Abstract
Superantigens are microbial proteins that strongly stimulate T cells. We described previously that the Epstein-Barr virus (EBV) transactivates a superantigen encoded by the human endogenous retrovirus, HERV-K18. We now report that the transactivation is dependent upon the EBV latent cycle proteins. Moreover, LMP-2A is sufficient for induction of HERV-K18 superantigen activity.
Superantigens are pathogen-derived proteins that elicit a strong primary T-cell response from the host (reviewed in references 25 and 27). Superantigens are presented to T cells by major histocompatibility complex (MHC) class II molecules on antigen-presenting cells. They differ from conventional peptide antigens by binding solely to the Vβ portion of the T-cell receptor (TCRBV) outside of the peptide-binding groove, thus forming a bridge between the T cell and the antigen-presenting cell (29). This bridging transduces a signal to the T cell, causing it to secrete cytokines that can further activate surrounding T cells. The hallmark of a superantigen response is the rapid and strong primary T-cell activation, which is MHC class II dependent and TCRBV restricted. In addition, antigen processing into peptides is not required. Both bacteria and viruses encode superantigens. The bacterial superantigens are mainly enterotoxins, which are secreted and bind externally to MHC class II molecules for presentation (34). In contrast, viral superantigens are glycosylated proteins that are endogenously produced in the infected cells.
There are three families of viruses that are associated with superantigen or superantigen-like activity: Retroviridae, Rhabdoviridae, and Herpesviridae. Retroviral superantigens were first depicted in the B-type virus group in mouse mammary tumor viruses and are found in both infectious mouse mammary tumor viruses and endogenous proviruses (14, 18, 39, 63). It has been previously shown that the env gene of HERV-K18, a defective human endogenous provirus located on chromosome 1, encodes a superantigen activity (51, 52). The HERV-K family is closely related to the B-type retroviruses based on amino acid similarity in the reverse transcriptase gene (48). HERV-K18 is a relatively recent integrant in the genome, as it is found in Old World primates but not in New World primates, indicating that it was acquired subsequent to the evolutionary divergence of these species (28). A few years ago, it was reported that Epstein-Barr virus (EBV) is associated with TCRBV13-specific superantigen activity, which is MHC class II dependent and not due to a recall antigen response (53). More recently, it was demonstrated that the superantigen activity is due to EBV transactivation of HERV-K18 env (52). We show here that this activity is dependent upon the major EBV latent gene transactivator EBNA-2, which upregulates most of the other EBV latent genes, all of which have the ability to transactivate host cell genes. In accordance with this finding, we show that the EBV latent membrane protein LMP-2A is sufficient for transactivation of HERV-K18 env.
EBV latent cycle genes are associated with transactivation of HERV-K18.
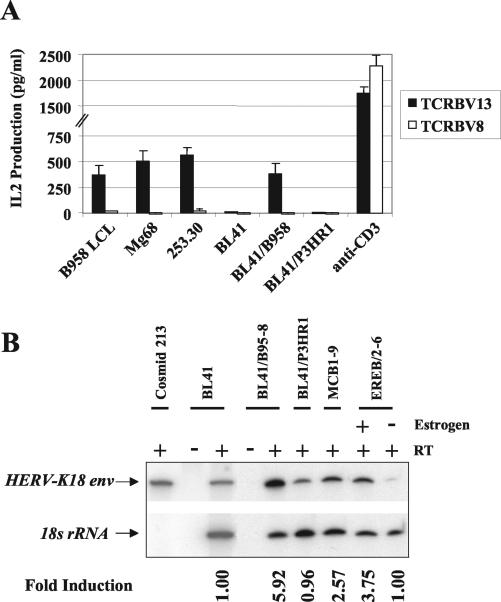
In order to map the EBV gene(s) responsible for transactivation of the HERV-K18 superantigen, we tested B cells infected with various EBV deletion mutants for their ability to preferentially stimulate interleukin-2 (IL-2) production from TCRBV13 T-cell hybridomas. We simultaneously assessed activation of TCRBV8 T-cell hybridomas as a specificity control. Hybridoma assays were performed as described previously (52, 53) by using the TCRBV13 T-cell hybridoma, hVβ13.1-1 (11, 12) and the TCRBV8 T-cell hybrid YLβ8#24 (53). Figure 1A depicts the results of a representative hybridoma assay with two lymphoblastoid cell lines (LCL), Mg68 and 253.30, that were derived by transformation with EBV deletion mutants lacking the majority of the lytic genes. A LCL transformed with the prototypic laboratory strain of EBV, B95-8, served as a positive control.
FIG. 1.
EBNA-2-dependent transactivation of the HERV-K18 superantigen. (A) LCL transformed by deletion mutant EBV (Mg68 and 253.30) or B95-8 EBV were tested for the ability to stimulate TCRBV13 and TCRBV8 T-cell hybridomas. EBV− BL-41 lymphoma cells were also tested and compared with BL-41 infected with B95-8 (BL-41/B95-8) or EBNA-2-deficient P3HR1 virus (BL-41/P3HR1). EBV cell lines used as superantigen-presenting cells were treated overnight with phorbol myristate acetate (10 ng/ml; Calbiochem) at 37°C. Cells were washed extensively in phosphate-buffered saline, counted, and resuspended with T-cell hybrids in 96-well round-bottom plates with 105 antigen-presenting cells and 2 × 104 T-cell hybrids per well. After 24 to 48 h at 37°C, the plates were frozen at −80°C to lyse the cells, and thawed supernatants were tested for the presence of IL-2 by HT-2 bioassay as previously described (53). As the positive control, the T-cell hybrids were cross-linked with plate-bound anti-CD3 (145 2C11). The mean IL-2 production for each T-cell hybrid measured in quadruplicate wells was expressed in picograms per milliliter of culture supernatant by comparison with values from a standard curve derived from recombinant IL-2 (R & D Systems). Error bars represent the difference measured between quadruplicate wells within a single experiment. Experiments were performed at least five times. (B) Semiquantitative RT-PCR for HERV-K18 read-through transcripts and 18S rRNA was performed on uninfected BL-41, BL-41/B95-8, BL-41/P3HR1, and BL-41/P3HR1 stably transfected with EBNA-2 (MCB1-9 cells). In addition, LCL transformed by recombinant EBV with estrogen-responsive EBNA-2 (EREB/2-6 cells) were tested in the presence or absence of estrogen. cDNA was prepared by random priming of total RNA that had been DNase I treated to remove contamination by genomic DNA. All samples were prepared in the presence (+) or absence (−) of RT. The PCR sense primer was 5′ TCCGAAGAGACAGTGACATCGA 3′, directed against a HERV-K18 env-specific sequence; the antisense primer was 5′ TGGCAATGCTGGCTATGTAAGT 3′, directed against a chromosome 1q23.1-q24.1 (GenBank accession number AL121985) sequence, located 127 bp downstream of the 3′ viral long terminal repeat. PCR was performed in the presence of [32P]α-dCTP, incorporating primers specific for 18S rRNA as an endogenous standard. Since the HERV-K18 read-through transcripts were extremely rare compared with the 18S rRNA, 18S Classic competimers (Ambion) were added at a primer-to-competimer ratio of 1:20. PCR was performed by using a hot start of 4 min at 94°C and then 25 cycles of 30 s at 94°C, 90 s at 72°C, and 60 s at 55°C, followed by a 7-min extension at 72°C, which yielded products within a linear range. PCR products were separated on a 6% denaturing polyacrylamide gel. HERV-K18 read-through transcripts were quantified by phosphorimaging (Molecular Dynamics), and induction (fold) was calculated after normalization against the 18S rRNA product; values are reported below each lane.
