have demonstrated the feasibility of using VHH for immunoprecipitation, eliminating interference by IgG bands in the subsequent western blot [19]. Furthermore, since VHH have been used as imaging agent to stain amyloid-β deposits in vivo in an Alzheimer disease mouse model [20], our iVHH could be an interesting in vivo imaging tool in HD models to visualize the htt protein.
Materials and methods
N-terminal htt fragments
A HTT reference sequence with 23 polyQs was used. For PCR primers see Table 1. PCR products were ligated directly into the pGEM-T easy vector (Promega, Madison, WI, USA), digested with NcoI and SalI (Fermentas, St. Leon-Rot, Germany), and ligated into a NcoI/XhoI (Fermentas) pre-digested pIVEX 1.3 vector. Clones were confirmed by sequence analysis. Htt protein fragments were produced with the RTS-100 wheat germ CECF kit (5 PRIME, Gaitersburg, MD, USA) using the ProteoMaster rapid translation system (Roche). To produce the N-terminal htt 1–148 Q46 protein fragment, the HD1955 pCI construct consisting of HTT nucleotides 1–1640 [21] was cloned into a pRP261 vector using the NcoI/SalI restriction sites, and re-cloned into the pET28 vector using the BamHI/SalI restriction sites. The HD1955-pET28 construct was digested with XhoI and self-religated, resulting in the HD828-pET28 construct. Production and purification were performed in BL21 codon+ E.coli cells as described for VHH. To prevent aggregation, dialysis was performed in PBS + 0.5 % Sarkosyl (Sigma–Aldrich).
Table 1.
Construction of N-terminal Htt fragments
| N-terminal htt fragment | Forward primer | Reverse primer |
|---|---|---|
| a.a. 1–318 (Q17/Q43) | TATGGCGACCCTGGAAA | GTCGACGAGCAGCACGCCAAGA |
| a.a. 15–378 (Q17/Q43) | CAAGTCCTTCCAGCAGCA | GTCGACGGCTCCGGTCACAACA |
| a.a. 49–415 (no polyQ) | GCCGCCTCCTCAGCTTC | GTCGACGCCACCAGACTCCTCCTT |
a.a. amino acid, Q17/Q43 polyQ stretch, Underscored SalI-site
VHH selection
Selections were performed as described [16]. The first selection round involved a direct coating of NUNC maxisorp plates (Thermo fisher Scientific, Rochester, NY, USA) with 10, 5, or 2.5 µg of antigen, or a pre-capturing of 10 µg antigen with 2, 1, or 0.5 µg of coated 1H6 antibody (Abnova, Taipei, Taiwan). Phage-VHH (P-VHH) from the first round were produced and purified as described [22] and subjected to a second round of selection with 5, 2.5, or 1 µg of directly-coated antigen. Purified P-VHH were stored at −20 °C in PBS containing 10 % glycerol. TG1 E. coli cells were infected with phage-VHH from the second selection round and plated on LB/Agar containing ampicillin. Ninety-four randomly selected clones were tested as described [10]. As secondary antibody for the screening ELISA, we used HRP conjugated mouse anti M13 (GE Healthcare, Buckinghamshire, UK). VHH DNA was purified using the Nucleospin Plasmid purification kit (Macherey–Nagel, Duren, Germany) according to manufacturer’s instructions. DNA was sequenced using primer M13REV (CAGGAAACAGCTATGAC). DNA to protein conversion: http://www.bioinformatics.picr.man.ac.uk/research/software/tools/sequenceconverter.html. VHH sequence alignment: http://www.ebi.ac.uk/Tools/msa/clustalW2.
