Abstract
Mouse strains are either resistant or susceptible to murine cytomegalovirus (MCMV). Resistance is determined by the Cmv1r (Ly49h) gene, which encodes the Ly49H NK cell activation receptor. The protein encoded by the m157 gene of MCMV has been defined as a ligand for Ly49H. To find out whether the m157 protein is the only Ly49H ligand encoded by MCMV, we constructed the m157 deletion mutant and a revertant virus. Viruses were tested for susceptibility to NK cell control in Ly49H+ and Ly49H− mouse strains. Deletion of the m157 gene abolished the viral activation of Ly49H+ NK cells, resulting in higher virus virulence in vivo. Thus, in the absence of m157, Ly49H+ mice react like susceptible strains. 129/SvJ mice lack the Ly49H activation NK cell receptor but express the inhibitory Ly49I NK cell receptor that binds to the m157 protein. The Δm157 inhibitory phenotype was weak because MCMV encodes a number of proteins that mediate NK inhibition, whose contribution could be shown by another mutant.
Infection of mice with murine cytomegalovirus (MCMV) is an established model for studying human cytomegalovirus infection. Immune control of MCMV infection is organized in a hierarchical and redundant manner by diverse components of the innate and adaptive immune response (19, 21, 32, 34). NK cells play an important role in the innate control of cytomegalovirus infection. This has been demonstrated in humans with a rare disorder characterized by complete absence of NK cells and in genetically deficient mice lacking NK cells or being depleted of these cells by treatment with anti-NK cell antibodies (4). On the basis of their susceptibility to MCMV infection, mouse strains are either susceptible (e.g., BALB/c mice) or resistant (e.g., C57BL/6 mice) (17, 37). MCMV titers in the spleens of mice inversely correlate with their ability to mount an effective NK cell response, which is controlled by the single dominant locus, named Cmv1, located in the natural killer gene complex on mouse chromosome 6 (10, 14, 37-39). The alleles of the Cmv1 locus can confer either susceptibility (Cmv1s, a recessive allele) or resistance (Cmv1r, a dominant allele) to MCMV (36, 39).
The Cmv1r (Ly49h) gene encodes the Ly49H receptor (5, 9, 23, 24), which belongs to the Ly49 family of NK cell receptors and is expressed on approximately 50% of NK cells in C57BL/6 mice (41, 43, 47). Unlike the inhibitory Ly49 receptors, Ly49H lacks the immunoreceptor tyrosine-based inhibition motif and is noncovalently coupled with DAP12 at the cell surface, allowing transduction of an activation signal into the cell via its immunoreceptor tyrosine-based activation motif (16, 43) that is required for resistance to MCMV (40). However, Ly49H does not define resistance to vaccinia virus and gammaherpesvirus 68 (2, 12).
Unlike other members of the Ly49 receptor family, which use major histocompatibility complex (MHC) class I molecules as their cellular ligands, Ly49H binds to at least one MCMV-encoded protein, the m157 gene product (2, 42). The m157 protein has structural homology to MHC class I molecules, similar to several other proteins encoded by MCMV m145 gene family members (42). An MCMV deletion mutant restricted Ly49H activation to 15 genes in the HindIII-E region (2). Isolated open reading frames (ORFs) from this region, with the exception of m157, failed to activate Ly49H. However, a similar contribution of other genes in this region cannot be excluded since certain cytomegalovirus proteins encoded by different genes can only be expressed as a complex (27). Therefore, it remained an open question whether m157 is the only viral gene that contributes to MCMV resistance defined by Ly49H.
To investigate the biological relevance of the m157 gene, we constructed an m157 deletion mutant, as well as the corresponding revertant virus. We studied the susceptibility of these recombinant viruses to control by NK cells in vivo in Ly49H+ and Ly49H− mouse strains. Loss of the m157 gene is associated with gain of virulence in Ly49H+ but not in Ly49H− mouse strains. Therefore, m157 is the only MCMV-encoded protein that activates Ly49H+ NK cells. The absence of the gene that encodes this protein in the m157 deletion mutant gave us the opportunity to reveal the function of viral genes that down-modulate NK cell activity in Cmv1r mice. Cmv1 has been defined as a locus of resistance to MCMV, influencing virus control mainly in the spleen (37). Furthermore, since we could define m157 as the only MCMV-encoded ligand for the Ly49H receptor, we could also address the question of NK cell control of infection at a different site of infection.
MATERIALS AND METHODS
Viruses and cells.
Bacterial artificial chromosome (BAC)-derived MCMV strain MW97.01 has previously been shown to be biologically equivalent to MCMV strain Smith (ATCC VR-194, recently reaccessed as VR-1399) and is here referred to as wild-type (w.t.) MCMV (50). The ΔMS94.5 virus, which possesses a deletion of 15 genes (m151 to m165) is described elsewhere (46). All viruses were propagated on third-passage BALB/c mouse embryonic fibroblasts (MEFs) and purified by sucrose cushion centrifugation. Tissue culture-grown virus preparations were used for mouse inoculations.
Cells of the mouse macrophage cell line IC-21 were obtained from the American Type Culture Collection (ATCC catalog no. TIB-186) and were grown in RPMI 1640 medium supplemented with 10% fetal calf serum.
Plasmid construction.
Plasmid pori6k-pA, which contains the Zeocin resistance gene, the poly(A) signal from BHG, the origin of replication (ori6k), and an additional 34-bp FRT site, was generated by ligation of a 353-bp KpnI/PvuII fragment from plasmid pCDNA4TO (Invitrogen) into the KpnI/EcoRV sites of plasmid pori6kZeo (A. Bubeck, M. Wagner, Z. Ruzsics, M. Iglesias, I. R. Singh, and U. H. Koszinowski, submitted for publication). With primers SpeI-nt215895-918 and SpeI-nt217226-250 and pSM3fr as the template DNA, the m157 gene and its putative promoter (nucleotide [nt] positions 215895 to 217250, as described in reference 33) were amplified by PCR and inserted into the SpeI site of plasmid pori6k-pA, thereby generating plasmid pori6k-m157-pA. The correct amplification of the m157 promoter and the m157 gene was confirmed by sequencing with primers M157-1 (5′-TGTTGACCGCCATCTGTTCTTGA), M157-2rev (5′-GGTAAGATTAATATTCAAGGATCA), and M157-3 (5′-GGATTGAAAATTGTTACAGCACG) (data not shown).
Insertion of an FRT site into MCMV BAC pSM3fr between genes m16 and m17.