Mg68 is an LCL transformed by recombinant EBV that has 18 kb of sequence deleted between EBNA-2 and EBNA-3A and 58 kb of lytic genes deleted between EBNA-1 and LMP-1 (49). The deletion virus was constructed by homologous recombination of cosmids containing overlapping portions of the B95-8 genome and was packaged by transformation-defective P3HR1 helper virus (49, 56). These deletions comprise 76 kb of the 172-kb genome, spanning the regions encoded from positions 68,928 to 87,030, and 117,609 to 163,415. Mg68 was originally produced in an effort to define the minimal transforming EBV genome (49). It is deleted for the majority of the lytic genes listed in Table 1. Mg68 contains all of the latent genes, as well as the major lytic gene transactivator BZLF-1. A detailed analysis of EBV gene expression in Mg68 has previously been reported (49). The cell line 253.30 is an LCL transformed by a mini-EBV plasmid, p1244.8a (32), packaged in transformation-defective HH514 helper virus (a subclone of P3HR1) (26). This plasmid was derived from 71 kb of noncontiguous portions of the B95-8 genome (163,477 to 19,359, 43,935 to 56,081, and 79,658 to 113,282) cloned into a recombinant F-factor-based Escherichia coli plasmid by a chromosome-building technique previously described in detail (32, 47). This cell line, like Mg68, is deficient for the majority of EBV lytic genes, containing an even larger deletion 101 kb in total size (Table 1). It expresses all EBV latent genes with the exception of EBNA-3A, which was mutated during the transformation event, and EBNA-LP is truncated, containing only 2 of the 11 W repeats (32). It should be noted that the 253.30 cell line was originally coinfected with helper virus but was consistently negative by PCR for helper virus at the time of the superantigen assay (32). Mg68 was also helper deficient (49).
TABLE 1.
Summary of EBV deletion mutant cell lines tested for superantigen activitya
| EBV cell linesb | Superantigen activityc | Sequence deletions | Genes affected |
|---|---|---|---|
| IM-1 LCL | + | ? Unknown | Wild type (isolated from an infectious mononucleosis patient) (a gift from M. A. Epstein) |
| B95-8 marmoset | − | 13.6 kb at 154,012 | Prototype lab strain (first virus entirely sequenced; has a 13.6-kb deletion relative to most other strains) |
| B95-8 LCL/BL | + | 13.6 kb at 154,012 | Prototype lab strain |
| Raji BL | + | 98,805-102,116, 163,978-166,635 | EBNA-3C (latent); BZLF-2, BALF-1-2, BARF-1 (lytic) |
| P3HR1 BL | − | 45,644-52,450 | EBNA-2, EBNA-LP (truncated) (latent) |
| Mg68 LCL | + | 68,928-87,030, 117,609-163,415 | BPLF-1, BOLF-1, BORF-1-2, BaRF-1, BMRF-1-2, BMLF-1, BSLF-1-2, BSRF-1, BBLF-1-2, BBRF-3, BGLF-1-5, BGRF-1, BDLF-1-4, BDRF-1, BcLF-1, BcRF-1, BTRF-1, BXLF-1-2, BXRF-1, BVRF-1-2, BdRF-1, BILF-1-2, BALF-2-5 (lytic) |
| 253.30 LCL | + | 19,360-43,934, 56,082-79,657, 113,283-163,476 | EBNA-3A, EBNA-LP (truncated) (latent); BFLF-1-2, BFRF-1-3, BPLF-1, BOLF-1, BORF-1-2, BaRF-1, BBLF-1-4, BBRF-1-3, BGLF-1-5, BGRF-1, BDLF-1-4, BDRF-1, BcLF-1, BcRF-1, BTRF-1, BXLF-1-2, BXRF-1, BVRF-1-2, BdRF-1, BILF-1-2, BALF-2-5 (lytic) |
Results obtained with the different cell lines tested for functional activation of the TCRBV13 T-cell hybridoma are shown (Fig. 1A). The results indicate that a deletion of a combined total of 115 kb of the 172-kb EBV genome comprising the majority of lytic genes does not affect superantigen activity, indicating that these genes are not responsible for transactivation of the superantigen. On the other hand, a deletion of EBNA-2, the key latent gene transactivator, abolished superantigen activity. In addition, the B95-8 marmoset cell line was negative (−) for superantigen activity; marmosets are New World primates which do not have the HERV-K18 provirus.
BL, Burkitt's lymphoma.
A + indicates a cell line that was positive for superantigen activity; a − indicates a cell line that was negative for superantigen activity.