VHH production
PCR was performed on M13 plasmid using primers iVHH-FW (CGGAATTCCTTTAGTTGTTCCA), and iVHH-Rev (CACATCATCATCACCATCACG), or nVHH-FW (CGCTGGATTGTTATTACTCGC) and nVHH-Rev (CCTCAGAACCCAAGACCA). PCR fragments were cloned into pUR5850 [23] by SfiI and BstEII (Fermentas) digestion and ligation. Clones were verified by sequence analysis and transferred into Neb5 E. coli for production. VHH were purified using the His6-tag with TALON metal affinity resin (Clontech, Mountain View CA, USA) according to the manufacturer’s instructions using 50 mM NaPO4, 0.3 M NaCl, pH7 as wash buffer. Wash buffer with 150 mM imidazole was used as elution buffer. Pooled eluates were dialyzed against PBS using Cellu-Sep dialyze-tube MWCO 3500 (Interchim, MontluÇon, France). VHH production was checked on Coomassie staining (PageBlue, Thermo-Scientific) and western blotting against the VSV-tag. VHH was quantified with bicinchoninic assay (BCA, Thermo-Scientific). VHH were stored in 5 % glycerol at −20 °C.
ELISA
A 96-well NUNC maxisorp plate was coated with 0.1 µg/well of antigen. Staining was performed using P-VHH diluted 1:20, followed by HRP conjugated mouse anti M13 (Millipore Billerica, MA, USA). ELISA signal was visualized with o-Phenylenediamine (OPD, Sigma–Aldrich). Optical density at λ = 490 nm was measured using a plate reader (Biotek, Winooski, USA). The huntingtin-specific antibody MAB5492 (Millipore) was used as positive control.
Western blot
N-terminal htt fragments were run on a 10 % SDS-PAGE gel and proteins were blotted onto nitrocellulose membrane (#170-4159, Bio-Rad, Hercules, CA, USA). Blots were blocked with 4 % non-fat milk (Nutricia, Schiphol, The Netherlands) in TBST. Primary antibodies: 3702-1 (Epitomics, Burlinggame, CA, USA), MAB5492 (Millipore), 1H6 (Abnova, Taipei city, Taiwan), P-VHH or 20 ng/µl VHH. Blots probed with VHH were subsequently incubated with mouse anti VSV (Cell signalling). Secondary antibodies: HRP conjugated goat anti-mouse, goat anti-rabbit (Santa-Cruz), or mouse anti-M13 (Millipore). Bands were detected with Amersham ECL (#RPN 2132, GE healthcare) and Hyperfilm ECL (#28906837, GE healthcare).
Immunoprecipitation assay
Human post-mortem brain tissue from the middle temporal gyrus of a 67 year old female HD subject (CAG1: 15, CAG2: 42) was obtained with the families full consent and the ethical approval of the various institutional Ethics Committees. Post-mortem delay was 9 h. VHH (15 µg) was bound to 10 µl bed volume of protein A sepharose beads (GE Healthcare), and incubated for 90 min with 100 µg of post-mortem human HD brain tissue lysate in PBS. Western blot analysis of the VHH-htt complexes was performed as described above, where SDS-PAGE was performed according to [24].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Electronic supplemental figure 1. Selection of P-iVHH by two rounds of panning against wild type N-terminal huntingtin. a TG1 E.coli cells infected with P-VHH were diluted as indicated and spotted on agar medium selective for presence of P-VHH. Round 1: P-VHH from the llama phage display bank panned against three different concentrations of htt a.a. 15-378 Q17. Most P-VHH are recovered using 5 µg of N-term htt (indicated in red). Inp 10−6 / −8 = cells infected with whole phage display bank library diluted 106 or 108 x. Round 2: P-VHH from the most efficient first round selection were enriched and panned for a second time against htt a.a. 15-378 Q17 at indicated concentrations. Most P-VHH are recovered at a htt fragment concentration of 5 µg (indicated in blue). - = cells infected with enriched first round P-VHH selected against a blank well (background). Inp 10−8 = cells infected with whole round 1 P-VHH diluted 108 x. b Screening ELISA of 94 individual P-VHH clones from round 2. There were 13 (14%) ELISA positive clones (AU490>0.4) detected. c High resolution melting curve analysis (HRMCA) of ELISA positive clones revealed two groups of similar clones; blue (7 clones) and red (3 clones), and three unique VHH (green, pink and grey) (EPS 9306 kb)
