Abstract
Many viral proteins limit host immune defenses, and their genes often originate from their hosts. CD200 (OX2) is a broadly distributed cell surface glycoprotein that interacts with a receptor on myeloid cells (CD200R) that is implicated in locally preventing macrophage activation. Distant, but recognizable, homologues of CD200 have been identified in many herpesviruses and poxviruses. Here, we show that the product of the K14 open reading frame from human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) interacts with human CD200R and is expressed at the surfaces of infected cells solely during the lytic cycle. Despite sharing only 40% primary sequence identity, K14 and CD200 interacted with CD200R with an almost identical and low affinity (KD = 0.5 μM), in contrast to other characterized viral homologue interactions. Cells expressing CD200 or K14 on the cell surface were able to inhibit secretion by activated macrophages of proinflammatory cytokines such as tumor necrosis factor alpha, an effect that could be specifically relieved by addition of monoclonal antibodies and soluble monomeric CD200 protein. We conclude that CD200 delivers local down-modulatory signals to myeloid cells through direct cell-cell contact and that the K14 viral homologue closely mimics this.
The immune system has evolved complex mechanisms to provide appropriate responses against rapidly evolving pathogens while controlling these responses to prevent damage to self. This is reflected in the large number of cell surface and secreted proteins involved in the fine control of immunological responses. One recently characterized interaction is that between the CD200 (OX2) membrane protein, which is expressed on a variety of cells, including activated T cells, B cells, follicular dendritic cells, neurons, and vascular endothelium (2, 7, 28, 41, 43), and its receptor (CD200R), which is restricted mainly to myeloid cells (42, 44). CD200 contains two extracellular immunoglobulin superfamily (IgSF) domains, a hydrophobic transmembrane sequence and a short cytoplasmic region that is devoid of any known signaling motifs (3, 12). CD200R also contains two extracellular IgSF domains but differs in that it has a substantial cytoplasmic region that contains tyrosine-based motifs that can be phosphorylated, suggesting that it is capable of signaling events within myeloid cells (44). In support of these biochemical data, the CD200 knockout mouse had changes in the myeloid compartment in tissues that normally expressed CD200 and its receptor. These included an increase in number and state of activation of myeloid cells in several tissues, as well as an increase in susceptibility to autoimmune disease models (19). In addition, CD200-Fc fusion proteins gave beneficial immunomodulatory effects in models of arthritis and allograft rejection (16, 17). Collectively, these data strongly indicate that the CD200-CD200R interaction is involved in the negative or restrictive control of myeloid cellular function in both healthy and disease states. Recent distribution data have shown that both the human and mouse CD200R are also expressed in T-cell subsets, indicating that some of these immunosuppressive effects may act through T cells (42).
Viruses of the Herpesviridae and Poxviridae families are highly adapted and successful pathogens that are characterized by asymptomatic lifelong infections in large immunocompetent populations. These viruses have large double-stranded genomes (150 to 200 kbp) and have evolved elaborate mechanisms to subvert the regulation of the host's immune system. These mechanisms include the down-regulation of MHC antigen expression and consequent inhibition of cytotoxic T-cell activity, production of cytokine and cytokine receptor homologues, and interference with programmed cell death (13, 33, 35, 39). Interestingly, many viral open reading frames (ORFs) have clear similarities with known host proteins and are therefore of cellular origin. Some of these ORFs have been shown to play an important role in controlling host immune responses to the virus, which must retain only those ORFs that lead to an increase in viral evolutionary fitness. CD200-like sequences have been identified in the genomes of several evolutionarily diverse viruses (Fig. 1), which include gamma- and betaherpesviruses and both yata- and leporipoxviruses. Recently, a CD200-like sequence has been reported for duck adenovirus. This virus has a smaller genome (33 kb) with fewer genes than the other viruses that contain CD200 homologues (Fig. 1). The capture of the host CD200 gene appears to have occurred independently in distinct viral families, indicating that it provides a strong selective advantage in viruses that have widely varying biologies and pathologies (25). Our working model, that the CD200-CD200R interaction provides restrictive local control of myeloid cells, fit with the idea that virally infected cells might also be able to down-regulate the activity of myeloid lineage cells via CD200R by displaying CD200-like proteins at the surfaces of infected cells.
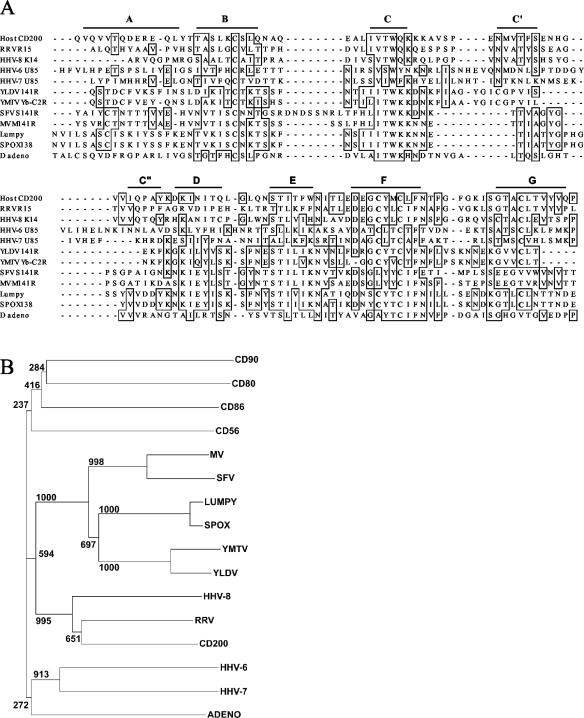
FIG. 1.
Alignment and phylogenetic analysis of CD200 and viral CD200-like ORFs. (A) Alignment of the predicted Ig V-like domains of CD200 and viral CD200 homologues. The sequences shown (with GenBank [unless stated] accession numbers) are as follows: human CD200 (Swissprot P41217), R15 from rhesus macaque rhadinovirus (RRV) (AF083501), K14 from HHV-8 (U75698), U85 from HHV-6 (X83413), U85 from HHV-7 (U43400), 141R from Yaba-like disease virus (YLDV) (NP_073526), Yb-C2R from Yaba monkey tumor virus (YMTV) (AB025319), LSDV138 from lumpy skin disease virus (Lumpy) (NP_150572), S141R from Shope fibroma virus (SFV) (AF170722), M141R from myxomavirus (MV) strain Lausanne (AF170726); sheep pox virus (SPOX) ORF138 (NP_659708), and duck adenovirus (D adeno) ORF4 (DAA00544). The bars predict the extent of the beta-strands characteristic of the Ig fold by comparison to solved structures. The cysteines that are thought to form an unusual F-to-G strand disulfide are in boldface, and residues identical in four or more sequences are boxed. (B) The N-terminal IgSF domains of the viral sequences in panel A, indicated by the virus name, were aligned to the N-terminal domains of human CD56, CD80, CD90, and CD200 by using ClustalW (38) before manual refinement and then were trimmed to remove the widely variant A strands. A neighbor-joining tree was constructed by using 1,000 bootstrap trials.