As can be seen in Fig. 1A, both Mg68 and 253.30 induce TCRBV13 T-cell stimulation similar to that of a B95-8 transformed LCL. These results indicate that superantigen activity is still present in Mg68 and 253.30, despite deletions cumulatively totaling 115 kb of sequence, comprising the genes listed in Table 1. Thus, the genes responsible for induction of superantigen activity map to within 57 kb of noncontiguous B95-8 sequence consisting mainly of the EBV latent cycle genes, which are the EBV nuclear antigens (EBNAs) and the latent membrane proteins (LMPs). Further evidence that the latent cycle genes play a role in transactivation of HERV-K18 env is provided by the finding that the EBV-negative (EBV−) Burkitt's lymphoma cell line BL-41 infected with P3HR1 EBV (7) had no superantigen activity, while BL-41 infected with B95-8 virus (7) was positive for superantigen activity (Fig. 1A). The P3HR1 virus is a replication-competent EBV that is deleted for the major latent gene transactivators EBNA-2 and EBNA-LP (3, 42). The P3HR1 deletion virus is incapable of transforming B cells (21), yet it expresses the full lytic gene repertoire and is frequently used as a packaging line for creating recombinant deletion viruses, like Mg68. The transforming genes EBNA-2 and EBNA-LP are the first genes expressed, aside from EBNA-1, subsequent to EBV infection of the B cell (1, 50). EBNA-2 interacts with the B-cell transcription factor RBPJκ (24, 64) and the ets members Pu.1 and Spi-B (31, 36), transactivating LMP-1 and the other EBV latent genes. EBNA-LP acts in concert with EBNA-2, increasing transactivation of LMP-1 (22, 46), which has oncogenic properties (2, 43, 61) and induces multiple cellular genes (20, 35, 62). EBNA-2 also directly transactivates a variety of cellular genes, such as CD23 (7, 45, 62), and represses immunoglobulin heavy chain gene (IgH) expression (30). Thus, the following question was posed. Could EBNA-2, either directly or indirectly, transactivate the HERV-K18 superantigen?
To assess this point, we determined the level of HERV-K18 transcription in the various EBV-infected cell lines. Since up to 8% of the human genome consists of HERV sequences, many of which are highly homologous, we designed a sensitive reverse transcriptase PCR (RT-PCR) assay for the detection of HERV-K18-specific read-through transcripts. This assay is based upon the observation that in up to 15% of proviral transcripts, the cellular RNA polymerase reads through the polyadenylation site in the 3′ long terminal repeat, transcribing adjacent chromosomal insertion sequences (54, 55). We therefore used RT-PCR primers specific for the upstream HERV-K18 env gene and the downstream chromosome 1 insertion site. Because of the length of the read-through transcripts, real-time PCR could not be used; thus, to render this assay semiquantitative, the PCR cycles were limited, keeping the product within the linear range, and primers specific for the 18S ribosomal subunit were included in each reaction as an endogenous standard. It has previously been reported that this assay correlates well with results obtained in an RNase protection assay for HERV-K18 transcription (52). Figure 1B shows that after infection with B95-8 virus, but not P3HR1, HERV-K18 transcription is strongly induced in BL-41 cells, confirming the functional results depicted in Fig. 1A. When EBNA-2 is provided in trans in BL-41 cells infected with P3HR1 (MCB1-9) (13), the level of HERV-K18 transcripts increases. Additional support for the hypothesis that EBNA-2 has a role in transactivating the HERV-K18 superantigen comes from an EBNA-2-conditional LCL, ER/EB2-6. This cell line is transformed with recombinant EBV in which the EBNA-2 gene was replaced with an estrogen-responsive EBNA-2 gene (32). LCL growth is dependent upon estrogen because EBNA-2 expression is required for expression of the other EBV-transforming genes. The removal of estrogen from these cells resulted in growth arrest (32) and, as can be seen in Fig. 1B, downregulation of HERV-K18 transcription. Since EBNA-2 transactivates all of the EBV latent genes, with the exception of EBNA-1, these results substantiate that a latent gene induces HERV-K18.
LMP-2A is sufficient for transactivation of HERV-K18 Env.
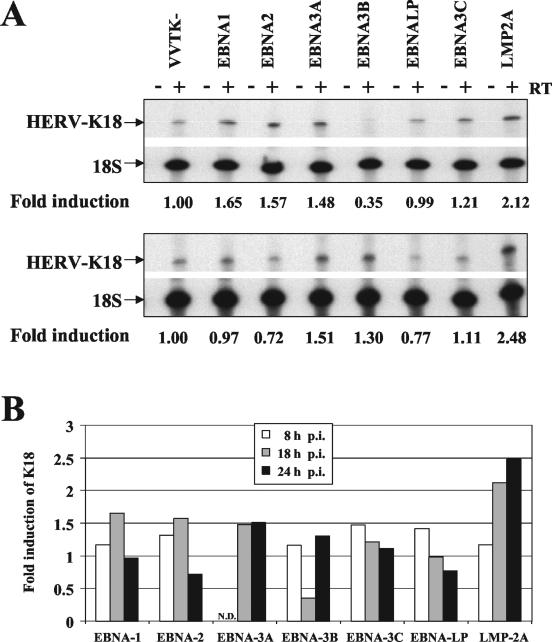
There are nine latent genes: EBNA-1, EBNA-2, EBNA-3A, EBNA-3B, EBNA-3C, EBNA-LP, LMP-1, LMP-2A, and LMP-2B. To define whether it was EBNA-2 itself or one of the other latent genes that induces HERV-K18, we obtained a panel of recombinant vaccinia viruses expressing various EBV latent genes (33, 44). We infected the EBV− BL-41 cell line with each of these recombinant viruses or a negative control virus, VVTK−, as previously described (33, 44), and we then looked for induction of HERV-K18 read-through transcripts by RT-PCR. At 8, 18, and 24 h postinfection, cells were lysed in Trizol (Invitrogen), and total RNA was isolated and subjected to RT-PCR analysis. Figure 2A shows results obtained in representative experiments after 18 h (upper panel) and 24 h (lower panel) of infection, and the severalfold induction of HERV-K18 at 8, 18, and 24 h postinfection is summarized in Fig. 2B, normalized against the VVTK− infected cells. The data indicate that only the LMP-2A virus consistently induced HERV-K18 transcripts more than twofold over time. These experiments were repeated twice, and in both cases, LMP-2A elevated HERV-K18 transcripts at 18 and 24 h.
FIG. 2.
Infection of BL-41 with LMP-2A vaccinia virus selectively induces HERV-K18 env. BL-41 cells were infected with a panel of recombinant vaccinia viruses containing different EBV latent genes, EBNA-1, EBNA-2, EBNA-3A, EBNA-3B, EBNA-3C, EBNA-LP, and LMP-2A, or control virus VVTK−. Eight, 18, or 24 h postinfection, cells were lysed, RNA was extracted, and semiquantitative RT-PCR for K18 and 18S transcripts was performed as described in the legend to Fig. 1B. (A) RT-PCR analysis at 18 h (top panel) and 24 h (bottom panel) postinfection. The ratio of HERV-K18 to 18S rRNA was measured by phosphorimager analysis, and the induction (fold) is reported below each lane. (B) Summary of RT-PCR analyses at 8, 18, and 24 h postinfection (p.i.) in one of two representative experiments. Induction (fold) of HERV-K18 was calculated by normalization with values obtained from VVTK− infected cells. N.D., not determined.