Electronic supplemental figure 2. Binding of P-VHH to N-terminal Htt fragment with elongated polyQ. Assays were performed on a recombinant N-terminal htt fragment consisting of amino acids 15 to 378 with a polyQ length of 43 (htt a.a. 15-378 Q43). Anti htt antibody MAB5492 served as positive control. Assays performed without P-VHH or the non-binding P-nVHH served as negative control. a ELISA with P-VHH on wells with (grey bars), or without (white bars) htt a.a. 15-378 Q43. Bars represent mean ELISA signal from two independent ELISA assays with standard deviation. Each assay was performed in triplicate. ELISA absorption units are measured at λ=490nm b Western blotting with P-VHH on htt a.a. 15-378 Q43. All blots were performed twice. kDa = running height in kilodalton (EPS 4686 kb)
Electronic supplemental figure 3. Epitope determination of 3702-1 and VHH antibodies. a Western blot on five different N-terminal htt fragments: htt a.a. 1 to 318 with wild type (Q17) and mutant (Q43) polyQ, htt a.a. 15 to 378 with wild type (Q17) and mutant (Q43) polyQ and htt a.a. 49-415 without the polyQ. MAB5492 (left bracket) binds all htt fragments. 3702-1 (right bracket) only binds htt a.a. 1 to 318 with either the wild type or mutant polyQ. b Epitope determination of P-iVHH1, 3 and 4. Fragments: I = N-terminal htt fragment with a.a. 1 to 148 with a mutant polyQ (Q46). II = N-terminal htt fragment with a.a. 15 to 378 with a wild type polyQ (Q17). III = htt fragment with a.a. 49 to 415 without polyQ stretch. - = no htt fragment. Blot performed with non-binding P-nVHH served as a negative control. All blots were performed twice (EPS 11320 kb)
Electronic supplemental figure 4. Immunoprecipitation of human full length htt with VHH. Input, -, nVHH, iVHH1-4 are shown in figure 4. VHH “X” corresponds to iVHH2 produced from the M13-vector. VHH produced from the M13-vector are less pure compared with VHH produced from pUR5850, hence the band intensity of VHH “X” is lower compared with iVHH2. Because the comparison between different VHH production vectors was outside the scope of this manuscript, we removed VHH X from figure 4 (EPS 4158 kb)
Acknowledgments
We thank Richard L. M. Faull and the Neurological Foundation of New Zealand Human Brain Bank for the HD post-mortem brain tissue. We thank Dr. Michael Hayden for the HD1955 (CAG44)-pCI construct. We thank Fausta Bankauskaite for technical assistance. This study was supported by a grant (WAR30808) from the Prinses Beatrix Spierfonds, and the Centre for Biomedical Genetics of the Netherlands.
Conflict of interest
The authors declare no competing financial interests.
References
- 1.(1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell 72(6): 971–983 [DOI] [PubMed]
- 2.Jacobsen JC, Gregory GC, Woda JM, et al. HD CAG-correlated gene expression changes support a simple dominant gain of function. Hum Mol Genet. 2011;20(14):2846–2860. doi: 10.1093/hmg/ddr195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolfgang WJ, Miller TW, Webster JM, et al. Suppression of Huntington’s disease pathology in Drosophila by human single-chain Fv antibodies. Proc Natl Acad Sci USA. 2005;102(32):11563–11568. doi: 10.1073/pnas.0505321102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snyder-Keller A, McLear JA, Hathorn T, Messer A. Early or late-stage anti-N-terminal Huntingtin intrabody gene therapy reduces pathological features in B6.HDR6/1 mice. J Neuropathol Exp Neurol. 2010;69(10):1078–1085. doi: 10.1097/NEN.0b013e3181f530ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamers-Casterman C, Atarhouch T, Muyldermans S, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363(6428):446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 6.van der Linden RH, Frenken LG, de Geus B, et al. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim Biophys Acta. 1999;1431(1):37–46. doi: 10.1016/S0167-4838(99)00030-8. [DOI] [PubMed] [Google Scholar]