Human herpesvirus 8 (HHV-8) belongs to the Gammaherpesviridae family of herpesviruses and has been etiologically linked to three distinct neoplasms: Kaposi's sarcoma, multicentric Castleman's disease, and a primary effusion lymphoma (18). Genome sequencing of HHV-8 revealed that it carried an ORF, K14, which was homologous to CD200 yet had only 40% sequence identity. Here, we identify the host K14 receptor as CD200R. By biochemically quantifying the interaction, we show that it binds the host CD200R with a low affinity (KD = 0.5 μM) and with binding kinetics that are indistinguishable from those of the host CD200. We also show that cells expressing cell surface host CD200 or K14 viral protein can restrict tumor necrosis factor alpha (TNF-α) production by activated macrophages and that this inhibition can be specifically relieved by either blocking monoclonal antibodies (MAbs) or a soluble monomeric (non-cross-linking) CD200 protein. These results contrast with a recent paper by Chung et al. (11), who reported that the HHV-8 protein K14 was able to deliver activating, proinflammatory signals to cells of the myeloid lineage through an uncharacterized receptor and thereby promote dissemination of the virus throughout the infected host. We suggest an explanation for the differences and conclude that the K14 protein, despite the low sequence similarity to CD200, has perfectly retained its biochemical binding properties for the CD200 receptor in order to directly mimic CD200 function and thereby deliver a localized down-regulatory signal to host myeloid cells.
MATERIALS AND METHODS
Cell lines and staining reagents.
The BC-3 cell line was grown in RPMI 1640 containing 20% fetal calf serum and supplemented with antibiotics. HHV-8 replication was induced by addition of sodium butyrate (2 mM). Debris was removed from all cultures by density gradient centrifugation on Ficoll-Hypaque (Pharmacia) and washing once in phosphate-buffered saline prior to MAb or bead staining. The MAbs (all mouse) used were OX104 (anti-human CD200, immunoglobulin G1 [IgG1]) (43), OX1 (anti-rat CD45, IgG1) (referenced in European Collection of Cell Cultures [http://www.ecacc.org/]), and OX112 (anti-HHV-8 K14, IgG1) (this study). In order to detect weak interactions at the cell surface, the soluble recombinant CD200R and CD200 proteins were made multivalent by coupling to fluorescent beads either through MAbs recognizing the CD4 part of the chimeric recombinant proteins (see below) (31) or by including a sequence in the chimeric protein that allows a biotin moiety to be added enzymatically (5).
Construction, expression, and purification of soluble fusion proteins.
The K14 ORF sequence predicts a protein of 348 amino acids, which is considerably longer than CD200 (278 amino acids) or the rhadinovirus homologue (253 amino acids). A second Met in the K14 ORF seems more likely to be used as the initiator Met, giving a protein of 271 amino acids with a typical signal-like sequence. This Met was used in the design of the construct to produce the recombinant protein (nucleotide A128116 in the sequence under GenBank accession number U75698). The sequence corresponding to the two IgSF domains was amplified from the K14 ORF from the BCP-1 cell line cloned into pGEM-T vector as a template (kindly provided by Chris Boshoff), using oligonucleotides 5′CGAGTGCCTCTAGAGGGACCATGTCTAGCCTCTTCATTTCATTACC (sense) and 5′CCTGGGTCGACGCAAGGTCATGGGCCAAGGGGC (antisense) (underlining indicates restriction sites). The products were digested with XbaI and SalI and ligated in the pEF-BOS vector to produce a chimeric protein with rat CD4 domains 3 and 4 (4). The construct sequence therefore starts with MSSLFI and ends with LAHDLASTSIT (the CD4 linker is in boldface). This construct was then subcloned into the expression vector pEE14, and a stably secreting CHO.K1 cell line was established. K14CD4d3 + 4 was purified from spent tissue culture medium by immunoaffinity chromatography with an OX68 MAb-Sepharose 4B column that recognizes the CD4 protein tag (4). The human CD200CD4d3 + 4 protein was prepared in an identical fashion and was described previously (43). Prior to BIAcore analysis, the purified CD200 and K14 soluble chimeric proteins were fractionated by gel filtration on Superdex S-200 (Pharmacia, Uppsala, Sweden) to exclude protein aggregates that are known to influence binding measurements (40). The extinction coefficients of both proteins were determined by amino acid analysis and were 40,534 M−1 cm−1 (human CD200CD4d3 + 4) and 51,304 M−1 cm−1 (K14CD4d3 + 4). The minimal fraction of each purified protein that was able to bind the CD200R was determined by depletion with avidin-Sepharose agarose beads (Sigma) coated with biotinylated rat CD200RCD4d3 + 4 compared to biotinylated CD4d3 + 4. Depleted and control fractions were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and densitometrically analyzed by using ImageQuant software. At least 90% of CD200 and 33% of the K14 proteins could be depleted by CD200R, and active protein concentrations were calculated by taking this into account. The soluble biotinylated forms of rat, mouse, and human CD200R were produced as described previously (42, 44).
Measurement of affinities by surface plasmon resonance.
Affinity and kinetic data were collected at 37°C as described previously (42, 44).
Production of MAbs that recognize the K14 ORF product.
BALB/c mice were immunized subcutaneously with 20 μg of purified K14CD4d3 + 4, initially in complete Freund's adjuvant and subsequently in incomplete Freund's adjuvant, and hybridomas were obtained by fusion with the NS-1 cell line according to standard procedures. Hybridoma supernatants were screened by flow cytometry on BC-3 cells incubated in 2 mM sodium butyrate for 36 h to induce the expression of K14. One hybridoma was selected, cloned, isotyped as a mouse IgG1, and named OX112.
Cloning and cell surface expression of full-length human CD200 and K14 proteins.
Full-length CD200 was amplified by PCR with oligonucleotides 5′GGATTCTAGAGGAGCAAGGATGGAG (sense) and 5′CGCGGATCCTTAGGGCTCTCGGTCCTGATT (antisense) (underlining indicates restriction sites) and IMAGE clone (reference 5299899; United Kingdom HGMP Resource Centre, Cambridge, United Kingdom) as a template. Full-length K14 was amplified by PCR from pGEM-T vector containing the K14 ORF (kindly provided by Chris Boshoff) by using oligonucleotides 5′CCGAGTGCCTCTAGAGGGACCATGTCTAGCCTCTTCATTTCATTACC (sense) and 5′TAGTAGGGATCCTCACTGGGTGGATAGGGGGT (antisense) (underlining indicates restriction sites). The product was digested with XbaI and BamHI and ligated into the expression vector pEE14 previously digested with XbaI and BclI. CHO-K1 cells were transfected by using Fugene transfection reagent (Roche, Mannheim, Germany) according to the manufacturer's instructions. Transfected cells were stained with OX104 (CD200) and OX112 (K14) and selected on a fluorescence-activated cell sorter (MoFlo; Cytomation, Fort Collins, Colo.). A CHO-K1 cell line transfected with pEE14 vector was also prepared similarly for use as a negative control in functional assays.
Isolation of human monocytes by density gradient and adherence.
Human monocytes were purified as described previously (23). Briefly, buffy coats from healthy donors were obtained from the National Blood Centre (Bristol, United Kingdom). Peripheral blood mononuclear cells were prepared by density gradient centrifugation on Ficoll Paque PLUS (Amersham Biosciences AB, Uppsala, Sweden). Purified peripheral blood mononuclear cells were resuspended in adhesion medium (X-VIVO 10; Bio-Whittaker, Walkersville, Md.) (4% autologous plasma), and monocytes were allowed to adhere to bacteriological plastic petri dishes coated with 2% endotoxin-free gelatin (Sigma) for 90 min at 37°C. Nonadherent cells were removed by washing five times with warm RPMI, and monocytes were lifted on the following day by repeatedly pipetting the medium onto the dish surface. Remaining monocytes were lifted by incubation with phosphate-buffered saline-EDTA (5 mM) for 10 min at 37°C and gentle pipetting. The monocytes were typically 95% CD14 positive. Expression of CD200R was tested by flow cytometry with a CD200R-specific MAb (OX108) and CD200-coated beads (42).