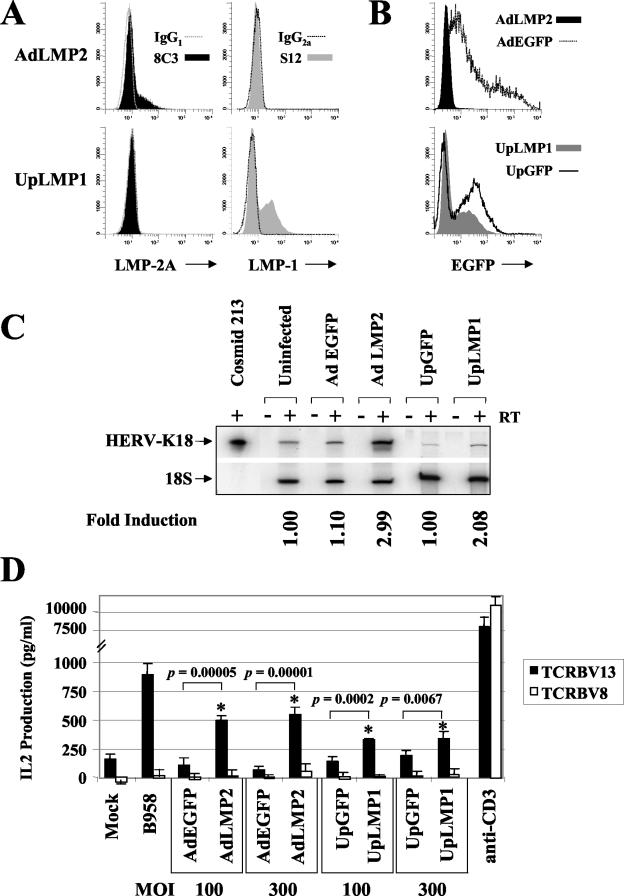
Because vaccinia virus rapidly takes over the cellular transcriptional machinery, it was necessary to confirm that LMP-2A actually transactivates HERV-K18. We did this by infecting BL-41 cells with an adenoviral vector containing LMP-2A driven by the cytomegalovirus (CMV) promoter, AdLMP2 (19). In order to test whether LMP-1 can transactivate HERV-K18, we created an adenoviral vector containing LMP-1 under control of the ubiquitin promoter, UpLMP1. The results obtained after infection of BL-41 with the LMP-2A and LMP-1 adenoviruses were compared with the results obtained after infection with their respective adenovirus vectors, AdEGFP (10) and UpGFP, which carry the enhanced green fluorescence protein (EGFP) gene driven by the CMV or the ubiquitin promoter. UpGFP virus was derived from the pSh-Up-Up-GFP shuttle vector (59, 60). UpLMP1 was created by subcloning the B95-8 LMP-1 cDNA into the KpnI-NotI sites of the polylinker of pSh-Up-Up-GFP. In this vector, LMP-1 is driven by the ubiquitin promoter, and there is an upstream ubiquitin promoter-EGFP cassette in tandem; thus, cells infected by UpLMP1 express both LMP-1 and EGFP. Recombinant adenovirus plasmids were produced by homologous recombination with the pAdEASY system (Quantum Biotechnologies) as described previously (10). Production, purification, and titering of recombinant adenoviruses was done in 293 cells as described previously (10). A total of 106 cells were infected with the adenoviral vectors at a multiplicity of infection (MOI) of 100 or 300 in 0.5 ml of medium in 24-well plates for 2 to 4 h at 37°C. Cells were then transferred to T25 flasks in 12 ml of medium and cultured for up to 96 h. At 72 or 96 h postinfection, infected cells were lysed in Trizol for RNA extraction or tested for superantigen activity in T-cell hybridoma studies.
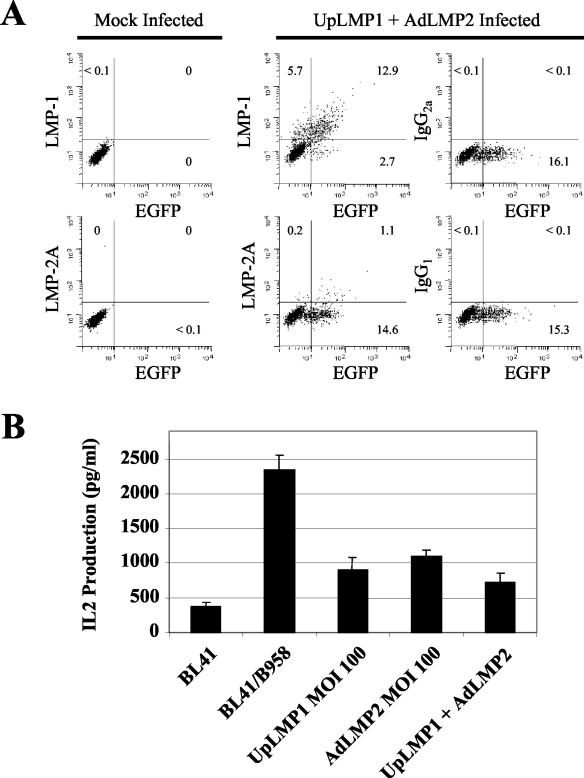
Figure 3A shows expression of LMP-2A and LMP-1 48 h after infection of BL-41 cells with AdLMP2 and UpLMP1 adenoviruses at an MOI of 100. At a higher MOI, the AdLMP2 virus appeared to have toxic effects on the infected cells, inhibiting growth while not showing any increase in specific staining of LMP-2A (19; data not shown). The cells were fixed and stained as previously described (19) with the 8C3 monoclonal antibody (MAb) specific for LMP-2A (19) or the S12 MAb specific for LMP-1 (BD Pharmingen), and each staining was compared with the isotype control MAb. Figure 3B depicts EGFP expression 48 h postinfection of BL-41 cells with each of the recombinant adenoviruses. Interestingly, while the ubiquitin promoter vectors yielded high expression of LMP-1 and EGFP in BL-41 cells, LMP-2A, which we also cloned into the ubiquitin promoter vector, was poorly expressed compared with its expression in the CMV promoter vector (data not shown). Furthermore, the expression of LMP-2A and LMP-1 after adenoviral infection of BL-41 was much higher than the expression of these genes in various LCL or B95-8-infected BL-41 (data not shown). In Fig. 3C, we demonstrate that AdLMP2 upregulates transcription of HERV-K18 read-through transcripts compared with AdEGFP, while LMP-1 upregulates HERV-K18 to a lesser extent compared with UpGFP. Figure 3D shows that after infection with AdLMP2 at MOI of 100 and 300, BL-41 cells preferentially stimulate the TCRBV13 T-cell hybridoma, similar to infection with B95-8 EBV, while infection with AdEGFP was nonstimulatory. This difference was highly significant (P = 0.00005 to 0.00001). UpLMP-1 gave a lower level stimulation that was also significantly elevated (P < 0.007) compared with UpGFP. We have performed these experiments repeatedly using various MOI between 50 and 1,000 and at various time points postinfection. Significant differences were consistently seen using MOI of 100 and 300, while an MOI of 1,000 often showed toxic effects on the cells. These results prove that LMP-2A, and to a lesser degree, LMP-1, is sufficient for induction of the HERV-K18 Env superantigen. It is possible that LMP-1 and/or other EBV latent genes act in synergy, increasing induction of HERV-K18 transcripts; however, we saw no evidence for synergy between LMP-1 and LMP-2A after coinfection of BL-41 with both adenoviral vectors (Fig. 4). In Fig. 4A, we show that both LMP-1 and LMP-2A are coexpressed after coinfection of BL-41 with the UpLMP1 and AdLMP2 viruses at an MOI of 100 for each. Figure 4B shows that superantigen activity is not increased by coinfection with both viruses at an MOI of 100. We have repeated these experiments at various time points postinfection (48 and 72 h) and using other MOI (50 and 300), but we failed to see synergy (data not shown). It is possible that the lack of synergism between the EBV latent membrane proteins in these experiments is due to the fact that both of these proteins are overexpressed compared with their levels in EBV-infected B cells. Alternatively, expression of other EBV nuclear antigens might modulate the transactivation.