Mutagenesis of the MCMV BACs was performed as previously described (49). The insertion of a 48-bp FRT site (5′-GAAGTTCCTATTCCGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTC-3′) into the intergenic region between MCMV ORFs m16 and m17 (nt positions 15678 to 15748) was achieved as follows. A linear PCR fragment containing a kanamycin resistance gene flanked by two 48-bp FRT sites and viral homologies to the noncoding region between ORFs m16 and m17 was generated by PCR with primers 5-m16-FRT-Kan-pCP15 (5′-CCCTCTTAATCATGACAATTATAAGTGTCTTATACGCAATACTTTTATCATAATTCGGGGGTGTCCAGGGTTTTCCC) and 3-m17-FRT-Kan-pCP15 (5′-GAGGAATAGGAATAACTCACCACCGATTTCAGCGTCTGCCCCAAGTCTGACTTCCGGCTCGTATGTTGTGTGG) and plasmid pCP15 (8). This fragment was inserted into pSM3fr by homologous recombination in Escherichia coli, thereby deleting 70 bp of the noncoding region between these two genes (nt positions 15678 to 15748). The kanamycin resistance-encoding gene was subsequently excised by FLP-mediated site-directed recombination as described previously (49), leaving only one FRT site, which can be used for site-directed insertion of any gene of interest into this site. The correct mutagenesis of resulting MCMV BAC pSM3fr-FRT was confirmed by restriction pattern analysis and sequencing of the m16-m17 genome region with primers MCMV-15461-down (5′-GAAGTCCATGTATCTCCTTCA) and MCMV-15939-up (5′-TCGGACAAATTCTAAACCTCG) (data not shown). The w.t.-FRT-MCMV strain generated from pSM3fr-FRT was shown to replicate to w.t. MCMV titers in NIH 3T3 fibroblasts and also in the lungs, spleens, and livers of BALB/c mice infected with 2 × 105 PFU at days 3 and 7 postinfection (data to be published elsewhere). This confirmed that the insertion of short sequences into this intergenic genome region does not significantly interfere with virus replication in vitro or in vivo.
Deletion of the m157 ORF in MCMV BACs pSM3fr and pSM3fr-FRT.
For deletion of the m157 ORF in the respective MCMV BACs, a linear DNA fragment was generated by PCR with primers 5-m157-Kan (5′-CGTGGTCAAGCCGGTCGTGTTGTACCAGAACTCGACTTCGGTCGCGTTCGATTTATTCAACAAAGCCACG) and 3-m157-Kan and plasmid pACYC177 as the template DNA. This fragment was subsequently inserted into MCMV BACs pSM3fr and pSM3fr-FRT, respectively, by homologous recombination in E. coli as described previously (49), generating recombinant MCMV BACs pΔm157 and pΔm157-FRT. These genomes lack most parts of the m157 gene, including the ATG start codon (nt positions 216291 to 216874).
Reinsertion of the m157 gene, including its promoter, at an ectopic position into the m157 deletion genome.
For reinsertion of the m157 ORF including its promoter and an additional poly(A) signal from BHG into MCMV BAC pΔm157-FRT at the ectopic position between genes m16 and m17, a linear DNA fragment that contains these elements and an additional Zeocin resistance gene was generated by PCR with primers 5-m157-Zeo-m16/17 (5′-CCCTCTTAATCATGACAATTATAAGTGTCTTATACGCAATACTTTTATCATAATACATGTGGAATTGTGAGC) and 3-m157-Zeo-m16/17 (5′-GAGGAATAGGAATAACTCACCACCGATTTCAGCGTCTGCCCCAAGTCTGATTAGCACGTGTCAGTCCT) and plasmid pori6k-m157-pA as the template DNA. After homologous recombination of this fragment with pΔm157-FRT in E. coli, revertant MCMV BAC pm157rev was generated. The correct insertion of the m157 gene, including its promoter, at this ectopic position was confirmed by restriction pattern analysis and sequencing with primers M157-1 (5′-TGTTGACCGCCATCTGTTCTTGA), M157-2rev (5′-GGTAAGATTAATATTCAAGGATCA), and M157-3 (5′-GGATTGAAAATTGTTACAGCACG) (data not shown).
Reconstitution of virus mutants from recombinant BACs.
By transfection of 2 μg of the corresponding MCMV BAC DNA into MEFs, mutants Δm157-MCMV and Δm157-FRT-MCMV and revertant virus m157Rev-MCMV were reconstituted as previously described (51).
Northern blot analysis.
NIH 3T3 fibroblasts were infected at a multiplicity of infection (MOI) of 3 with Δm157, w.t. MCMV, or m157Rev, and total RNA was isolated 6 and 24 h postinfection with the TriZol reagent (Invitrogen) in accordance with the manufacturer's instructions. An [α-32P]dCTP-labeled DNA probe specific for 528 bp of the m157 gene (nt positions 216350 to 216878, according to reference 33) was generated with the nick translation kit from Amersham (Amersham Biosciences Europe) in accordance with the manufacturer's instructions. Three micrograms of total RNA from each sample of virus-infected cells was separated on a denaturing formaldehyde gel, blotted to a nylon membrane, and hybridized with the [α-32P]dCTP-labeled DNA probe for detection of m157-specific mRNA transcripts.
Animals.
BXD-8/Ty (H-2b), 129/SvJ (H-2b), and C57BL/6 RAG1−/− (H-2b) mice were purchased from The Jackson Laboratory (Bar Harbor, Maine). All of the mice used in this study, including congenic inbred BALB.B6-Cmv1r (H-2d) and inbred C57BL/6 (H-2b) and BALB/c (H-2d) mice, were housed and bred under specific-pathogen-free conditions at the Central Animal Facility of the Medical Faculty, University of Rijeka, in accordance with the guidelines contained in the International Guiding Principles for Biomedical Research Involving Animals. The Ethical Committee at the University of Rijeka approved all of the animal experiments described here. C57BL/6 RAG1−/− (H-2b) mice were maintained under specific-pathogen-free conditions in the animal facility of the Washington University School of Medicine, and the experiments conducted with these mice were in accordance with institutional guidelines for animal care and use. Six- to eight-week-old female mice were used in all of the experiments.
Infection conditions, detection of infectious MCMV in tissues, and statistical evaluation.
Mice were injected intravenously with 5 × 105 (Ly49H+ mice) or 2 × 105 (Ly49H− mice) PFU of tissue culture-grown recombinant virus or w.t. MCMV in a volume of 500 μl of diluents. Organs were collected 3 days after infection, and viral titers were determined with a standard assay of viral plaque formation on MEFs (34). Each experiment shown here is representative of at least three independent experiments. The statistical significance of differences between experimental groups was determined by the Mann-Whitney exact rank test. Viral titers (from groups x and y) were considered significantly different for P (x versus y) < α = 0.05 (one sided), where P is the observed probability value and α is a selected significance level.
Depletion of NK cell subsets in vivo.
Depletion of NK1.1+ cells was done with monoclonal antibody (MAb) PK136 (20) at a concentration of 1 mg per mouse by intraperitoneal inoculation 24 to 2 h before infection. The efficacy of depletion was assessed by cytofluorometric analysis of spleen cells with phycoerythrin (PE)-conjugated antibodies to mouse NK1.1 (BD Bioscience Pharmingen, San Diego, Calif.). Depletion of Ly49C/I+ NK cell subsets was performed with MAb 5E6 (31) at a concentration of 150 μg per mouse, and depletion of Ly49C/H/I+ NK cell subsets was performed with MAb 1F8 (9) at a concentration of 150 μg per mouse by intraperitoneal inoculation 24 h before infection.