Detection of cytokines.
Purified monocytes (7.5 × 105/ml) were cultivated in X-VIVO 10 supplemented with 4% autologous plasma in 24-well plates at 37°C for 5 to 7 days, allowing them to mature into macrophages. CHO-CD200, CHO-K14, or CHO-pEE14 cells were irradiated (1,450 rads) prior to addition (7.5 × 105/ml) to macrophages simultaneously stimulated with gamma interferon (IFN-γ) (200U/ml) and lipopolysaccharide (LPS) (20 ng/ml) in the presence or absence of CD200CD4d3 + 4 recombinant protein (4 μM). Supernatants were collected after 20 h and tested for TNF-α by enzyme-linked immunosorbent assay (PharMingen, Oxford, United Kingdom). In selected experiments a panel of 14 cytokines was analyzed by flow cytometry with the Bio-Plex system (Bio-Rad Laboratories) and a Luminex 100 apparatus. Cytokines assayed were interleukin 1β (IL-1β), IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-13, IL-17, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor, monocyte chemoattractant protein 1 (MCP-1), macrophage inflammatory protein 1β, and TNF-α.
RESULTS
CD200 homologues are found in Herpesviridae and Poxviridae.
A variety of ORFs from several disparate viruses, including herpesviruses, poxviruses, and an adenovirus, all contain regions with amino acid sequence similarity to the membrane distal IgSF receptor binding domain (31) of human CD200 (Fig. 1A). The viral ORFs contain the sequence patterns typical of the IgSF, including the canonical disulfide that links beta strands B to F (3). Importantly, several of the virus ORFs contained two cysteine residues that are predicted to form a novel disulfide bond between beta strands F and G (Fig. 1A). This unusual cysteine pattern is highly characteristic of the CD200 protein family but was absent from the CD200-like ORFs from Shope fibroma virus, myxomavirus, and the adenovirus. As more viral genomes are sequenced, a significant difference is apparent between the CD200-like proteins from the two viral families: within the Herpesviridae they code for proteins with two extracellular IgSF domains, a transmembrane region, and a short cytoplasmic segment, whereas within the Poxviridae and duck adenovirus, they encode a secreted protein with a single IgSF domain and no transmembrane region.
In order to quantify the similarities of the potential viral homologues to CD200, their N-terminal domains were compared to those of CD200 and three other leukocyte proteins by constructing a neighbor-joining tree (Fig. 1B). The herpesvirus sequences clustered with CD200 even though their extracellular regions show only about 40% identities with CD200. The poxviruses formed a cluster, and the leukocyte proteins other than CD200 formed a separate cluster from CD200 and the viral homologues.
HHV-8 K14 protein binds human CD200R with kinetics indistinguishable from those of the host CD200 ligand.
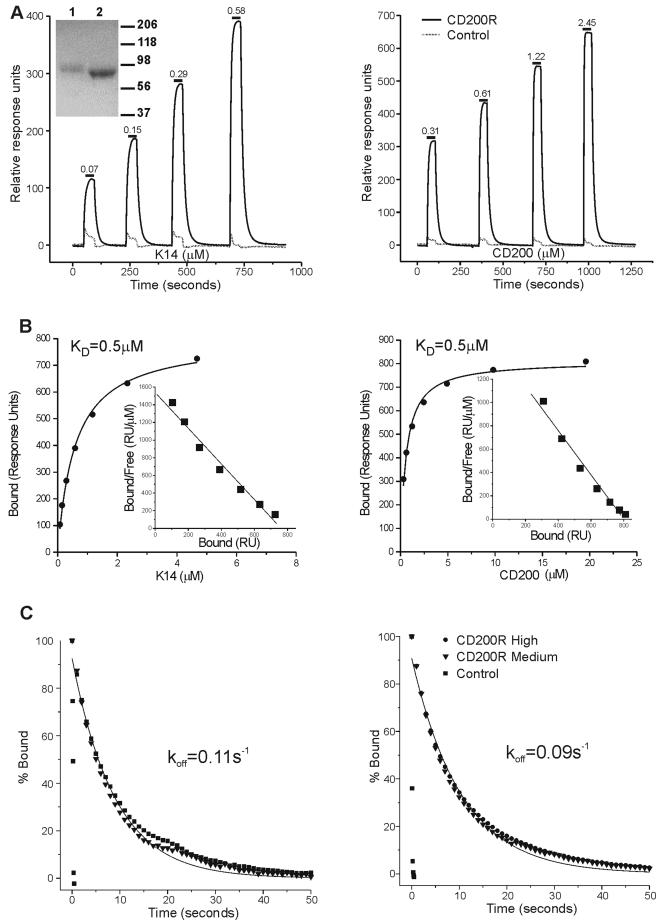
Despite the low sequence identity between CD200 and HHV-8 K14, it seemed feasible that the CD200 viral homologues would bind CD200R. This was tested at the protein level by using recombinant K14 protein from HHV-8. A chimeric soluble recombinant protein consisting of the extracellular part of K14 was engineered onto rat CD4d3 + 4 as an antigenic tag and expressed in CHO cells. This expression system has consistently allowed the production of correctly processed, antigenically active extracellular domains that retain their biochemical binding properties from proteins belonging to the IgSF (5, 42). A comparable construct for the CD200R protein containing a peptide sequence at the C terminus that allowed enzymatic biotinylation was also produced (5, 29, 42). The biochemical interactions of K14 and host CD200 with CD200R were detected and quantified in real time by using surface plasmon resonance in a BIAcore apparatus. CD200RCD4d3 + 4biotin protein and a negative control (CD4d3 + 4biotin) were immobilized in separate flow cells on a streptavidin-coated sensor chip before injection of purified soluble CD200 and K14 proteins. Both CD200 and K14 ligands bound to the CD200R flow cell in comparison to the control reference cell (Fig. 2A). This binding was quantified by calculating the difference in response units observed in the CD200R and control flow cells once equilibrium had been reached and was plotted as a binding curve (Fig. 2B). The equilibrium binding affinities (KD) were calculated by both nonlinear curve fitting (Fig. 2B) and Scatchard transformations (Fig. 2B, inset) of the binding data, giving similar values. Remarkably, the host CD200 and viral K14 interacted with the human CD200R with almost identical equilibrium binding affinities (KD of ∼0.5 μΜ at 37°C) (Fig. 2B). Kinetic analysis of the interactions yielded similar off rates (koff of ∼0.1 s−1, equivalent to a half-life of 7 s) at different levels of CD200R immobilization, indicating that kinetic measurements were not grossly affected by rebinding or mass transport effects (Fig. 2C). In order to provide data for HHV-8 infection models within rodents, these measurements were repeated with soluble biotinylated forms of both the rat and mouse CD200R produced in an identical manner, and the data are presented in Table 1. This revealed that the K14 showed almost identical cross-species binding to both mouse and rat CD200R, although the former was weaker, with about a 10-fold higher dissociation rate.
FIG. 2.