FIG. 3.
Infection of BL-41 with AdLMP2 transactivates the HERV-K18 superantigen. Adenovirus vectors containing LMP-2A (AdLMP2), LMP-1 (UpLMP1), or EGFP (AdEGFP, UpGFP, and UpLMP1) were used to infect BL-41 cells at an MOI of 100. (A) AdLMP2- and UpLMP1-infected cells were stained 48 h postinfection with MAbs specific for LMP-2A (8C3) and LMP-1 (S12) or isotype-matched control antibody. (B) EGFP expression in BL-41 48 h postinfection with each adenovirus. (C) Adenovirus-infected cells were lysed 96 h postinfection and subjected to RT-PCR analysis for K18 and 18S rRNA transcripts as described in the legend to Fig. 1B. Alternatively, cells infected at an MOI of 100 or 300 were tested for their ability to stimulate the TCRBV13 and TCRBV8 T-cell hybridomas, as described in the legend to Fig. 1A. (D) The mean IL-2 production for each T-cell hybrid was measured in quadruplicate wells by enzyme-linked immunosorbent assay and expressed in picograms per milliliter of culture supernatant by comparison with values from a standard curve derived from recombinant IL-2 (R & D Systems). Error bars represent the difference measured between quadruplicate wells in one representative experiment. Experiments were performed at least three times. The response of the hybrids to BL-41/B95-8 and anti-CD3 cross-linkage was included as a positive control.
FIG. 4.
Lack of synergy between latent membrane proteins after coinfection of BL-41 with UpLMP1 and AdLMP2. (A) BL-41 cells were mock infected or coinfected with UpLMP1 and AdLMP2 at an MOI of 100 for each. Cells were stained 48 h postinfection with MAb specific for LMP-2A (8C3) and LMP-1 (S12) or isotype-matched control antibody shown on the y axis, and EGFP is shown on the x axis by flow cytometry. (B) Cells infected at an MOI of 100 were tested for superantigen activity by the TCRBV13 T-cell hybridoma, as described in the legend to Fig. 1A.
In this paper, we have demonstrated that the EBV latent membrane protein LMP-2A is sufficient for transactivation of the HERV-K18 superantigen. This superantigen induction results in TCRBV13-specific T-cell activation. It was previously shown that, in addition to EBV, the cytokine alpha interferon (IFN-α) strongly induces HERV-K18 expression in the peripheral blood (51). LMP-2A and IFN-α are not known to share signaling pathways; thus, it is puzzling that both can transactivate this particular HERV. LMP-2A is a membrane protein with intracellular domains containing immunoreceptor tyrosine-based activation motifs (ITAMs). Studies from LMP-2A transgenic mice suggest that LMP-2A signaling mimics B-cell antigen receptor (BCR) signal transduction. In these mice, LMP-2A+ B cells lacking surface Ig exit the bone marrow and enter the circulation. Since Ig-negative B cells normally do not survive, these data indicate that LMP-2A signaling rescues the cells (5, 6). In EBV-infected B cells, the tyrosine kinases Syk and Lyn bind to the phosphorylated ITAMs on LMP-2A through their SH2 domains. In this manner, they are sequestered away from the BCR, resulting in phospholipase C-γ2 activation (5, 6, 38, 40, 41). Mutation of the Syk-binding ITAM on LMP-2A abolishes the B-cell survival signal in the mice (40). In addition, it was shown that LMP-2A signals mediate tyrosine phosphorylation of the SH2-containing adaptor protein SLP-65, leading to complex formation with the adaptor protein CrkL and ultimately resulting in phosphorylation of Cbl and C3G (16). IFN-α signaling, on the other hand, is well known to occur through the JAK-STAT pathway (reviewed in reference 4). It has been reported that IFN-α binding to its receptor activates the Janus family kinase Tyk-2, signaling Lyn to bind through its SH2 domain to the phosphorylated ITAM on Tyk-2 (57). Thus, the tyrosine kinase Lyn appears in both the IFN-α receptor pathway and the LMP-2A and BCR signaling pathways, suggesting a possible link. In addition, while STAT1 and STAT2 proteins have long been considered requisites for type I IFN receptor signaling, recent data suggest that STAT5 is also important (17, 58). Furthermore, it was shown that the CrkL adaptor protein is required for IFN-α-dependent gene transcription, presumably via CrkL complex formation with STAT5, which allows DNA binding to gamma-activated sites (GAS) (37, 58). Thus, CrkL is activated in both LMP-2A and IFN-α signaling pathways, implying a possible role of this adaptor in the induction of HERV-K18.