- 7.Arbabi Ghahroudi M, Desmyter A, Wyns L, et al. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett. 1997;414(3):521–526. doi: 10.1016/S0014-5793(97)01062-4. [DOI] [PubMed] [Google Scholar]
- 8.Verheesen P, de Kluijver A, van Koningsbruggen S, et al. Prevention of oculopharyngeal muscular dystrophy-associated aggregation of nuclear polyA-binding protein with a single-domain intracellular antibody. Hum Mol Genet. 2006;15(1):105–111. doi: 10.1093/hmg/ddi432. [DOI] [PubMed] [Google Scholar]
- 9.Chartier A, Raz V, Sterrenburg E, et al. Prevention of oculopharyngeal muscular dystrophy by muscular expression of Llama single-chain intrabodies in vivo. Hum Mol Genet. 2009;18(10):1849–1859. doi: 10.1093/hmg/ddp101. [DOI] [PubMed] [Google Scholar]
- 10.Pepers BA, Schut MH, Vossen RH, et al. Cost-effective HRMA pre-sequence typing of clone libraries; application to phage display selection. BMC Biotechnol. 2009;9:50. doi: 10.1186/1472-6750-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu B, Huang L, Sihlbom C, et al. Towards proteome-wide production of monoclonal antibody by phage display. J Mol Biol. 2002;315(5):1063–1073. doi: 10.1006/jmbi.2001.5276. [DOI] [PubMed] [Google Scholar]
- 12.Lunkes A, Lindenberg KS, Ben-Haiem L, et al. Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Mol Cell. 2002;10(2):259–269. doi: 10.1016/S1097-2765(02)00602-0. [DOI] [PubMed] [Google Scholar]
- 13.Leavy O. Therapeutic antibodies: past, present and future. Nat Rev Immunol. 2010;10(5):297. doi: 10.1038/nri2763. [DOI] [PubMed] [Google Scholar]
- 14.Rutgers KS, Nabuurs RJ, van den Berg SA, et al. Transmigration of beta amyloid specific heavy chain antibody fragments across the in vitro blood-brain barrier. Neuroscience. 2011;190:37–42. doi: 10.1016/j.neuroscience.2011.05.076. [DOI] [PubMed] [Google Scholar]
- 15.Li T, Bourgeois JP, Celli S, et al. Cell-penetrating anti-GFAP VHH and corresponding fluorescent fusion protein VHH-GFP spontaneously cross the blood-brain barrier and specifically recognize astrocytes: application to brain imaging. FASEB J. 2012;26(10):3969–3979. doi: 10.1096/fj.11-201384. [DOI] [PubMed] [Google Scholar]
- 16.Verheesen P, Roussis A, de Haard HJ, et al. Reliable and controllable antibody fragment selections from Camelid non-immune libraries for target validation. Biochim Biophys Acta. 2006;1764(8):1307–1319. doi: 10.1016/j.bbapap.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Qin ZH, Wang Y, Sapp E, et al. Huntingtin bodies sequester vesicle-associated proteins by a polyproline-dependent interaction. J Neurosci. 2004;24(1):269–281. doi: 10.1523/JNEUROSCI.1409-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26(3):300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 19.Landles C, Sathasivam K, Weiss A, et al. Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J Biol Chem. 2010;285(12):8808–8823. doi: 10.1074/jbc.M109.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nabuurs RJ, Rutgers KS, Welling MM, et al. In vivo detection of amyloid-beta deposits using heavy chain antibody fragments in a transgenic mouse model for Alzheimer’s disease. PLoS ONE. 2012;7(6):e38284. doi: 10.1371/journal.pone.0038284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martindale D, Hackam A, Wieczorek A, et al. Length of huntingtin and its polyglutamine tract influences localization and frequency of intracellular aggregates. Nat Genet. 1998;18(2):150–154. doi: 10.1038/ng0298-150. [DOI] [PubMed] [Google Scholar]
- 22.Marks JD, Hoogenboom HR, Bonnert TP, et al. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222(3):581–597. doi: 10.1016/0022-2836(91)90498-U. [DOI] [PubMed] [Google Scholar]