The viral HHV-8 K14 protein interacts with human CD200R with equilibrium affinity and kinetics similar to those of the host CD200 protein. The left panels show data for K14 (viral ligand), and the right panels show data for human CD200 (host ligand). (A) The indicated active concentrations of viral ligand K14CD4d3 + 4 (left panel) or host ligand CD200CD4d3 + 4 (right panel) were injected at 15 μl/min, for the durations indicated by the short bars, through flow cells with 1,815 response units (RU) of CD200RCD4d3 + 4-biotin (CD200R) or 2,185 RU of CD4d3 + 4biotin (control) immobilized. The amount of CD200CD4d3 + 4 or K14CD4d3 + 4 that bound at each concentration was calculated as the difference between the response at equilibrium in the CD200RCD4d3 + 4 and control flow cells, and these are plotted as binding curves in panel B. The inset shows sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the two chimeric proteins K14CD4d3 + 4 (lane 1) and CD200CD4d3 + 4 (lane 2) used in these studies. (B) The curved lines in the main plots are nonlinear curve fits to the data and correspond to an affinity of 0.5 μM for both the host and viral ligand. Scatchard transformations of the same binding data are shown in the insets, and the linear fit shown corresponds to a KD of 0.5 μΜ for both ligands. (C) The dissociation rate constants of the CD200- and K14-CD200R interactions were measured by injecting 10 μl of soluble CD200CD4d3 + 4 (2.0 μM) or K14CD4d3 + 4 (0.5 μM) at 100 μl/min over immobilized CD200RCD4d3 + 4biotin at high (1,815 RU) and medium (895 RU) levels and also over a negative control, CD4d3 + 4-biotin (2,185 RU), and data were collected at 10 Hz. The data were then normalized (100% at the start of the dissociation phase), and first-order exponential decay curves were fitted to the dissociation data (CD200R medium in both cases) and yielded the indicated koff values. For clarity, only every 10th data point is shown. Data for the human CD200R-CD200 interaction are from reference 42.
TABLE 1.
Kinetic data for human CD200 and HHV-8 K14 proteins binding human, rat and mouse CD200Ra
| CD200R |
KD (μM)
|
koff
|
||||
|---|---|---|---|---|---|---|
| Host CD200 | Viral K14 | Host CD200
|
Viral K14
|
|||
| s−1 | t1/2 (s) | s−1 | t1/2 (s) | |||
| Human | 0.49 ± 0.08 | 0.49 ± 0.09 | 0.09 ± 0.004 | 7.7 | 0.11 ± 0.008 | 6.1 |
| Rat | 0.59 ± 0.07 | 0.67 ± 0.07 | 0.24 ± 0.03 | 2.9 | 0.17 ± 0.03 | 4.1 |
| Mouse | 7.0 ± 0.33 | 9.2 ± 0.98 | 2.10 ± 0.2 | 0.3 | 1.7 ± 0.3 | 0.4 |
The KD values were determined by averaging those calculated from each of the three different CD200R immobilization levels obtained by nonlinear curve fitting (note that the curves shown in Fig. 2B are from high levels of human CD200R immobilization alone). The koff values were determined by averaging the values obtained by fitting the dissociation phases of the binding curves to a simple 1:1 binding model (see Materials and Methods), using two different concentrations of ligand over three levels of receptor immobilization (i.e., n = 6). In all cases, the level of receptor immobilization had negligible effects on koff, indicating that mass transport and rebinding effects were minimal. Data for CD200-CD200R interaction are from reference 42. Results are shown as means ± standard deviations.
K14 protein expressed at the cell surface binds CD200R.
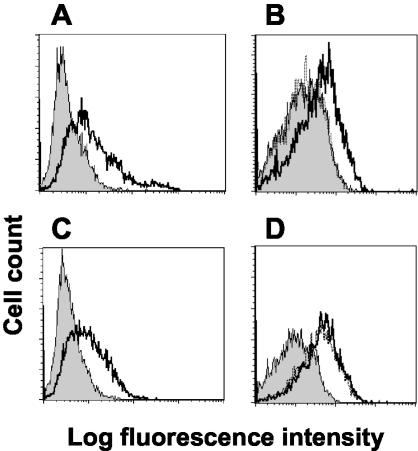
The CD200 receptor was first identified by using an assay with beads coated with rat CD200 recombinant protein to provide a multimeric array of the protein in order to detect weak (KD of ∼1 μM; half-life of less than 1 s) interactions (5, 6, 40, 44) and mimic events that might occur during cell contact. The equivalent interaction was later shown in humans by using human CD200R protein at the protein level and also in cell binding assays by using cells transfected with CD200R (42). To see if the K14 ORF encoded a membrane protein that could interact with CD200R, CHO cells were transfected with full-length CD200 and K14. K14 was always expressed at low levels, and in order to have comparable levels of CD200 for functional analysis, cells expressing equivalent levels of CD200 were separated by cell sorting. Cell surface expression of CD200 was shown with MAb OX104 (Fig. 3A) and K14 expression with a MAb, OX112, which was raised against purified K14 recombinant protein (Fig. 3C). The CD200- or K14-transfected CHO cells were tested for their ability to bind CD200R by interacting with CD200RCD4d3 + 4biotin protein presented as a multimeric array on avidin-coated fluorescent beads. CD200R beads bound cells transfected with CD200, and this binding could be blocked by prior incubation of the cells with MAb OX104 (Fig. 3B). In addition, CD200R-bound K14 transfected cells, but in this case, the K14 MAb (OX112) did not block, in agreement with binding studies with purified protein and the BIAcore (Fig. 3D and data not shown).
FIG. 3.
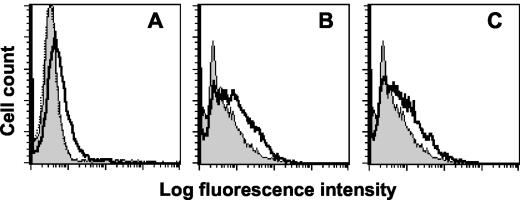
CD200RCD4d3 + 4-coated beads bind CHO cells transfected with CD200 or K14. (A) Flow cytometry shows expression of CD200 on transfected CHO cells stained with MAb OX104 (thick line) compared to an isotype-matched control MAb, OX1 (shaded histogram). (B) CD200R-coated fluorescent beads bind CHO-CD200 cells (thick line) compared to beads coated with negative control CD4d3 + 4 (shaded histogram). The binding was blocked when CHO-CD200 cells were incubated with OX104 (dotted line [indistinguishable from the shaded histogram]). (C) Flow cytometry shows expression of K14 on transfected CHO cells stained with MAb OX112 (thick line) compared to an isotype-matched control MAb, OX1 (shaded histogram). (D) CD200R-coated fluorescent beads bind CHO-K14 cells (thick line) compared to beads coated with negative control CD4d3 + 4 (shaded histogram). The binding was not blocked with MAb OX112, which is known to be nonblocking (dotted line is indistinguishable from thick line).
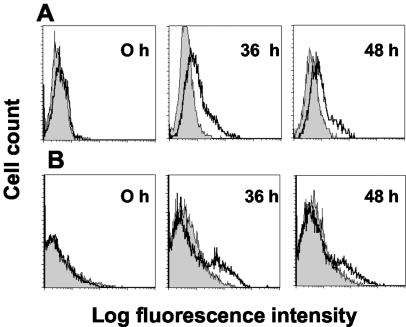
A cell line harboring HHV-8 expresses the K14 protein and can bind CD200R upon lytic induction.