Staining of intracellular IFN-γ.
An in vitro assay to detect gamma interferon (IFN-γ) production was performed as previously described (42). Briefly, C57BL/6 RAG1−/− splenocytes were cocultured for 12 h with IC-21 cells that were either uninfected or infected for 24 h at an MOI of 5 with Δm157 mutant, m157Rev, or w.t. MCMV. Brefeldin A (BD Bioscience Pharmingen) was added for the last 11 h of coincubation. Cells were first surface stained with biotinylated 3D10 (α-Ly49H) (41) and then stained with PE-streptavidin and allophycocyanin-PK136. Cells were fixed and permeabilized with the Cytofix/Cytoperm kit (BD Bioscience Pharmingen) in accordance with the manufacturer's instructions. Intracellular IFN-γ was stained with fluorescein isothiocyanate-XMG1.2 (BD Bioscience Pharmingen) in the permeabilization buffer. Cells were analyzed with a FACScalibur cytometer (BD Biosciences, San Jose, Calif.) gating on the NK1.1+ CD3− populations.
RESULTS
Generation of MCMV mutants.
To investigate the significance of the m157 gene product for virus control in vivo, two independent MCMV mutants were constructed. First, mutants with a targeted deletion of the m157 gene (Δm157 and Δm157-FRT) were prepared (Fig. 1). In vitro and in vivo testing in the lungs, spleens, and livers of BALB/c and C57BL/6 mice at day 3 postinfection revealed no difference between Δm157 and Δm157-FRT (data not shown). In the experiments described here, only Δm157 was used. Thereafter, a revertant virus (m157Rev) was constructed. Rather than reconstructing the w.t. situation, in the revertant virus the m157 gene was reintroduced at an ectopic position between ORFs m16 and m17 of the MCMV genome to selectively study the effect of the m157 ORF. All mutants were derived from parental MCMV BAC pSM3fr, which has w.t. MCMV properties in vitro and in vivo (50). The correct mutagenesis of the resulting Δm157 and Δm157-FRT strains, as well of the m157Rev MCMV strain, was confirmed by restriction pattern analysis and sequencing of the m157 genome region.
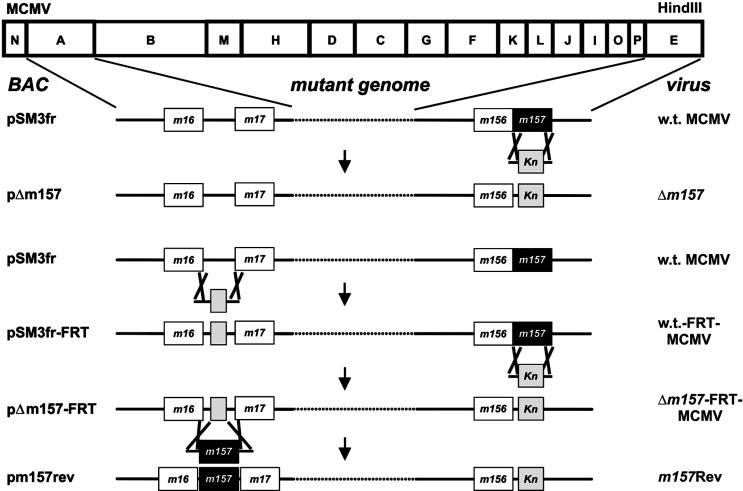
FIG. 1.
Genome structure of recombinant MCMVs. The HindIII cleavage map of the MCMV (Smith strain) genome is shown at the top. Deletion and insertion of the m157 gene, respectively, were achieved by homologous recombination between the parental MCMV BACs and a linear DNA fragment containing the desired sequence, a kanamycin resistance gene (Kn), and flanking homologies to the target site in the viral genome. The deletion genome pΔm157 was generated by homologous recombination of the linear DNA fragment and w.t. MCMV BAC pSM3fr. An independent Δm157 genome was constructed as follows. First, a 48-bp FRT site (grey box) was inserted into the intergenic region between genes m16 and m17, generating pSM3fr-FRT, to prove that insertion of sequences at this positions does not interfere with virus replication. In a second mutagenesis step, the m157 gene was deleted from pSM3fr-FRT, generating the second independent m157 deletion genome, pΔm157-FRT. Finally, the m157 gene, including its native promoter, was reinserted into pΔm157-FRT at the ectopic positions between genes m16 and m17, thereby removing the FRT site. The names of the MCMV BACs are indicated on the left. Recombinant viruses were reconstituted by transfection of the recombinant MCMV BACs into permissive MEFs. The names of the corresponding reconstituted viruses are given on the right.
No phenotype of Δm157 and m157Rev in cell culture.
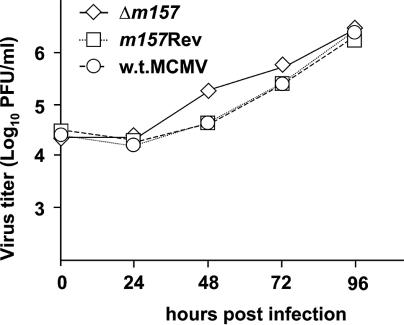
Multistep growth curves of recombinant and w.t. MCMVs served to assess whether deletion of the m157 gene and its ectopic reinsertion affect virus growth in cell culture. After infection of primary BALB/c MEFs at 0.01 PFU per cell, the replication of Δm157 and the m157Rev was indistinguishable from that of w.t. MCMV (Fig. 2). These results indicated that the m157 gene product is dispensable for virus growth in fibroblasts and that ectopic reinsertion of this gene between MCMV ORFs m16 and m17 does not compromise viral growth in vitro.
FIG. 2.
In vitro growth of recombinant viruses. BALB/c MEFs were infected with Δm157, m157Rev, or w.t. MCMV at 0.01 PFU per cell. Supernatants were harvested at the indicated time points after infection, and virus titers were determined.
Δm157 gains virulence in Ly49H+ mice through loss of NK cell-mediated control.