- 23.Verheesen P, ten Haaft MR, Lindner N, et al. Beneficial properties of single-domain antibody fragments for application in immunoaffinity purification and immuno-perfusion chromatography. Biochim Biophys Acta. 2003;1624(1–3):21–28. doi: 10.1016/j.bbagen.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Hu J, Matsui M, Gagnon KT, et al. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat Biotechnol. 2009;27(5):478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabat E, Wu TT, Perry H et al. (1991) United States Public Health Services Publication No. 91-3242 (National Institutes of Health, Bethesda, MD)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electronic supplemental figure 1. Selection of P-iVHH by two rounds of panning against wild type N-terminal huntingtin. a TG1 E.coli cells infected with P-VHH were diluted as indicated and spotted on agar medium selective for presence of P-VHH. Round 1: P-VHH from the llama phage display bank panned against three different concentrations of htt a.a. 15-378 Q17. Most P-VHH are recovered using 5 µg of N-term htt (indicated in red). Inp 10−6 / −8 = cells infected with whole phage display bank library diluted 106 or 108 x. Round 2: P-VHH from the most efficient first round selection were enriched and panned for a second time against htt a.a. 15-378 Q17 at indicated concentrations. Most P-VHH are recovered at a htt fragment concentration of 5 µg (indicated in blue). - = cells infected with enriched first round P-VHH selected against a blank well (background). Inp 10−8 = cells infected with whole round 1 P-VHH diluted 108 x. b Screening ELISA of 94 individual P-VHH clones from round 2. There were 13 (14%) ELISA positive clones (AU490>0.4) detected. c High resolution melting curve analysis (HRMCA) of ELISA positive clones revealed two groups of similar clones; blue (7 clones) and red (3 clones), and three unique VHH (green, pink and grey) (EPS 9306 kb)
Electronic supplemental figure 2. Binding of P-VHH to N-terminal Htt fragment with elongated polyQ. Assays were performed on a recombinant N-terminal htt fragment consisting of amino acids 15 to 378 with a polyQ length of 43 (htt a.a. 15-378 Q43). Anti htt antibody MAB5492 served as positive control. Assays performed without P-VHH or the non-binding P-nVHH served as negative control. a ELISA with P-VHH on wells with (grey bars), or without (white bars) htt a.a. 15-378 Q43. Bars represent mean ELISA signal from two independent ELISA assays with standard deviation. Each assay was performed in triplicate. ELISA absorption units are measured at λ=490nm b Western blotting with P-VHH on htt a.a. 15-378 Q43. All blots were performed twice. kDa = running height in kilodalton (EPS 4686 kb)
Electronic supplemental figure 3. Epitope determination of 3702-1 and VHH antibodies. a Western blot on five different N-terminal htt fragments: htt a.a. 1 to 318 with wild type (Q17) and mutant (Q43) polyQ, htt a.a. 15 to 378 with wild type (Q17) and mutant (Q43) polyQ and htt a.a. 49-415 without the polyQ. MAB5492 (left bracket) binds all htt fragments. 3702-1 (right bracket) only binds htt a.a. 1 to 318 with either the wild type or mutant polyQ. b Epitope determination of P-iVHH1, 3 and 4. Fragments: I = N-terminal htt fragment with a.a. 1 to 148 with a mutant polyQ (Q46). II = N-terminal htt fragment with a.a. 15 to 378 with a wild type polyQ (Q17). III = htt fragment with a.a. 49 to 415 without polyQ stretch. - = no htt fragment. Blot performed with non-binding P-nVHH served as a negative control. All blots were performed twice (EPS 11320 kb)
Electronic supplemental figure 4. Immunoprecipitation of human full length htt with VHH. Input, -, nVHH, iVHH1-4 are shown in figure 4. VHH “X” corresponds to iVHH2 produced from the M13-vector. VHH produced from the M13-vector are less pure compared with VHH produced from pUR5850, hence the band intensity of VHH “X” is lower compared with iVHH2. Because the comparison between different VHH production vectors was outside the scope of this manuscript, we removed VHH X from figure 4 (EPS 4158 kb)