The K14 ORF mRNA is induced during the lytic cycle as shown by RNA analysis (20) (but see Discussion). A HHV-8 harboring a primary effusion lymphoma cell line (BC-3) was induced into the lytic cycle of viral replication by the addition of 2 mM sodium butyrate and then subsequently tested for the expression of K14 with the specific MAb OX112 or CD200R-coated beads at different time points. Figure 4 shows that upon induction, the OX112 MAb labeled increasing numbers of cells and this correlated with the ability to bind beads coated with human CD200RCD4d3 + 4biotin. Analysis by fluorescence microscopy showed viable cells clearly labeled with CD200R beads (data not shown). The possibility that these cells expressed CD200 (and hence bind CD200R) was excluded by showing no labeling with OX104, a human CD200 MAb (data not shown). As expected, the K14 MAb did not block the binding of CD200R to BC3 cells (Fig. 3 and data not shown). The combined biochemical and cellular evidence demonstrates that the K14 protein is expressed at the cell surfaces of HHV-8-harboring cells during the lytic cycle and can interact with CD200R on host cells.
FIG. 4.
Detection of the K14 protein on HHV-8-harboring cells undergoing lytic induction. (A) Flow cytometry of BC-3 cells undergoing the lytic cycle and stained with MAb OX112 (bold line) compared to an isotype-matched control MAb, OX1 (shaded histogram). (B) CD200R-coated fluorescent beads bind BC-3 cells (thick line) in increasing amounts following induction of lytic replication of HHV-8 compared to a negative control (CD4d3 + 4-coated beads) (shaded histogram). The cells became larger and more granular, consistent with lytic induction. The cells shown in the labeling were gated on the viable population. Results from one typical experiment of five are shown.
Cell surface CD200 and K14 deliver a down-regulatory signal to activated macrophages via CD200R.
What type of signal could CD200 and K14 be giving to receptor-positive cells? Evidence from the mouse knockout studies (19) and functional studies using CD200 fusion proteins (16, 17) and antibodies (44) suggests that CD200 delivers a down-regulatory signal to activated myeloid cells. As the affinity and kinetics of the viral K14 protein were virtually indistinguishable from those of the host CD200, it seemed likely that the viral protein would also down-regulate macrophages through CD200R. Although MAbs and recombinant proteins binding cell surface proteins can give valuable information about the way a protein signals, the actual signals between interacting membrane proteins at the cell surface will be more subtle. In order to test this, an assay was devised to culture activated macrophages in the presence of cells expressing either CD200 or K14 at their cell surface and then monitor macrophage activity through cytokine production. CHO cell lines expressing full-length CD200 and K14 were produced. The K14 was not expressed at high levels compared to CD200, and the CD200-expressing cell line was sorted to obtain lines with similar levels of expression of CD200 and K14 as determined by flow cytometry and MAb binding. The lower levels are more likely to reflect physiological levels and represent normal signals. These cells could also bind CD200R-coated beads (Fig. 3). Human monocytes, which are known to express CD200R (42), were allowed to adhere and mature into macrophages for 5 to 7 days. At this time, the cells expressed CD200R as detected by both MAb OX108 (Fig. 5A) binding and their ability to bind CD200- and K14-coated beads (Fig. 5B and C). The macrophages were activated with a combination of IFN-γ and LPS to give moderate activation of the cells. CHO cells expressing CD200 or K14 protein or cells that were transfected with empty vector were irradiated and then added to the macrophage cultures, and supernatants were assayed for TNF-α secretion. The CD200 and K14 lines produced a reduction in the TNF-α secreted by activated macrophages, but there was some variation, not only between different donors but also in the levels produced with various control cell lines (data not shown). This might be expected if the cell lines had different levels of other surface proteins that might affect macrophage activity; several cell surface proteins on CHO cells have been shown to affect T-cell assays (15). A system was therefore set up whereby the effects of the CD200 protein could be isolated; this was achieved by specifically blocking the CD200-CD200R interaction with the OX104 MAb (Fig. 6A). The results show that in contrast to the case for the controls (Fig. 6B), by preventing the CD200-CD200R interaction, the inhibition of TNF-α production was relieved.
FIG. 5.
Macrophages express CD200R and are able to bind CD200- and K14-coated beads. (A) Flow cytometry shows expression of CD200R on human macrophages stained with MAb OX108 (thick line) compared to an isotype-matched control MAb, OX1 (shaded histogram). Macrophages did not express CD200 when stained with MAb OX104 (dotted line [but indistinguishable from the shaded histogram]). (B) CD200-CD4d3 + 4-coated fluorescent beads bind macrophages (thick line) compared to beads coated with negative control CD4d3 + 4 (shaded histogram). (C) K14-CD4d3 + 4-coated fluorescent beads bind macrophages (thick line) compared to beads coated with negative control CD4d3 + 4 (shaded histogram).
FIG. 6.
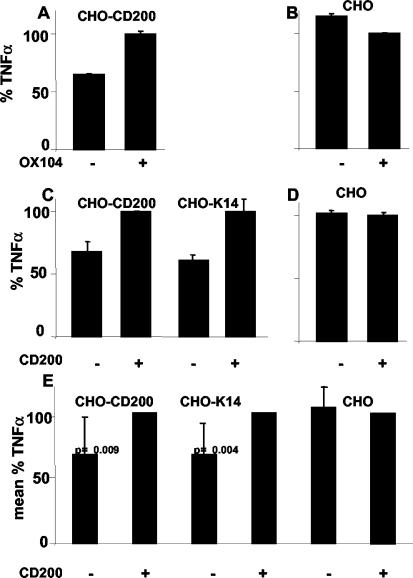
CD200 and the viral HHV-8 K14 expressed at the surface of CHO cells deliver a regulatory signal to activated macrophages. (A) CHO cells expressing CD200 inhibit TNF-α production, and this inhibition can be specifically overcome by blocking CD200-CD200R interaction with MAb OX104 recognizing CD200. Error bars indicate standard deviations. (B) The OX104 MAb had a minimal effect on TNF-α production in the control assay where CHO cells transfected with empty vector were added to activated macrophages. (C and D) The inhibition of TNF-α production by activated macrophages with CHO-CD200 cells can be relieved by the addition of purified monomeric CD200CD4d3 + 4 protein. An increase in TNF-α production is seen only upon the addition of soluble CD200 to those assays that have either CD200- or K14-expressing cells (C) and not in control experiments (D). This shows that the effect was the result of blocking the CD200-CD200R interaction and was not due to any other interaction. (E) Summary of eight independent experiments showing the inhibitory effects of CD200 and K14 on activated macrophages. The cells expressing either CD200 or K14 give a clear reduction in TNF-α production that can be overcome by the addition of the CD200 protein, whereas there is no CD200 protein effect on the control cells. Results are expressed as a mean percentage of TNF-α. Statistical analysis was done with Student's t test. Probabilities are indicated for each cell line. TNF-α levels (which ranged between 150 and 750 pg/ml) were normalized for each cell line where the CD200-CD200R interaction was blocked with either with MAb (A and B) or soluble monomeric CD200 protein (C, D, and E).