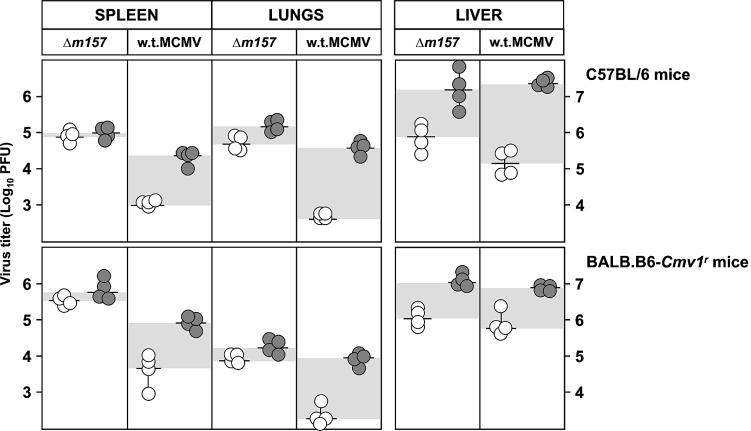
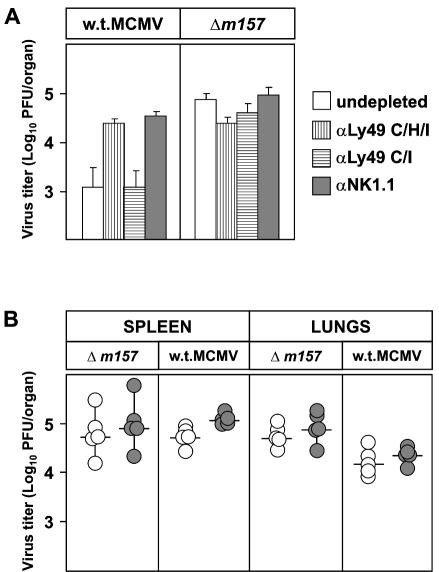
Replication of MCMV in vivo during the early period after infection inversely correlates with the ability of mouse strains to mount an effective NK cell response, which is controlled by the Ly49h locus (15). The MCMV protein encoded by the m157 gene is the only ligand for the Ly49H receptor that has been identified so far (2, 42). To test whether the m157 protein is the only MCMV protein interacting with Ly49H, we used the Δm157 virus. C57BL/6 (Ly49H+) mice (Fig. 3, top) and congenic BALB.B6-Cmv1r mice (Fig. 3, bottom) were injected with Δm157 or w.t. MCMV, and virus titers in their organs were determined 3 days later. The spleens and lungs of mice infected with Δm157 showed significantly higher virus titers than did those of mice infected with w.t. MCMV. Consistent with published data (6, 52, 53), depletion of NK cells by anti-NK1.1 MAbs increased the virus titers in mice infected with w.t. MCMV, whereas it had no effect on the virus titers in the spleens and lungs of mice infected with Δm157. We concluded that Δm157 virus is resistant to NK cell control in vivo and that there is no other viral ligand for Ly49H encoded by the MCMV genome.
FIG. 3.
Deletion of the m157 gene abolishes MCMV susceptibility to NK cell-mediated control in Ly49H+ mice. Undepleted (open circles) or NK1.1-depleted (shaded circles) C57BL/6 and congenic BALB.B6-Cmv1r mice were injected intravenously with 5 × 105 PFU of Δm157 or w.t. MCMV. Three days after infection, the virus titers in the spleen, lungs, and liver were determined by standard plaque assay. Titers in individual mice (circles) and median values (horizontal bars) are shown. Virus titers were calculated per organ in the spleen and lungs and per gram of liver. The differences in viral titers between undepleted and NK cell-depleted groups of mice are shown by shaded areas. There were significant differences (P < 0.025) in virus titers between the groups of undepleted mice infected with Δm157 and w.t. MCMV in the spleen and lungs.
Remarkably, NK cells were efficient in limiting virus replication in liver irrespective of the presence or absence of the m157 gene. This finding is in line with previously published data on the protective effect of the Cmv1 (Ly49h) locus in the spleen but not in the liver (5, 37, 44). Interestingly, contrary to the MCMV titer in the liver and similar to that in the spleen, the MCMV titer in the lungs was also controlled by the Ly49H NK cell activation receptor.
Reintroduction of the m157 gene reverses the susceptibility of MCMV to NK cells.
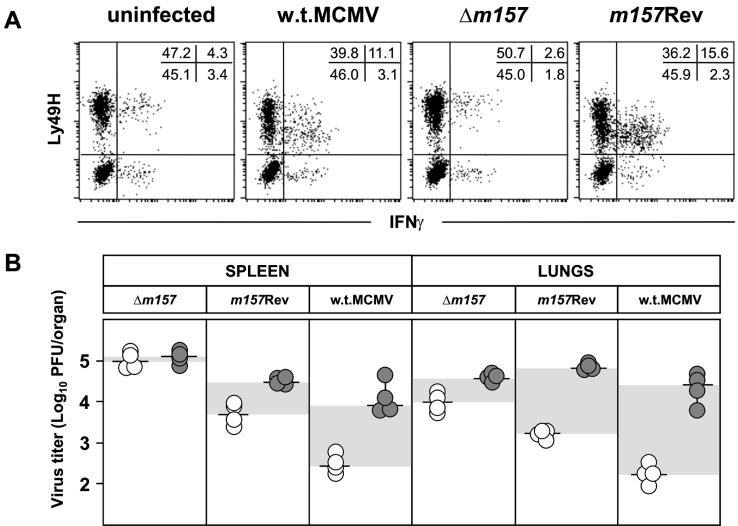
To confirm that resistance of mutant virus to NK cells in vivo is solely due to lack of the m157 protein and no other unwanted effect in Δm157, we compared the abilities of Δm157, m157Rev, and w.t. MCMVs to induce activation of Ly49H+ NK cells. Splenocytes derived from C57BL/6 RAG1−/− mice were incubated for 12 h with MCMV-infected IC-21 cells, and the frequency of Ly49H+ IFN-γ-secreting cells was measured (Fig. 4 A). The incubation of splenocytes with w.t. MCMV- or m157Rev-infected IC-21 cells resulted in a comparable expansion of Ly49H+ IFN-γ-secreting cells (11.1 and 15.6%, respectively). IC-21 cells infected with Δm157 failed to activate Ly49H+ cells, and the percentage of activated Ly49H+ NK cells was the same as in cultures incubated with uninfected IC-21 cells. These findings confirm previously published data, obtained by anti-Ly49H blockade and reporter cell assays, that showed that the isolated m157 protein is crucial for the activation of Ly49H+ NK cells (2, 42). The results presented here extend the above studies to the situation found during virus infection. As shown in Fig. 4B, reintroduction of the m157 gene into the Δm157 genome also reversed the sensitivity of the virus to NK cell control in vivo. This is illustrated by the fact that the titers of the m157Rev virus in C57BL/6 mice were significantly lower than those of Δm157. Furthermore, the depletion of NK cells increased the titer of m157Rev whereas, consistent with the data presented in Fig. 3, Δm157 remained resistant to NK cell control. The reconstitution of viral sensitivity to NK cells essentially confirmed the role of the m157 gene. Still, the in vivo function of the gene in m157Rev was not fully restored. This is probably due to the fact that the bona fide promoter of m157 was transferred together with the ORF. Therefore, other sequences modulating gene expression may be lacking, and indeed, Northern blot analysis revealed that the m157 transcripts appear earlier in m157Rev-infected cells and peak later in w.t.-infected cells (see supplemental material). This difference in the activity of ectopic m157 might explain the variance in virus sensitivity to NK cells in vivo.
FIG. 4.