Our data indicate that LMP-1 transactivates HERV-K18 less efficiently than does LMP-2A. LMP-1 is known to activate the mitogen-activated protein kinase and NF-κB pathways (reviewed in reference 15). Interestingly, there is an NF-κB site in a designated EBV-inducible enhancer sequence upstream of the CD48 promoter on chromosome 1, within easy distance of HERV-K18, which resides in the first CD48 intron (23). It has been postulated that the CD48 enhancer may boost transcription of HERV-K18 (52). Moreover, recent evidence suggests that, like IFN-α, LMP-1 also signals through the JAK-STAT pathway, phosphorylating both STAT3 and STAT5 in different cell types (8, 9). In EBV-infected B cells, LMP-1 and LMP-2A are frequently coexpressed, often in the presence of other EBV transcription factors. While we have shown here that LMP-2A by itself is sufficient to transactivate the HERV-K18 superantigen in EBV− Burkitt's lymphoma cells, it is likely that in the complex situation of EBV infection where multiple viral genes are expressed, other genes, such as LMP-1, modulate transcription of the HERV.
In this paper, we have shown that the EBV latent genes transactivate a host cell superantigen. In terms of EBV biology, we propose that EBV elicits superantigen-activated T cells to supply requisite signals to the EBV-infected B cells, allowing them to differentiate into memory cells, the site of long-term viral persistence in the host. It is well known that B-cell differentiation to the memory stage is T cell dependent; therefore, we postulate that EBV induces the superantigen to facilitate entry into the memory B-cell compartment, where it can persist for the lifetime of the healthy host. In the immunosuppressed host, superantigen-activated T cells might instead cause viral reactivation and/or enhanced survival of EBV+ tumor cells.
Acknowledgments
This work was supported by the NIH (R37 AI14910), an NCI Howard Temin Award (K01 CA095443), the Eshe Foundation, the Natalie V. Zucker Women's Scholar Fund, the Erle P. Charlton Research Fund, and a Center grant to GRASP.
We are most grateful to David Thorley-Lawson, Bettina Kempkes, Georg Bornkamm, Gilbert Lenoir, Richard Longnecker, and Erle Robertson for constructive scientific advice and donation of EBV cell lines. We thank Denis Moss for the panel of recombinant vaccinia viruses and Premlata Shankar for help in amplifying and titering the vaccinia viruses. We are grateful to Cliona Rooney for the LMP-2A adenoviral vector, James DeGregori for the UpGFP adenoviral vector, and Wei Li for the AdEGFP vector and for advice on adenovirus technology. In addition, we thank Elisabeth Kremmer for the LMP-2A MAb. We also thank Philippa Marrack and Rafick-Pierre Sekaly for the TCRBV T-cell hybridomas.
REFERENCES
- 1.Allday, M. J., D. H. Crawford, and B. E. Griffin. 1989. Epstein-Barr virus latent gene expression during the initiation of B cell immortalization. J. Gen. Virol. 70:1755-1764. [DOI] [PubMed] [Google Scholar]
- 2.Baichwal, V. R., and B. Sugden. 1988. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene 2:461-467. [PubMed] [Google Scholar]
- 3.Bornkamm, G. W., J. Hudewentz, U. K. Freese, and U. Zimber. 1982. Deletion of the nontransforming Epstein-Barr virus strain P3HR-1 causes fusion of the large internal repeat to the DSL region. J. Virol. 43:952-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brierley, M. M., and E. N. Fish. 2002. Review: IFN-alpha/beta receptor interactions to biologic outcomes: understanding the circuitry. J. Interferon Cytokine Res. 22:835-845. [DOI] [PubMed] [Google Scholar]
- 5.Caldwell, R. G., R. C. Brown, and R. Longnecker. 2000. Epstein-Barr virus LMP2A-induced B-cell survival in two unique classes of EmuLMP2A transgenic mice. J. Virol. 74:1101-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldwell, R. G., J. B. Wilson, S. J. Anderson, and R. Longnecker. 1998. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity 9:405-411. [DOI] [PubMed] [Google Scholar]
- 7.Calender, A., M. Billaud, J. P. Aubry, J. Banchereau, M. Vuillaume, and G. M. Lenoir. 1987. Epstein-Barr virus (EBV) induces expression of B-cell activation markers on in vitro infection of EBV-negative B-lymphoma cells. Proc. Natl. Acad. Sci. USA 84:8060-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, H., L. Hutt-Fletcher, L. Cao, and S. D. Hayward. 2003. A positive autoregulatory loop of LMP1 expression and STAT activation in epithelial cells latently infected with Epstein-Barr virus. J. Virol. 77:4139-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, H., J. M. Lee, Y. Zong, M. Borowitz, M. H. Ng, R. F. Ambinder, and S. D. Hayward. 2001. Linkage between STAT regulation and Epstein-Barr virus gene expression in tumors. J. Virol. 75:2929-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J., B. T. Huber, R. J. Grand, and W. Li. 2001. Recombinant adenovirus coexpressing covalent peptide/MHC class II complex and B7-1: in vitro and in vivo activation of myelin basic protein-specific T cells. J. Immunol. 167:1297-1305. [DOI] [PubMed] [Google Scholar]
- 11.Choi, Y. W., A. Herman, D. DiGiusto, T. Wade, P. Marrack, and J. Kappler. 1990. Residues of the variable region of the T-cell-receptor β-chain that interact with S. aureus toxin superantigens. Nature 346:471-473. [DOI] [PubMed] [Google Scholar]
- 12.Choi, Y. W., B. Kotzin, J. Lafferty, J. White, M. Pigeon, R. Kubo, J. Kappler, and P. Marrack. 1991. A method for production of antibodies to human T-cell receptor beta-chain variable regions. Proc. Natl. Acad. Sci. USA 88:8357-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordier, M., A. Calender, M. Billaud, U. Zimber, G. Rousselet, O. Pavlish, J. Banchereau, T. Tursz, G. Bornkamm, and G. M. Lenoir. 1990. Stable transfection of Epstein-Barr virus (EBV) nuclear antigen 2 in lymphoma cells containing the EBV P3HR1 genome induces expression of B-cell activation molecules CD21 and CD23. J. Virol. 64:1002-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyson, P. J., A. M. Knight, S. Fairchild, E. Simpson, and K. Tomonari. 1991. Genes encoding ligands for deletion of V beta 11 T cells cosegregate with mammary tumour virus genomes. Nature 349:531-532. [DOI] [PubMed] [Google Scholar]
