Abstract
The study was conducted to analyze the genetic parameters of somatic cell score (SCS) of Holstein cows, which is an important indicator to udder health. Test-day records of somatic cell counts (SCC) of 305-day lactation design from first to fifth lactations were collected on Holsteins in Korea during 2000 to 2012. Records of animals within 18 to 42 months, 30 to 54 months, 42 to 66 months, 54 to 78 months, and 66 to 90 months of age at the first, second, third, fourth and fifth parities were analyzed, respectively. Somatic cell scores were calculated, and adjusted for lactation production stages by Wilmink’s function. Lactation averages of SCS (LSCS1 through LSCS5) were derived by further adjustments of each test-day SCS for five age groups in particular lactations. Two datasets were prepared through restrictions on number of sires/herd and dams/herd, progenies/sire, and number of parities/cow to reduce data size and attain better relationships among animals. All LSCS traits were treated as individual trait and, analyzed through multiple-trait sire models and single trait animal models via VCE 6.0 software package. Herd-year was fitted as a random effect. Age at calving was regressed as a fixed covariate. The mean LSCS of five lactations were between 3.507 and 4.322 that corresponded to a SCC range between 71,000 and 125,000 cells/mL; with coefficient of variation from 28.2% to 29.9%. Heritability estimates from sire models were within the range of 0.10 to 0.16 for all LSCS. Heritability was the highest at lactation 2 from both datasets (0.14/0.16) and lowest at lactation 5 (0.11/0.10) using sire model. Heritabilities from single trait animal model analyses were slightly higher than sire models. Genetic correlations between LSCS traits were strong (0.62 to 0.99). Very strong associations (0.96 to 0.99) were present between successive records of later lactations. Phenotypic correlations were relatively weaker (<0.55). All correlations became weaker at distant lactations. The estimated breeding values (EBVs) of LSCS traits were somewhat similar over the years for a particular lactation, but increased with lactation number increment. The lowest EBV in first lactation indicated that selection for SCS (mastitis resistance) might be better with later lactation records. It is expected that results obtained from these multi-trait lactation model analyses, being the first large scale SCS data analysis in Korea, would create a good starting step for application of advanced statistical tools for future genomic studies focusing on selection for mastitis resistance in Holsteins of Korea.
Keywords: Somatic Cell Score, Sire Model, Animal Model, Multiple-trait, Genetic Parameters, Holstein
INTRODUCTION
Most dairy breeding programs aim at the improvement in milk production by selection which consequently lead to decreased udder health, reproduction performances of cows (Dematawewa and Berger, 1998; Negussie et al., 2008) and impose increasing production costs on milk. Mastitis, as an udder disease, always captures the most interest being the highly frequent and costly dairy disease and, eventually leads to an involuntary and premature culling of milking cows for defective udder characteristics and decreased milk yield.
Given the economic importance of mastitis, selection for udder health or resistance to mastitis becomes very important consideration in dairy breeding programs. But genetic evaluation of mastitis is particularly difficult as it is a low heritable trait and categorical in nature (Rupp and Boichard, 1999; Carlen et al., 2004; Negussie et al., 2008; Bloemhof at al., 2009). Instead, the somatic cell count (SCC) is considered to be the most suitable indicator trait for mastitis resistance in view of its medium to high genetic correlation with mastitis (0.36 to 0.98) and its greater heritability than mastitis (Mrode and Swanson, 1996; Poso and Mantysaari, 1996; Rupp and Boichard, 1999; Carlen et al., 2004; Negussie et al., 2008; Bloemhof et al., 2009). The SCC is also easy to record. The selection for lower SCC in milk has a positive effect against the incidence of mastitis (Rupp and Boichard, 1999; Nash et al., 2000). Selection against high SCC also does not deteriorate the immune system of cattle and decreases a risk of infection at the same time (Rupp et al., 2000; Boettcher et al., 2002; Odegard et al., 2003; Madsen et al., 2008). However, the genetic evaluation of this trait is mostly based on somatic cell score (SCS), a logarithmic transformation of SCC to achieve normality of distribution (Ali and Shook, 1980).
In Korea, Holsteins are the major source of domestic milk production. Recently, an active participation of Korea in the INTERBULL dairy evaluation programs for Holstein cattle greatly emphasizes the analysis of large scale lactation data. Local breeders and producers are also little aware about the genetic contribution for the deterioration in udder health of Holsteins. Except some studies for milk production traits, no study has reported a large scale evaluation on SCS yet. For this reason, as a priori, the present study was aimed to estimate the genetic parameters for SCS of Holstein cows using multi-trait lactation models.
MATERIALS AND METHODS
Animals, data recording and data transformation
A total of 1,409,052 test-day lactation SCC raw records were collected from first five parities of Holstein cows in South Korea from 2000 and 2012. Records of SCC less than 6 thousand or greater than 3 million cells/mL of milk were discarded from the original dataset. Also, the SCC with test dates recorded before 5 days in milk (DIM) and after 305 DIM, and less than 5 SCC records/lactation of a cow were removed. Initial edits on the data also included SCC records from cows that have calved from 18 to 42 mo, 30 to 54 mo, 42 to 66 mo, 54 to 78 mo, and 66 to 90 mo of age in the first, second, third, fourth, and fifth calving, respectively.