NK cell activation and reversal of susceptibility to NK cell control by reintroduction of the m157 gene. (A) C57BL/6 RAG1−/− splenocytes were incubated for 12 h (1 h without brefeldin A and 11 h with brefeldin A) with IC-21 cells that were either uninfected or infected (MOI = 5, 24 h) with Δm157, m157Rev, or w.t. MCMV. Cells were stained with biotinylated anti-Ly49H, followed by PE-streptavidin and allophycocyanin-anti-NK1.1. The cells were fixed, permeabilized, and stained with fluorescein isothiocyanate-anti-IFN-γ antibody. Data represent NK1.1+ cells. The values are the percentages of cells within the quadrants. (B) Undepleted (open circles) or NK1.1-depleted (shaded circles) C57BL/6 mice were injected intravenously with 5 × 105 PFU of Δm157, m157Rev, or w.t. MCMV, and virus titers in the spleen and lungs were determined 3 days after infection. Titers in individual mice (circles) and median values (horizontal bars) are shown. The differences in viral titers between undepleted and NK cell-depleted groups of mice are shown by shaded areas. There were significant differences (P < 0.025) in virus titers between the groups of undepleted mice infected with Δm157 and m157Rev and between the groups of undepleted mice infected with Δm157 and w.t. MCMV.
Δm157 lacks a phenotype in C57BL/6 mice depleted of Ly49H+ NK cells and in mice lacking the Ly49H receptor.
To address the role of the Ly49H+ NK cell subset in the control of MCMV infection and the role of the m157 protein, C57BL/6 mice were injected with MAb 5E6 (αLy49C/I), 1F8 (αLy49C/H/I), or PK136 (αNK1.1) prior to infection with either w.t. or Δm157 MCMV. Depletion of 1F8+ cells, but not 5E6+ cells, in mice infected with w.t. MCMV led to an increase in virus titers comparable to the effect of NK1.1+ cell depletion. This indicates that other NK cell subsets contribute very little, if at all, to virus control (Fig. 5A). In accordance with the data presented in Fig. 3, in mice infected with Δm157, depletion of the Ly49H+ subset did not increase the virus titers. Therefore, the m157 effect on NK cell activation was mediated solely by the Ly49H+ NK cell subset, thus confirming previous studies (5, 9).
FIG. 5.
Ly49H+ NK cells play no role in control of Δm157 virus. (A) C57BL/6 mice were depleted with MAb 5E6 (αLy49 C/I), 1F8 (αLy49 C/H/I), or PK136 (αNK1.1) and injected intravenously with 5 × 105 PFU of Δm157 and w.t. MCMV. Virus titers in the lungs were determined 3 days after infection. Data represent the mean value ± the standard deviation of four or five mice. Depletion with MAb 1F8 or PK136 resulted in a significant increase (P < 0.025) in the w.t. MCMV titer. (B) Recombinant BXD-8 mice were injected with 2 × 105 PFU of Δm157 and w.t. MCMV, and virus titers in the spleen and lungs were determined 3 days after infection. Titers in individual mice (circles) and median values (horizontal bars) are shown.
The BXD-8 mouse strain is a recombinant inbred strain derived from C57BL/6 (Ly49H+) and DBA/2 (Ly49H−) progenitor strains (45). Although BXD-8 mice possess the entire natural killer gene complex from C57BL/6 mice, they are susceptible to MCMV infection because of a lesion in the Ly49h gene (5, 23). For that reason the m157 protein should play no role in NK cell-mediated virus control in these mice. Indeed, 3 days after infection, no differences in virus titers between the group of mice infected with Δm157 and that infected with w.t. MCMV were observed (Fig. 5B). Furthermore, NK depletion had no influence on virus titers, confirming that in the absence of Ly49H the m157 protein plays no role in virus control.
Weak phenotype of Δm157 in BALB/c and 129/SvJ mice.
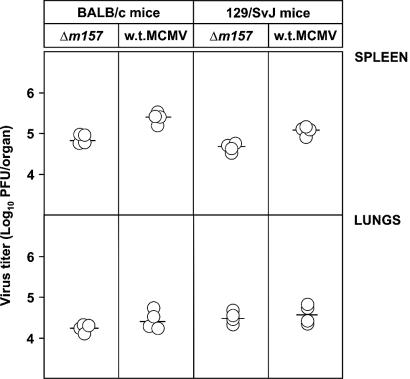
We tested the role of the m157 protein in mice lacking the Ly49H but expressing the Ly49I NK cell receptor. 129/J mice do not contain Ly49H or other NK cell activation receptors that recognize m157 (2). However, 129/J mice express the inhibitory NK cell receptor Ly49I (26). The m157 protein binds to 129/J NK cells but not to NK cells of BALB/c mice in vitro (2). If the in vitro conditions reflect the situation in vivo, Δm157 should be more attenuated in 129/J mice than in BALB/c mice. To test this, BALB/c and 129/SvJ mice were injected with Δm157 or w.t. MCMV and virus titers were tested. Remarkably, there was no difference in the reactivities of these two mouse strains. However, there was a small but statistically significant difference (P < 0.025) between Δm157 and w.t. MCMV control in the spleen, indicating a certain degree of attenuation (Fig. 6). Nevertheless, we concluded that the presence or absence of the m157 gene has no strong phenotype in mice expressing the Ly49I NK receptor.
FIG. 6.
Control of Δm157 virus in BALB/c and 129/SvJ mice. BALB/c and 129/SvJ mice were injected with 2 × 105 PFU of Δm157 and w.t. MCMV, and virus titers in the spleen and lungs were determined 3 days later. Titers in individual mice (circles) and median values (horizontal bars) are shown. There were significant differences (P < 0.025) in virus titers in spleen between groups of mice infected with Δm157 and w.t. MCMV.
NK response to Δm157 in C57BL/6 mice is prevented by viral evasion genes.
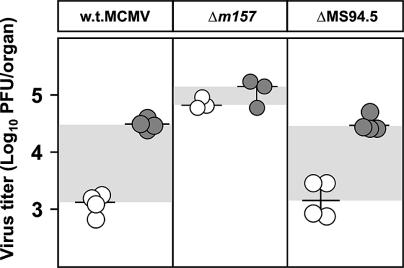
MCMV has several NK silencing genes, of which only m152 and m144 have been characterized (13, 22, 25). To show the impact of viral silencing genes in a Ly49H+ strain, we compared Δm157 with w.t. MCMV and a virus in which in addition to m157, 14 other genes, including m152, were deleted (Fig. 7). W.t. MCMV is strictly controlled by NK cells, and the virus grows only after NK cell depletion. Δm157 lacks the NK activation via m157-Ly49H, and therefore NK depletion has no phenotype. The ΔMS94.5 virus lacks m157 and, in addition, 14 other genes, including m152, that prevent NK cell activation by down-modulating NKG2D ligands (22, 25). We now have evidence that at least one more MCMV gene, in addition to m152, can down-modulate ligands for the NKG2D receptor (S.J., unpublished data). This inhibitory effect is present in the Δm157 virus, and therefore the Δm157 virus grows to a higher titer than the ΔMS94.5 virus, which has lost this NK silencing gene(s). Deletion of NK cells abolishes this type of control, resulting in identical titers of the ΔMS94.5 and Δm157 viruses. This situation is comparable to the situation in Cmv1s mice after infection in the absence or presence of the m152 gene (22). Therefore, in the presence of a number of additional viral silencing genes the effect of the m157-Ly49I interaction is not expected to have a strong impact.