- 15.Eliopoulos, A. G., and L. S. Young. 2001. LMP1 structure and signal transduction. Semin. Cancer Biol. 11:435-444. [DOI] [PubMed] [Google Scholar]
- 16.Engels, N., M. Merchant, R. Pappu, A. C. Chan, R. Longnecker, and J. Wienands. 2001. Epstein-Barr virus latent membrane protein 2A (LMP2A) employs the SLP-65 signaling module. J. Exp. Med. 194:255-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson, S., S. Matikainen, L. Thyrell, O. Sangfelt, I. Julkunen, S. Einhorn, and D. Grander. 2002. Interferon-alpha inhibits Stat5 DNA-binding in IL-2 stimulated primary T-lymphocytes. Eur. J. Biochem. 269:29-37. [DOI] [PubMed] [Google Scholar]
- 18.Frankel, W. N., C. Rudy, J. M. Coffin, and B. T. Huber. 1991. Linkage of Mls genes to endogenous mammary tumour viruses of inbred mice. Nature 349:526-528. [DOI] [PubMed] [Google Scholar]
- 19.Gahn, B., F. Siller-Lopez, A. D. Pirooz, E. Yvon, S. Gottschalk, R. Longnecker, M. K. Brenner, H. E. Heslop, E. Aguilar-Cordova, and C. M. Rooney. 2001. Adenoviral gene transfer into dendritic cells efficiently amplifies the immune response to LMP2A antigen: a potential treatment strategy for Epstein-Barr virus-positive Hodgkin's lymphoma. Int. J. Cancer 93:706-713. [DOI] [PubMed] [Google Scholar]
- 20.Hammarskjold, M. L., and M. C. Simurda. 1992. Epstein-Barr virus latent membrane protein transactivates the human immunodeficiency virus type 1 long terminal repeat through induction of NF-κB activity. J. Virol. 66:6496-6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammerschmidt, W., and B. Sugden. 1989. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature 340:393-397. [DOI] [PubMed] [Google Scholar]
- 22.Harada, S., and E. Kieff. 1997. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 71:6611-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasuike, S., K. Miura, O. Miyoshi, T. Miyamoto, N. Niikawa, Y. Jinno, and M. Ishikawa. 1999. Isolation and localization of an IDDMK1,2-22-related human endogenous retroviral gene, and identification of a CA repeat marker at its locus. J. Hum. Genet. 44:343-347. [DOI] [PubMed] [Google Scholar]
- 24.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science 265:92-95. [DOI] [PubMed] [Google Scholar]
- 25.Herman, A., J. W. Kappler, P. Marrack, and A. M. Pullen. 1991. Superantigens: mechanism of T-cell stimulation and role in immune responses. Annu. Rev. Immunol. 9:745-772. [DOI] [PubMed] [Google Scholar]
- 26.Heston, L., M. Rabson, N. Brown, and G. Miller. 1982. New Epstein-Barr virus variants from cellular subclones of P3J-HR-1 Burkitt lymphoma. Nature 295:160-163. [DOI] [PubMed] [Google Scholar]
- 27.Huber, B. T., P. N. Hsu, and N. Sutkowski. 1996. Virus-encoded superantigens. Microbiol. Rev. 60:473-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes, J. F., and J. M. Coffin. 2001. Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat. Genet. 29:487-489. [DOI] [PubMed] [Google Scholar]
- 29.Jardetzky, T. S., J. H. Brown, J. C. Gorga, L. J. Stern, R. G. Urban, Y. Chi, C. Stauffacher, J. L. Strominger, and D. C. Wiley. 1994. Three-dimensional structure of a human class II histocompatibility molecule complexed with superantigen. Nature 368:711-718. [DOI] [PubMed] [Google Scholar]
- 30.Jochner, N., D. Eick, U. Zimber-Strobl, M. Pawlita, G. W. Bornkamm, and B. Kempkes. 1996. Epstein-Barr virus nuclear antigen 2 is a transcriptional suppressor of the immunoglobulin mu gene: implications for the expression of the translocated c-myc gene in Burkitt's lymphoma cells. EMBO J. 15:375-382. [PMC free article] [PubMed] [Google Scholar]
- 31.Johannsen, E., E. Koh, G. Mosialos, X. Tong, E. Kieff, and S. R. Grossman. 1995. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J. Virol. 69:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kempkes, B., D. Pich, R. Zeidler, and W. Hammerschmidt. 1995. Immortalization of human primary B lymphocytes in vitro with DNA. Proc. Natl. Acad. Sci. USA 92:5875-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khanna, R., S. R. Burrows, M. G. Kurilla, C. A. Jacob, I. S. Misko, T. B. Sculley, E. Kieff, and D. J. Moss. 1992. Localization of Epstein-Barr virus cytotoxic T cell epitopes using recombinant vaccinia: implications for vaccine development. J. Exp. Med. 176:169-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozono, H., D. Parker, J. White, P. Marrack, and J. Kappler. 1995. Multiple binding sites for bacterial superantigens on soluble class II MHC molecules. Immunity 3:187-196. [DOI] [PubMed] [Google Scholar]
- 35.Laherty, C. D., H. M. Hu, A. W. Opipari, F. Wang, and V. M. Dixit. 1992. The Epstein-Barr virus LMP1 gene product induces A20 zinc finger protein expression by activating nuclear factor kappa B. J. Biol. Chem. 267:24157-24160. [PubMed] [Google Scholar]
- 36.Laux, G., B. Adam, L. J. Strobl, and F. Moreau-Gachelin. 1994. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-J kappa interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 13:5624-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lekmine, F., A. Sassano, S. Uddin, B. Majchrzak, O. Miura, B. J. Druker, E. N. Fish, A. Imamoto, and L. C. Platanias. 2002. The CrkL adapter protein is required for type I interferon-dependent gene transcription and activation of the small G-protein Rap1. Biochem. Biophys. Res. Commun. 291:744-750. [DOI] [PubMed] [Google Scholar]
- 38.Longnecker, R., and C. L. Miller. 1996. Regulation of Epstein-Barr virus latency by latent membrane protein 2. Trends Microbiol. 4:38-42. [DOI] [PubMed] [Google Scholar]
- 39.Marrack, P., E. Kushnir, and J. Kappler. 1991. A maternally inherited superantigen encoded by a mammary tumour virus. Nature 349:524-526. [DOI] [PubMed] [Google Scholar]