CD200 has a short (19-amino-acid) cytoplasmic region that contains no known signaling motifs, and therefore the CD200 MAb is unlikely to signal to the CHO cells by cross-linking CD200 and thereby cause changes in TNF-α production by the human macrophages. The MAb recognizing K14 did not block the interaction between CD200R beads and CD200 (Fig. 3) and therefore could not be tested in the same manner. It could be argued that the clustered Fc regions of MAb on the surfaces of the CHO cells could provide stimuli to the activated macrophages via cell surface Fc receptors. To exclude this possibility and to assay for effects of the K14-expressing cells, we attempted to block the weak CD200-CD200R and K14-CD200R interactions by using soluble monomeric CD200CD4d3 + 4 protein. To ensure a constant high level of receptor occupancy, the CD200CD4d3 + 4 protein was added at a concentration of 4 μM, a concentration that is well above the equilibrium dissociation constant of 0.5 μM (Fig. 2B). The addition of purified CD200 overcame the inhibition of TNF-α production by CD200 and K14 cell lines (Fig. 6C). The amount of TNF-α produced varied between donors. Indeed, in assays that resulted in high levels of TNF-α production, no blocking effects could be detected. This is presumably due to a very potent macrophage activation signal that was too strong to be overcome by the CD200R inhibitory signal. In eight independent experiments, however, inhibition of TNF-α production could be observed with the transfected cell lines, which could in turn be overcome with recombinant CD200 protein. A summary of these results with appropriate controls is presented in Fig. 6E. In order to compare results and avoid any variation due to differences in the cell lines other than CD200 or K14, each set of experiments were normalized with respect to TNF-α production in the presence of blocking protein, and then the effects of the different cell lines were compared. In the control lines, addition of recombinant protein had no effect, but clear effects were found with both CD200- and K14-expressing cell lines, showing that the phenomena observed were due to the ligand interaction under study and not to other, uncharacterized interactions in the assay (e.g., between the monocytes and CHO cells or between the monocytes themselves). In two experiments that gave clear inhibition of TNF-α production, we screened for 14 cytokines by using a multiplex assay. This assay confirmed the effects previously found on TNF-α. As expected, T-cell cytokines such as IL-2, IL-4, IL-5, IL-7, IL-13, and IL-17 were produced at very low levels (1 to 20 pg/ml). The myeloid cytokines were stimulated at different levels, IL-1β was poorly stimulated (10 to 50 pg/ml) even in the presence of CD200- or K14-transfected cells, while macrophage inflammatory protein 1β, IL-6, and IL-8 were induced greatly by LPS and IFN-γ and effects with CD200 or K14 were not observed. G-CSF production was inhibited to about the same degree as TNF-α and MCP-1 production, by about 25% (data not shown). We conclude that both CD200 and the HHV-8 viral homologue, K14, can deliver a down-regulatory signal to activated macrophages.
DISCUSSION
A large body of both biochemical (44) and genetic (19) data have suggested that CD200 is able to locally deliver a restrictive signal to myeloid cells via a receptor (CD200R) expressed on these cell types. This work provides, for the first time, a molecular explanation for the CD200-CD200R interaction by demonstrating that cell surface engagement of CD200R can inhibit TNF-α secretion from activated macrophages. These new data fit with the hypothesis that the widely distributed CD200 locally regulates the function of activated macrophages by direct cell contact in a variety of tissues. The use of a soluble monomeric CD200 protein to block the CD200-CD200R interaction in our functional assays minimizes possible signaling artifacts due to CD200 cross-linking or through Fc receptors if MAbs or Fc fusion proteins are used.
The finding that both the host CD200 and the HHV-8 K14 proteins share only 40% sequence identity and yet bind CD200R with identical affinity and kinetics and have similar functional effects is strong evidence that the virus directly mimics the function of the host CD200 protein. This also implies that the characteristics of receptor binding are important for CD200 function, since the rapid generation time of the virus would allow significant opportunity for the biochemical interaction characteristics to evolve and increase virus fitness. The observation that the K14 protein is able to locally restrict macrophage activation by inhibiting TNF-α production provides a clear immunomodulatory mechanism whereby the virus is able to subvert the host immune system.
For mice, proteins related to CD200R, but not binding CD200, that have an activating phenotype through association with DAP12 have been described (42). Although there is a comparable gene in humans, we could not express it as a recombinant soluble protein, and it does not appear to be expressed. This is probably because of mutations in key cysteine residues, as discussed previously (42), and therefore the gene is not a candidate for a receptor for K14.
These results contrast with those recently published by Chung et al. (11), who showed that recombinant K14 fusion protein has an activating effect on macrophages through an undefined receptor. Strangely, however, they observed no effect with the equivalent CD200 fusion protein. This is highly surprising in light of our biochemical analysis that demonstrates the identical binding properties of the CD200 and K14 proteins with CD200R. They suggested that K14 might interact with a myeloid-restricted receptor and that by inducing a different conformational change to CD200, this would then alter the polarity of the delivered signal by an undefined mechanism. This explanation seems unlikely; it is without precedent within this family of cell adhesion molecules, which have been shown to exhibit rigid body association (22). One possible explanation for this discrepancy may be the inherent instability of the recombinant K14 protein: we found that even freshly prepared K14CD4d3 + 4 chimeric protein contained a significant proportion that would not bind CD200R, despite retaining antigenic activity for CD4. It appeared that this protein tended to contain more misfolded protein (60%) than many recombinant proteins, including CD200 (10%) (see Materials and Methods). The presence of this denatured material would make functional experiments hard to confidently interpret. Chung et al. (11) also found that K14 expressed on a transfected B-cell line caused U937 myeloid cells to produce inflammatory cytokines. The control, however, was simply untransfected cells. Making transfected cells can lead to changes in their ability to stimulate, as discussed above, which are probably due to changes in expression of cell surface accessory molecules. In addition, we found that U937 did not express CD200R (unpublished data). The experiments reported here show down-regulation of TNF-α by using cell lines expressing active K14 protein, as shown through MAb and ligand binding, and thereby closely resembling the native viral protein. Further specificity was shown by blocking with the stable monomeric CD200 protein. By using this blocking approach, we can eliminate variation due to heterogeneity between transfected cell lines. In some experiments we showed that CD200 and K14 were able to down-regulate other proinflammatory cytokines such as G-CSF and MCP-1. In contrast with the results of Chung et al. (11), very low levels of IL-1β were produced with IFN-γ and LPS and the CD200- or K14-expressing cell lines. Our data fit with indications from many other functional studies on CD200 (19, 42, 44).
How do these new data fit in with what is known about the biology and sarcomagenesis of HHV-8 infection? Interestingly, the K14 ORF is encoded on a ∼2.7-kb bicistronic transcript that also encodes ORF74, a constitutively active G-protein-coupled receptor (21, 37). This transcript has been classified as a lytic gene based upon in vitro chemical induction of viral gene expression from infected B cells from a primary effusion lymphoma (36). In an elegant study that set out to identify the viral mediators of sarcomagenesis (27), Montaner et al. found that ORF74 alone was sufficient to induce tumors in a mouse model whereas latently expressed genes lacked this ability. The demonstration that a lytically expressed gene was the transforming factor was difficult to reconcile with a simple latent-lytic model of viral gene expression. Indeed, many HHV-8-infected tissues and cell lines contain a small proportion (usually around 1%) of cells expressing “lytic” genes, which has led to the suggestion that these genes contribute to sarcomagenesis through downstream paracrine mechanisms (8, 10, 30). ORF74 is an excellent candidate for this function; it constitutively activates mitogen-activated protein kinase signaling and induces the secretion of proangiogenic factors from infected cells (1, 34), thereby promoting vascularization of the tumors, and “unmasks” the tumorigenic potential of the latently expressed viral proteins (27). We suggest that the virus coexpresses K14 with ORF74 to locally restrict activation of macrophages, which are abundant within the lesions (27), and thereby prevent a host response against the cells within the lesion that are exerting these paracrine effects that are essential for tumorigenesis.