FIG. 7.
NK cell response against Δm157 in C57BL/6 mice is prevented by viral evasion genes. Undepleted (open circles) or NK1.1-depleted (shaded circles) C57BL/6 mice were injected intravenously with 5 × 105 PFU of w.t., Δm157, or ΔMS94.5 MCMV, and virus titers in the lungs were determined 3 days after infection. Titers in individual mice (circles) and median values (horizontal bars) are shown. The differences in viral titers between undepleted and NK cell-depleted groups of mice are shown by shaded areas. There were significant differences (P < 0.025) in virus titers between the groups of undepleted mice infected with Δm157 and ΔMS94.5 and between the groups of undepleted mice infected with Δm157 and w.t. MCMV.
DISCUSSION
Viruses affect NK cell control by providing viral ligands that bind NK cell receptors. Depending on whether these receptors are activating or inhibitory, the resulting NK function is modulated (29). The second type of NK cell modulation affects cellular ligands for NK cell receptors that are regulated by infection or stress (3, 22, 25). Two MCMV genes are thought to encode ligands for NK cell receptors, m144 and m157, but only the cognate NK cell receptor for the m157 protein, Ly49H, has been identified (2, 42). The interaction between Ly49H and the m157 protein is a unique situation and contributes to MCMV resistance in C57BL/6 mice, which express the Ly49H NK cell activation receptor. In previous studies the ligand-receptor interaction was demonstrated by expressing the isolated viral ligand (2, 42). By deleting m157 from the MCMV genome, we show that m157 indeed is the only MCMV ligand for Ly49H. Accordingly, in congenic mice lacking the Ly49H receptor (BXD-8), as well as in mice depleted of Ly49H+ NK cells, deletion of this gene from the virus has no phenotype. Ly49H+ mice, after infection with Δm157, lose their MCMV-encoded resistance phenotype.
m157 also serves as a ligand for the inhibitory NK receptor Ly49I in the MCMV-susceptible 129/J strain, and it also binds to NK cells of other MCMV-sensitive strains (1). Therefore, m157 is seen as an inhibitory NK cell ligand and Ly49H+ mice are considered an exception to the rule. However, no experiments in which NK cell function is blocked through m157-Ly49I have been published. Loss of NK cell activation can be studied in the context of virus infection when the virus either expresses or lacks the gene of interest. However, when we studied the effect of m157 deletion in the 129/SvJ strain no vigorous phenotype became apparent. A certain degree of virus attenuation was noticed in both BALB/c and 129/SvJ mice. Considering that the m157 protein binds to the Ly49I allelic form of 129/J but not of BALB/c mice (2), it is not clear whether this effect is related to the m157-Ly49I interaction. A strong signal was not to be expected, since in comparison to NK cells from the C57BL/6 strain only a minority of NK cells from 129/J mice binds m157 but does not lyse m157+ targets in vitro (2).
The absence of NK activation in Ly49H+ mice infected with Δm157 provided the opportunity to demonstrate the function of other viral genes inhibiting NK cell function. These genes act by down-modulating NKG2D ligands similarly to the function of m152 (22, 25). There is another not yet identified gene(s) in the left end of the genome (28), and we have evidence of at least one additional gene elsewhere (U.H.K. and S.J., unpublished data). Most NK cells express NKG2D receptors (18). Therefore, the loss of one silencing signal from a chorus of several is difficult to detect. This situation is similar to that of the genes modulating MHC class I expression, in which identification of the three genes involved (m04, m06, and m152) had to precede the construction of virus mutants that lack or express class I modulating functions in all possible combinations (49). In addition, deletion of the viral gene that down-modulates a ligand for an activating NK cell receptor has an immediate effect on NK control, since virus infection upregulates stress-induced ligands (7, 11). On the other hand, deletion of a viral ligand for an inhibitory NK cell receptor should not have a strong phenotype, especially if the stress-induced activating ligands are down-modulated at the same time.
Our study also confirms and extends the known complexity of NK cell control in different organs (30, 44). The Cmv1 locus has been described as a host resistance locus that regulates NK cell responses during acute MCMV infection in the spleen (37). Accordingly, Δm157 lacks this type of control. In addition, we show here for the first time that, akin to virus control in the spleen, Ly49H+ NK cells also mediate MCMV control in the lungs. Additional evidence for this is provided by C57BL/6 mice depleted of Ly49H+ NK cells, in which the titer of w.t. MCMV in the lungs reached a level comparable to that after depletion of NK1.1+ NK cells. In contrast, MCMV control in the liver appears to be independent of m157-Ly49H interaction. This finding is in accordance with the minimal effects of anti-Ly49H MAbs on the virus titer in the liver (5), pointing at a different type of NK control of MCMV in that organ (30, 44). After MCMV infection, NK cells pass through two different stages of activation. The first, nonspecific phase, during the first 2 days after MCMV infection, is characterized by IFN-γ production and NK cell proliferation, irrespective of Ly49H expression, while in the second, specific phase, there is a selective proliferation of Ly49H+ NK cells (12). This early activation of NK cells may be sufficient to control virus infection in the liver but not in the spleen and lungs. The mode by which NK cells mediate their antiviral effector function also differs between the liver and spleen. While in the spleen NK cells act via a cytolytic mechanism, in the liver MCMV is controlled by cytokines including IFN-γ produced by NK and NKT cells (30, 44). In perforin-deficient C57BL/6 mice, no differences between Δm157 and w.t. MCMV titers in the spleen were observed, which suggests that Ly49H-positive cells mediate their effect almost exclusively via a perforin-dependent pathway (I.B., unpublished data).
The MCMV genome harbors genes, in addition to m152 (22), whose products down-modulate NKG2D ligands, also in mice expressing Cmv1r (Ly49h) (S.J., unpublished). However, this inhibition of NK cell activation is overridden by NK activation through m157, which makes these mice an exception to the rule. It has been proposed that natural isolates from wild mice, depending on the presence or absence of the Ly49H receptor, are expected to be variable in the m157 gene (1, 35). Indeed, this has been recently demonstrated by Voigt et al., who reported that most of the MCMV strains they isolated from wild mice possessed a specific mutation of the m157 gene (48). Furthermore, this study provides evidence that NK cells can exert sufficient immunological pressure on MCMV that it undergoes rapid and specific mutation in the m157 gene region. Accordingly, we have been studying immunodeficient C57BL/6 mice that rely on NK functions to survive MCMV infection. Under these selective conditions, virus mutants arise that indeed do not respond to the Ly49H receptor (R. A. French, T. J. Pingel, M. Wagner, I. Bubic, L. Yang, S. Kim, U. H. Koszinowski, S. Jonjic, and W. M. Yokoyama, submitted for publication).
Supplementary Material
Acknowledgments
We thank J. R. Ortaldo and M. Bennett for generously providing the 5E6 and 1F8 MAbs. We also thank D. Rumora for technical assistance and T. Andreanszky for organizing the mouse facility.