- 40.Merchant, M., R. G. Caldwell, and R. Longnecker. 2000. The LMP2A ITAM is essential for providing B cells with development and survival signals in vivo. J. Virol. 74:9115-9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, C. L., J. H. Lee, E. Kieff, A. L. Burkhardt, J. B. Bolen, and R. Longnecker. 1994. Epstein-Barr virus protein LMP2A regulates reactivation from latency by negatively regulating tyrosine kinases involved in sIg-mediated signal transduction. Infect. Agents Dis. 3:128-136. [PubMed] [Google Scholar]
- 42.Miller, G., J. Robinson, L. Heston, and M. Lipman. 1974. Differences between laboratory strains of Epstein-Barr virus based on immortalization, abortive infection, and interference. Proc. Natl. Acad. Sci. USA 71:4006-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moorthy, R. K., and D. A. Thorley-Lawson. 1993. All three domains of the Epstein-Barr virus-encoded latent membrane protein LMP-1 are required for transformation of Rat-1 fibroblasts. J. Virol. 67:1638-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murray, R. J., M. G. Kurilla, H. M. Griffin, J. M. Brooks, M. Mackett, J. R. Arrand, M. Rowe, S. R. Burrows, D. J. Moss, E. Kieff, et al. 1990. Human cytotoxic T-cell responses against Epstein-Barr virus nuclear antigens demonstrated by using recombinant vaccinia viruses. Proc. Natl. Acad. Sci. USA 87:2906-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murray, R. J., L. S. Young, A. Calender, C. D. Gregory, M. Rowe, G. M. Lenoir, and A. B. Rickinson. 1988. Different patterns of Epstein-Barr virus gene expression and of cytotoxic T-cell recognition in B-cell lines infected with transforming (B95.8) or nontransforming (P3HR1) virus strains. J. Virol. 62:894-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nitsche, F., A. Bell, and A. Rickinson. 1997. Epstein-Barr virus leader protein enhances EBNA-2-mediated transactivation of latent membrane protein 1 expression: a role for the W1W2 repeat domain. J. Virol. 71:6619-6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Connor, M., M. Peifer, and W. Bender. 1989. Construction of large DNA segments in Escherichia coli. Science 244:1307-1312. [DOI] [PubMed] [Google Scholar]
- 48.Ono, M., T. Yasunaga, T. Miyata, and H. Ushikubo. 1986. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J. Virol. 60:589-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robertson, E., and E. Kieff. 1995. Reducing the complexity of the transforming Epstein-Barr virus genome to 64 kilobase pairs. J. Virol. 69:983-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rooney, C., J. G. Howe, S. H. Speck, and G. Miller. 1989. Influence of Burkitt's lymphoma and primary B cells on latent gene expression by the nonimmortalizing P3J-HR-1 strain of Epstein-Barr virus. J. Virol. 63:1531-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stauffer, Y., S. Marguerat, F. Meylan, C. Ucla, N. Sutkowski, B. Huber, T. Pelet, and B. Conrad. 2001. Interferon-alpha-induced endogenous superantigen. A model linking environment and autoimmunity. Immunity 15:591-601. [DOI] [PubMed] [Google Scholar]
- 52.Sutkowski, N., B. Conrad, D. A. Thorley-Lawson, and B. T. Huber. 2001. Epstein-Barr virus transactivates the human endogenous retrovirus HERV-K18 that encodes a superantigen. Immunity 15:579-589. [DOI] [PubMed] [Google Scholar]
- 53.Sutkowski, N., T. Palkama, C. Ciurli, R. P. Sekaly, D. A. Thorley-Lawson, and B. T. Huber. 1996. An Epstein-Barr virus-associated superantigen. J. Exp. Med. 184:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swain, A., and J. M. Coffin. 1992. Mechanism of transduction by retroviruses. Science 255:841-845. [DOI] [PubMed] [Google Scholar]
- 55.Swain, A., and J. M. Coffin. 1989. Polyadenylation at correct sites in genome RNA is not required for retrovirus replication or genome encapsidation. J. Virol. 63:3301-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomkinson, B., E. Robertson, R. Yalamanchili, R. Longnecker, and E. Kieff. 1993. Epstein-Barr virus recombinants from overlapping cosmid fragments. J. Virol. 67:7298-7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uddin, S., I. M. Grumbach, T. Yi, O. R. Colamonici, and L. C. Platanias. 1998. Interferon alpha activates the tyrosine kinase Lyn in haemopoietic cells. Br. J. Haematol. 101:446-449. [DOI] [PubMed] [Google Scholar]
- 58.Uddin, S., F. Lekmine, A. Sassano, H. Rui, E. N. Fish, and L. C. Platanias. 2003. Role of Stat5 in type I interferon-signaling and transcriptional regulation. Biochem. Biophys. Res. Commun. 308:325-330. [DOI] [PubMed] [Google Scholar]
- 59.Wan, Y. Y., and J. DeGregori. 2003. The survival of antigen-stimulated T cells requires NFkappaB-mediated inhibition of p73 expression. Immunity 18:331-342. [DOI] [PubMed] [Google Scholar]
- 60.Wan, Y. Y., R. P. Leon, R. Marks, C. M. Cham, J. Schaack, T. F. Gajewski, and J. DeGregori. 2000. Transgenic expression of the coxsackie/adenovirus receptor enables adenoviral-mediated gene delivery in naive T cells. Proc. Natl. Acad. Sci. USA 97:13784-13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, D., D. Liebowitz, and E. Kieff. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831-840. [DOI] [PubMed] [Google Scholar]
- 62.Wang, F., C. Gregory, C. Sample, M. Rowe, D. Liebowitz, R. Murray, A. Rickinson, and E. Kieff. 1990. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J. Virol. 64:2309-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woodland, D. L., F. E. Lund, M. P. Happ, M. A. Blackman, E. Palmer, and R. B. Corley. 1991. Endogenous superantigen expression is controlled by mouse mammary tumor proviral loci. J. Exp. Med. 174:1255-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zimber-Strobl, U., L. J. Strobl, C. Meitinger, R. Hinrichs, T. Sakai, T. Furukawa, T. Honjo, and G. W. Bornkamm. 1994. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 13:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]