Genes with IgSF domains are among the most common in the human genome, and many are involved in the fine-tuning and regulation of the immune system; e.g., about one-third of leukocyte membrane proteins contain IgSF domains (3). Genes with IgSF domains, however, are relatively rare in viruses (32), and this may reflect that they are of particular value only in viruses such as poxviruses and herpesviruses, which can coexist with the host for long periods and therefore require more subtle control of the immune system than other viruses. Viruses that contain CD200-like ORFs are evolutionarily very diverse. For example, it has been estimated that the subfamilies Betaherpesviridae (HHV-6 and -7) and Gammaherpesviridae (rhesus macaque rhadinovirus and HHV-8) diverged approximately 200 million years ago (24). In addition, the locations of these ORFs differ within the genome of each virus subfamily. Together, this is compelling evidence that the CD200-like viral ORFs have been acquired independently in these viruses (26). It would appear that the ability to negatively control macrophage activation by capture of the host CD200 protein is desirable for many viruses and therefore makes a significant contribution to viral selective fitness. It is worth mentioning that although a regulatory role seems most likely, it is possible that the K14-CD200R interaction provides a mechanism of entry into myeloid cells, and it could be involved in cell-cell spread of virus as observed for herpes simplex virus (14).
One other host-virus interaction that is superficially similar to the K14-CD200R interaction is that between the UL18, an MHC class I-like protein from human cytomegalovirus, and the NK inhibitory receptor LIR-1. In this case, however, the viral UL18 interaction has about 1,000-fold-higher affinity than the normal MHC class I-LIR-1 interaction (9). It is proposed that UL18 provides a highly efficient way of engaging the inhibitory receptor to compensate for other effects of human cytomegalovirus, thus leading to down-regulation of MHC class I levels. The striking difference in the UL18 and K14 interactions is that the former has a much higher affinity, whereas the latter is indistinguishable from the host interaction.
In summary, a large body of data indicates that the role of CD200 is to provide localized and tissue-specific negative regulation of myeloid lineage cells via CD200R. We have extended these studies, providing a molecular explanation for these observations and showing that CD200R signaling inhibits TNF-α production by activated macrophages. We also show that despite low levels of sequence identity, a homologue of CD200 present in a human herpesvirus, K14, binds to host CD200R with identical affinity and kinetics and is also able to inhibit TNF-α production. The presence of CD200 homologues in viruses of many different families suggests that the ability to restrictively control host macrophages is a common viral immunomodulatory strategy. These data also suggest that the CD200-CD200R interaction offers a potential therapeutic mechanism for locally controlling unwanted immune reactions.
Acknowledgments
We are grateful to Chris Boshoff for the BC-3 line, K14 ORF, and helpful discussions; to Ruth Goddard for producing the K14CD4d3 + 4 stable line; to Alan Johnstone for organizing cytokine assays; to Han-Joo Lee, Geoff Smith, and Patrick Whelan for discussions of unpublished data; to Tony Willis for amino acid analysis; to Nigel Rust for cell sorting; and to Steve Dewhurst, Mansun Law, and Anton van der Merwe for valuable discussions.
This research was supported by the Medical Research Council.
REFERENCES
- 1.Bais, C., B. Santomasso, O. Coso, L. Arvanitakis, E. G. Raaka, J. S. Gutkind, A. S. Asch, E. Cesarman, M. C. Gershengorn, E. A. Mesri, and M. C. Gerhengorn. 1998. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature 391:86-89. [DOI] [PubMed] [Google Scholar]
- 2.Barclay, A. N. 1981. Different reticular elements in rat lymphoid tissue identified by localization of Ia, Thy-1 and MRC OX 2 antigens. Immunology 44:727-736. [PMC free article] [PubMed] [Google Scholar]
- 3.Barclay, A. N., M. H. Brown, S. K. A. Law, A. J. McKnight, M. G. Tomlinson, and P. A. van der Merwe. 1997. Leucocyte antigens factsbook, 2nd ed. Academic Press, London, United Kingdom.
- 4.Brown, M. H., and A. N. Barclay. 1994. Expression of immunoglobulin and scavenger receptor superfamily domains as chimeric proteins with domains 3 and 4 of CD4 for ligand analysis. Prot. Eng. 7:515-521. [DOI] [PubMed] [Google Scholar]
- 5.Brown, M. H., K. Boles, P. A. van der Merwe, V. Kumar, P. A. Mathew, and A. N. Barclay. 1998. 2B4, the natural killer and T cell immunoglobulin superfamily surface protein, is a ligand for CD48. J. Exp. Med. 188:2083-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, M. H., S. Preston, and A. N. Barclay. 1995. A sensitive assay for detecting low-affinity interactions at the cell surface reveals no additional ligands for the adhesion pair rat CD2 and CD48. Eur. J. Immunol. 25:3222-3228. [DOI] [PubMed] [Google Scholar]
- 7.Bukovsky, A., J. Presl, J. Zidovsky, and P. Mancal. 1983. The localization of Thy-1.1, MRC OX 2 and Ia antigens in the rat ovary and fallopian tube. Immunology 48:587-596. [PMC free article] [PubMed] [Google Scholar]
- 8.Cesarman, E., E. A. Mesri, and M. C. Gershengorn. 2000. Viral G protein-coupled receptor and Kaposi's sarcoma: a model of paracrine neoplasia? J. Exp. Med. 191:417-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman, T. L., A. P. Heikeman, and P. J. Bjorkman. 1999. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity 11:603-613. [DOI] [PubMed] [Google Scholar]
- 10.Chiou, C. J., L. J. Poole, P. S. Kim, D. M. Ciufo, J. S. Cannon, C. M. ap Rhys, D. J. Alcendor, J. C. Zong, R. F. Ambinder, and G. S. Hayward. 2002. Patterns of gene expression and a transactivation function exhibited by the vGCR (ORF74) chemokine receptor protein of Kaposi's sarcoma-associated herpesvirus. J. Virol. 76:3421-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung, Y. H., R. E. Means, J. K. Choi, B. S. Lee, and J. U. Jung. 2002. Kaposi's sarcoma-associated herpesvirus OX2 glycoprotein activates myeloid-lineage cells to induce inflammatory cytokine production. J. Virol. 76:4688-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark, M. J., J. Gagnon, A. F. Williams, and A. N. Barclay. 1985. MRC OX-2 antigen: a lymphoid/neuronal membrane glycoprotein with a structure like a single immunoglobulin light chain. EMBO J. 4:113-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosman, D., N. Fanger, and L. Borges. 1999. Human cytomegalovirus, MHC class I and inhibitory signalling receptors: more questions than answers. Immunol. Rev. 168:177-185. [DOI] [PubMed] [Google Scholar]