This work was supported by Croatian Ministry of Science grants 0062004 (S. Jonjic) and 0062007 (A. Krmpotic), Deutsche Forschungsgemeinschaft SFB 455, and the Bayerische Forschungsstiftung (U. H. Koszinowski). W. M. Yokoyama is supported by NIH grants, the Barnes-Jewish Hospital Foundation, and the Howard Hughes Medical Institute.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Arase, H., and L. L. Lanier. 2002. Virus-driven evolution of natural killer cell receptors. Microbes Infect. 4:1505-1512. [DOI] [PubMed] [Google Scholar]
- 2.Arase, H., E. S. Mocarski, A. E. Campbell, A. B. Hill, and L. L. Lanier. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296:1323-1326. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, S., V. Groh, J. Wu, A. Steinle, J. H. Phillips, L. L. Lanier, and T. Spies. 1999. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285:727-729. [DOI] [PubMed] [Google Scholar]
- 4.Biron, C. A., K. B. Nguyen, G. C. Pien, L. P. Cousens, and T. P. Salazar-Mather. 1999. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu. Rev. Immunol. 17:189-220. [DOI] [PubMed] [Google Scholar]
- 5.Brown, M. G., A. O. Dokun, J. W. Heusel, H. R. Smith, D. L. Beckman, E. A. Blattenberger, C. E. Dubbelde, L. R. Stone, A. A. Scalzo, and W. M. Yokoyama. 2001. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science 292:934-937. [DOI] [PubMed] [Google Scholar]
- 6.Bukowski, J. F., B. A. Woda, and R. M. Welsh. 1984. Pathogenesis of murine cytomegalovirus infection in natural killer cell-depleted mice. J. Virol. 52:119-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerwenka, A., and L. L. Lanier. 2003. NKG2D ligands: unconventional MHC class I-like molecules exploited by viruses and cancer. Tissue Antigens 61:335-343. [DOI] [PubMed] [Google Scholar]
- 8.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 9.Daniels, K. A., G. Devora, W. C. Lai, C. L. O'Donnell, M. Bennett, and R. M. Welsh. 2001. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J. Exp. Med. 194:29-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Depatie, C., E. Muise, P. Lepage, P. Gros, and S. M. Vidal. 1997. High-resolution linkage map in the proximity of the host resistance locus Cmv1. Genomics 39:154-163. [DOI] [PubMed] [Google Scholar]
- 11.Diefenbach, A., and D. H. Raulet. 1999. Natural killer cells: stress out, turn on, tune in. Curr Biol. 9:R851-R853. [DOI] [PubMed] [Google Scholar]
- 12.Dokun, A. O., S. Kim, H. R. Smith, H. S. Kang, D. T. Chu, and W. M. Yokoyama. 2001. Specific and nonspecific NK cell activation during virus infection. Nat. Immunol. 2:951-956. [DOI] [PubMed] [Google Scholar]
- 13.Farrell, H. E., H. Vally, D. M. Lynch, P. Fleming, G. R. Shellam, A. A. Scalzo, and N. J. Davis-Poynter. 1997. Inhibition of natural killer cells by a cytomegalovirus MHC class I homologue in vivo. Nature 386:510-514. [DOI] [PubMed] [Google Scholar]
- 14.Forbes, C. A., M. G. Brown, R. Cho, G. R. Shellam, W. M. Yokoyama, and A. A. Scalzo. 1997. The Cmv1 host resistance locus is closely linked to the Ly49 multigene family within the natural killer cell gene complex on mouse chromosome 6. Genomics 41:406-413. [DOI] [PubMed] [Google Scholar]
- 15.French, A. R., and W. M. Yokoyama. 2003. Natural killer cells and viral infections. Curr. Opin. Immunol. 15:45-51. [DOI] [PubMed] [Google Scholar]
- 16.Gosselin, P., L. H. Mason, J. Willette-Brown, J. R. Ortaldo, D. W. McVicar, and S. K. Anderson. 1999. Induction of DAP12 phosphorylation, calcium mobilization, and cytokine secretion by Ly49H. J. Leukoc. Biol. 66:165-171. [DOI] [PubMed] [Google Scholar]
- 17.Grundy, J. E., J. S. Mackenzie, and N. F. Stanley. 1981. Influence of H-2 and non-H-2 genes on resistance to murine cytomegalovirus infection. Infect. Immun. 32:277-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jamieson, A. M., A. Diefenbach, C. W. McMahon, N. Xiong, J. R. Carlyle, and D. H. Raulet. 2002. The role of the NKG2D immunoreceptor in immune cell activation and natural killing. Immunity 17:19-29. [DOI] [PubMed] [Google Scholar]
- 19.Jonjic, S., I. Pavic, P. Lucin, D. Rukavina, and U. H. Koszinowski. 1990. Efficacious control of cytomegalovirus infection after long-term depletion of CD8+ T lymphocytes. J. Virol. 64:5457-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koo, G. C., and J. R. Peppard. 1984. Establishment of monoclonal anti-Nk-1.1 antibody. Hybridoma 3:301-303. [DOI] [PubMed] [Google Scholar]
- 21.Koszinowski, U. H., M. J. Reddehase, and S. Jonjic. 1993. The role of T-lymphocyte subsets in the control of cytomegalovirus infection, viruses and the cellular immune response. Marcel Dekker, Inc., London, United Kingdom.
- 22.Krmpotic, A., D. H. Busch, I. Bubic, F. Gebhardt, H. Hengel, M. Hasan, A. A. Scalzo, U. H. Koszinowski, and S. Jonjic. 2002. MCMV glycoprotein gp40 confers virus resistance to CD8+ T cells and NK cells in vivo. Nat. Immunol. 3:529-535. [DOI] [PubMed] [Google Scholar]
- 23.Lee, S. H., S. Girard, D. Macina, M. Busa, A. Zafer, A. Belouchi, P. Gros, and S. M. Vidal. 2001. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat. Genet. 28:42-45. [DOI] [PubMed] [Google Scholar]
- 24.Lee, S. H., A. Zafer, Y. de Repentigny, R. Kothary, M. L. Tremblay, P. Gros, P. Duplay, J. R. Webb, and S. M. Vidal. 2003. Transgenic expression of the activating natural killer receptor Ly49H confers resistance to cytomegalovirus in genetically susceptible mice. J. Exp. Med. 197:515-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lodoen, M., K. Ogasawara, J. A. Hamerman, H. Arase, J. P. Houchins, E. S. Mocarski, and L. L. Lanier. 2003. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J. Exp. Med. 197:1245-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makrigiannis, A. P., A. T. Pau, A. Saleh, R. Winkler-Pickett, J. R. Ortaldo, and S. K. Anderson. 2001. Class I MHC-binding characteristics of the 129/J Ly49 repertoire. J. Immunol. 166:5034-5043. [DOI] [PubMed] [Google Scholar]
- 27.Menard, C., M. Wagner, Z. Ruzsics, K. Holak, W. Brune, A. E. Campbell, and U. H. Koszinowski. 2003. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J. Virol. 77:5557-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira, S. A., S.-H. Park, P. Lee, A. Bendelac, and T. E. Shenk. 2002. Murine cytomegalovirus m02 gene family protects against natural killer cell-mediated immune surveillance. J. Virol. 76:885-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orange, J. S., M. S. Fassett, L. A. Koopman, J. E. Boyson, and J. L. Strominger. 2002. Viral evasion of natural killer cells. Nat. Immunol. 3:1006-1012. [DOI] [PubMed] [Google Scholar]
- 30.Orange, J. S., B. Wang, C. Terhorst, and C. A. Biron. 1995. Requirement for natural killer cell-produced interferon gamma in defense against murine cytomegalovirus infection and enhancement of this defense pathway by interleukin 12 administration. J. Exp. Med. 182:1045-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ortaldo, J. R., R. Winkler-Pickett, A. T. Mason, and L. H. Mason. 1998. The Ly-49 family: regulation of cytotoxicity and cytokine production in murine CD3+ cells. J. Immunol. 160:1158-1165. [PubMed] [Google Scholar]
- 32.Polic, B., H. Hengel, A. Krmpotic, J. Trgovcich, I. Pavic, P. Luccaronin, S. Jonjic, and U. H. Koszinowski. 1998. Hierarchical and redundant lymphocyte subset control precludes cytomegalovirus replication during latent infection. J. Exp. Med. 188:1047-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reddehase, M. J., F. Weiland, K. Munch, S. Jonjic, A. Luske, and U. H. Koszinowski. 1985. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J. Virol. 55:264-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scalzo, A. A. 2002. Successful control of viruses by NK cells—a balance of opposing forces? Trends Microbiol. 10:470-474. [DOI] [PubMed] [Google Scholar]
- 36.Scalzo, A. A., M. G. Brown, D. T. Chu, J. W. Heusel, W. M. Yokoyama, and C. A. Forbes. 1999. Development of intra-natural killer complex (NKC) recombinant and congenic mouse strains for mapping and functional analysis of NK cell regulatory loci. Immunogenetics 49:238-241. [DOI] [PubMed] [Google Scholar]
- 37.Scalzo, A. A., N. A. Fitzgerald, A. Simmons, A. B. La Vista, and G. R. Shellam. 1990. Cmv-1, a genetic locus that controls murine cytomegalovirus replication in the spleen. J. Exp. Med. 171:1469-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scalzo, A. A., N. A. Fitzgerald, C. R. Wallace, A. E. Gibbons, Y. C. Smart, R. C. Burton, and G. R. Shellam. 1992. The effect of the Cmv-1 resistance gene, which is linked to the natural killer cell gene complex, is mediated by natural killer cells. J. Immunol. 149:581-589. [PubMed] [Google Scholar]
- 39.Scalzo, A. A., P. A. Lyons, N. A. Fitzgerald, C. A. Forbes, W. M. Yokoyama, and G. R. Shellam. 1995. Genetic mapping of Cmv1 in the region of mouse chromosome 6 encoding the NK gene complex-associated loci Ly49 and musNKR-P1. Genomics 27:435-441. [DOI] [PubMed] [Google Scholar]
- 40.Sjolin, H., E. Tomasello, M. Mousavi-Jazi, A. Bartolazzi, K. Karre, E. Vivier, and C. Cerboni. 2002. Pivotal role of KARAP/DAP12 adaptor molecule in the natural killer cell-mediated resistance to murine cytomegalovirus infection. J. Exp. Med. 195:825-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, H. R., H. H. Chuang, L. L. Wang, M. Salcedo, J. W. Heusel, and W. M. Yokoyama. 2000. Nonstochastic coexpression of activation receptors on murine natural killer cells. J. Exp. Med. 191:1341-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, H. R., J. W. Heusel, I. K. Mehta, S. Kim, B. G. Dorner, O. V. Naidenko, K. Iizuka, H. Furukawa, D. L. Beckman, J. T. Pingel, A. A. Scalzo, D. H. Fremont, and W. M. Yokoyama. 2002. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. USA 99:8826-8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, K. M., J. Wu, A. B. Bakker, J. H. Phillips, and L. L. Lanier. 1998. Ly-49D and Ly-49H associate with mouse DAP12 and form activating receptors. J. Immunol. 161:7-10. [PubMed] [Google Scholar]
- 44.Tay, C. H., and R. M. Welsh. 1997. Distinct organ-dependent mechanisms for the control of murine cytomegalovirus infection by natural killer cells. J. Virol. 71:267-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor, B. A., C. Wnek, B. S. Kotlus, N. Roemer, T. MacTaggart, and S. J. Phillips. 1999. Genotyping new BXD recombinant inbred mouse strains and comparison of BXD and consensus maps. Mamm. Genome 10:335-348. [DOI] [PubMed] [Google Scholar]
- 46.Thale, R., U. Szepan, H. Hengel, G. Geginat, P. Lucin, and U. H. Koszinowski. 1995. Identification of the mouse cytomegalovirus genomic region affecting major histocompatibility complex class I molecule transport. J. Virol. 69:6098-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomasello, E., L. Olcese, F. Vely, C. Geourgeon, M. Blery, A. Moqrich, D. Gautheret, M. Djabali, M. G. Mattei, and E. Vivier. 1998. Gene structure, expression pattern, and biological activity of mouse killer cell activating receptor-associated protein (KARAP)/DAP-12. J. Biol. Chem. 273:34115-34119. [DOI] [PubMed] [Google Scholar]
- 48.Voigt, V., C. A. Forbes, J. N. Tonkin, M. A. Degli-Esposti, H. R. Smith, W. M. Yokoyama, and A. A. Scalzo. 2003. Murine cytomegalovirus m157 mutation and variation leads to immune evasion of natural killer cells. Proc. Natl. Acad. Sci. USA 100:13483-13488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner, M., A. Gutermann, J. Podlech, M. J. Reddehase, and U. H. Koszinowski. 2002. Major histocompatibility complex class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J. Exp. Med. 196:805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner, M., D. Michel, P. Schaarschmidt, B. Vaida, S. Jonjic, M. Messerle, T. Mertens, and U. Koszinowski. 2000. Comparison between human cytomegalovirus pUL97 and murine cytomegalovirus (MCMV) pM97 expressed by MCMV and vaccinia virus: pM97 does not confer ganciclovir sensitivity. J. Virol. 74:10729-10736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welsh, R. M., J. O. Brubaker, M. Vargas-Cortes, and C. L. O'Donnell. 1991. Natural killer (NK) cell response to virus infections in mice with severe combined immunodeficiency. The stimulation of NK cells and the NK cell-dependent control of virus infections occur independently of T and B cell function. J Exp. Med 173:1053-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welsh, R. M., C. L. O'Donnell, and L. D. Shultz. 1994. Antiviral activity of NK1.1+ natural killer cells in C57BL/6 scid mice infected with murine cytomegalovirus. Nat. Immun. 13:239-245. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.