- 14.Dingwell, K. S., and D. C. Johnson. 1998. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J. Virol. 72:8933-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaglia, J. L., A. Mattoo, E. A. Greenfield, G. J. Freeman, and V. K. Kuchroo. 2001. Characterization of endogenous Chinese hamster ovary cell surface molecules that mediate T cell costimulation. Cell. Immunol. 213:83-93. [DOI] [PubMed] [Google Scholar]
- 16.Gorczynski, R. M., M. S. Cattral, Z. Chen, J. Hu, J. Lei, W. P. Min, G. Yu, and J. Ni. 1999. An immunoadhesin incorporating the molecule OX-2 is a potent immunosuppressant that prolongs allo- and xenograft survival. J. Immunol. 163:1654-1660. [PubMed] [Google Scholar]
- 17.Gorczynski, R. M., Z. Chen, K. Yu, and J. Hu. 2001. CD200 immunoadhesin suppresses collagen-induced arthritis in mice. Clin. Immunol. 101:328-334. [DOI] [PubMed] [Google Scholar]
- 18.Gruffat, H., A. Sergeant, and E. Manet. 2000. Kaposi's sarcoma-associated herpesvirus and Kaposi's sarcoma. Microbes Infect. 2:671-680. [DOI] [PubMed] [Google Scholar]
- 19.Hoek, R. M., S. R. Ruuls, C. A. Murphy, G. J. Wright, R. Goddard, S. M. Zurawski, B. Blom, M. E. Homla, W. J. Streit, M. H. Brown, A. N. Barclay, and J. D. Sedgwick. 2000. Down-regulation of the macrophage lineage through interaction with OX2 (CD200). Science 290:1768-1771. [DOI] [PubMed] [Google Scholar]
- 20.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirshner, J. R., K. Staskus, A. Haase, M. Lagunoff, and D. Ganem. 1999. Expression of the open reading frame 74 (G-protein-coupled receptor) gene of Kaposi's sarcoma (KS)-associated herpesvirus: implications for KS pathogenesis. J. Virol. 73:6006-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maenaka, K., T. Juji, T. Nakayama, J. R. Wyer, G. F. Gao, T. Maenaka, N. R. Zaccai, A. Kikuchi, T. Yabe, K. Tokunaga, K. Tadokoro, D. I. Stuart, E. Y. Jones, and P. A. van der Merwe. 1999. Killer cell immunoglobulin receptors and T cell receptors bind peptide-major histocompatibility complex class I with distinct thermodynamic and kinetic properties. J. Biol. Chem. 274:28329-28334. [DOI] [PubMed] [Google Scholar]
- 23.Mahoney, J. A., B. Ntolosi, R. P. DaSilva, S. Gordon, and A. J. McKnight. 2001. Cloning and characterization of CPVL, a novel serine carboxypeptidase, from human macrophages. Genomics 72:243-251. [DOI] [PubMed] [Google Scholar]
- 24.McGeoch, D. J., S. Cook, A. Dolan, F. E. Jamieson, and E. A. Telford. 1995. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 247:443-458. [DOI] [PubMed] [Google Scholar]
- 25.McGeoch, D. J., and A. J. Davison. 1999. The descent of human herpesvirus 8. Semin. Cancer Biol. 9:201-209. [DOI] [PubMed] [Google Scholar]
- 26.McGeoch, D. J., A. Dolan, and A. C. Ralph. 2000. Towards a comprehensive phylogeny for mammalian and avian herpesviruses. J. Virol. 74:10401-10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montaner, S., A. Sodhi, A. Molinolo, T. H. Bugge, E. T. Sawai, Y. He, Y. Li, P. E. Ray, and J. S. Gutkind. 2003. Endothelial infection with KSHV genes in vivo reveals that vGPCR initiates Kaposi's sarcomagenesis and can promote the tumorigenic potential of viral latent genes. Cancer Cell 3:23-36. [DOI] [PubMed] [Google Scholar]
- 28.Morris, R. J., and J. N. Beech. 1987. Sequential expression of OX2 and Thy-1 glycoproteins on the neuronal surface during development. An immunohistochemical study of rat cerebellum. Dev. Neurosci. 9:33-44. [DOI] [PubMed] [Google Scholar]
- 29.O'Callaghan, C., A., M. F. Byford, J. R. Wyer, B. E. Willcox, B. K. Jakobsen, A. J. McMichael, and J. I. Bell. 1999. BirA enzyme: production and application in the study of membrane receptor-ligand interactions by site-specific biotinylation. Anal. Biochem. 266:9-15. [DOI] [PubMed] [Google Scholar]
- 30.Pati, S., M. Cavrois, H. G. Guo, J. S. Foulke, Jr., J. Kim, R. A. Feldman, and M. Reitz. 2001. Activation of NF-κB by the human herpesvirus 8 chemokine receptor ORF74: evidence for a paracrine model of Kaposi's sarcoma pathogenesis. J. Virol. 75:8660-8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Preston, S., G. J. Wright, K. Starr, A. N. Barclay, and M. H. Brown. 1997. The leukocyte/neuron cell surface antigen OX2 binds to a ligand on macrophages. Eur. J. Immunol. 27:1911-1918. [DOI] [PubMed] [Google Scholar]
- 32.Schultz, J., R. R. Copley, T. Doerks, C. P. Ponting, and P. Bork. 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28:231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, G. L., J. A. Symons, A. Khanna, A. Vanderplasschen, and A. Alcami. 1997. Vaccinia virus immune evasion. Immunol. Rev. 159:137-154. [DOI] [PubMed] [Google Scholar]
- 34.Sodhi, A., S. Montaner, V. Patel, M. Zohar, C. Bais, E. A. Mesri, and J. S. Gutkind. 2000. The Kaposi's sarcoma-associated herpes virus G protein-coupled receptor up-regulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1alpha. Cancer Res. 60:4873-4880. [PubMed] [Google Scholar]
- 35.Spriggs, M. K. 1996. One step ahead of the game: viral immunomodulatory molecules. Annu. Rev. Immunol. 14:101-130. [DOI] [PubMed] [Google Scholar]
- 36.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talbot, S. J., R. A. Weiss, P. Kellam, and C. Boshoff. 1999. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257:84-94. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tortorella, D., B. E. Gewurz, M. H. Furman, D. J. Schust, and H. L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861-926. [DOI] [PubMed] [Google Scholar]
- 40.van der Merwe, P. A., and A. N. Barclay. 1996. Analysis of cell-adhesion molecule interactions using surface plasmon resonance. Curr. Opin. Immunol. 8:257-261. [DOI] [PubMed] [Google Scholar]
- 41.Webb, M., and A. N. Barclay. 1984. Localisation of the MRC OX-2 glycoprotein on the surfaces of neurones. J. Neurochem. 43:1061-1067. [DOI] [PubMed] [Google Scholar]
- 42.Wright, G. J., H. Cherwinski, M. Foster-Cuevas, G. Brooke, M. J. Puklavec, M. Bigler, Y. Song, M. Jenmalm, D. Gorman, T. McClanahan, M. R. Liu, M. H. Brown, J. D. Sedgwick, J. H. Phillips, and A. N. Barclay. 2003. Characterization of the CD200 receptor family in mice and humans and their interactions with CD200. J. Immunol. 171:3034-3046. [DOI] [PubMed] [Google Scholar]
- 43.Wright, G. J., M. Jones, M. J. Puklavec, M. H. Brown, and A. N. Barclay. 2001. The unusual distribution of the rat lymphoid/neuronal OX2 glycoprotein is conserved in humans (CD200). Immunology 102:173-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright, G. J., M. J. Puklavec, A. C. Willis, R. M. Hoek, J. D. Sedgwick, M. H. Brown, and A. N. Barclay. 2000. Lymphoid/neuronal cell surface OX2 glycoprotein recognizes a novel receptor on macrophages implicated in the control of their function. Immunity 13:233-242. [DOI] [PubMed] [Google Scholar]