As SCC data usually have a non-normal distribution, a logarithmic scaling of SCC was carried out, as suggested by earlier studies, to achieve a data with more normal distribution, increased heritability, and reduced heterogeneity of within-herd and within-sire variances (Shook, 2006). Each of the test-day SCC was transformed to SCS as SCS = Log2 (SCC/100)+4. Instead of a constant 3 which most studies used, a constant 4 was used to avoid the problem of negative value in the data (Martins et al., 2011). Each of the test-day SCS was adjusted by Wilmink’s (1987) function for milking stage of the cow due to the changing pattern of SCS within its production trajectory. The adjustment function used was yij=a0+a1tij +a2exp(−0.05tij) +eij, where yij is a record of SCS for ith animal at jth test-day, eij is the residual errors, tij is the difference in days between jth test-date and calving-date of the ith animal, and a0, a1, and a2 are the regression coefficients of SCS (yij) on tij and exp(−0.05tij) estimated within each lactation stage. An average lactation SCS (LSCS) was obtained from all test-day SCSs which were finally adjusted for lactation age groups (5 groups/parity; at 5 months interval each).
Finally, two datasets were created applying restrictions for herd, number of sires and dams, and number of LSCS records/animal. For both datasets, record of a herd that had at least 3 selected sires and 3 dams were included. And a sire was selected if it contributed minimum 10 progeny records from 5 herds. Also, an animal was included if it had at least two (dataset 1) or three consecutive parity records (dataset 2). Above restrictions were implemented repeatedly through an iterative process until a dataset was stabilized. Above basic edits were performed on original dataset to obtain a computational feasibility and a better linked data structure for genetic variance components.
Animal pedigree
Animal pedigrees were prepared separately for animal and sire models according to the datasets. A total of 3,641 and 3,152 animals were found for the sire model analyses of dataset 1 and 2, respectively. Respective pedigrees were traced back to 16 and 15 generations as well. For animal models, pedigrees sizes for dataset 1 and 2 were 252,917 and 151,589 animals, respectively, with a pedigree depth of 20 generations for each. About 32% to 36% inbred were found among pedigree files. Most of the inbred (86% to 95%), which generally found to be in the parent generations, had inbreeding coefficient less than 0.05 among pedigree sets. Average inbreeding coefficients among pedigree datasets varied from 0.007 to 0.009.
Data analysis
The datasets for analyses contained a total of 264,716 and 81,955 animal records, respectively. Sire model and animal model analyses were carried out using both datasets. In the analysis each LSCS record was considered as individual traits. The herd-year (HY) classes were created by merging respective herd number and calving year, and fitted as a random effect in various models. Age at calving of animals was regressed with the response variable as a fixed covariate. All factors were fitted parity-wise to their respective LSCS using either multivariate sire models or univariate animal models that employed the restricted maximum likelihood approach, to estimates the components of variances, covariances and genetic parameters of each trait. Genetic variances and covariances were estimated using the VCE 6.0 software package (Neumaier and Groeneveld, 1998). The genetic model applied to calculate additive genetic and residual variances, in matrix notation, was as follows:
where y is the vector of observations, b is the vector of fixed effects and covariates, u is the vector of random additive effects, q is the vector of random HY effects, e is the vector of random residual effects, and X, Z, and W are known design matrices relating observations to the fixed and random effects b, u and q respectively. The assumptions for distribution of y, u, q and e can be described as
where G is the variance-covariance matirx of the random effects vector u, H is the variance-covariance matrix of the random effects vector q, and R is the matrix of residual variances and covariances. The G, H, and R matrices are described as G = A⊗G0, where A is the animal relationship matrix, G0 is the additive genetic variance-covariance matrix between traits, and ⊗ is the direct product operator, H = I⊗H0 and, R = I⊗R0, where I is the identity matrix of the same order as y, H0 is the matrix of HY variance-covariance between traits, R0 is the matrix of residual variance-covariance between traits, 0 is the null vector, and Φ is the null matrix.
Next, we calculated heritabilities (h2) for traits using (co)variance estimates of sire and animal models. Genetic and phenotypic correlations were calculated from the (co)variance estimates of the multivariate sire models only. To obtain h2 from sire model, the estimated sire variance for a trait was multiplied by 4, as it is known that sire model can obtain only 1/4th of the total genetic variances. Genetic trends for LSCS in dataset 1 were obtained from estimated breeding values (EBV) for all animals using (co)variances obtained by sire-models, which were derived by PEST software package (Groeneveld et al., 1990).
RESULTS AND DISCUSSION
Descriptive statistics of the datasets
Summary data on lactation average SCSs in dataset 1 and 2 are shown in Table 1 and 2. Number of records from fifth parity was comparatively lower than earlier lactations. In dataset 1, the average LSCS among parities varied from 3.507±1.09 to 4.322±1.48, corresponding to a range between 71,000 to 125,000 cells/mL. Dataset 2 also derived similar average LSCS in Holsteins. Average LSCS were increased with the increment of lactations number in the cows, although differences in means were little (Figure 1). We obtained generally low to moderate coefficient of variation (CV) for LSCS traits which ranged from 28.2% to 29.9%.
Table 1.
Structure of the somatic cell score (SCS) datasets of the Holstein cows lactation-wise1
| Item | Dataset | LSCS1 | LSCS2 | LSCS3 | LSCS4 | LSCS5 |
|---|---|---|---|---|---|---|
| No. of records | 1 | 146,738 | 153,680 | 89,135 | 46,299 | 20,976 |
| 2 | 77,065 | 80,601 | 81,954 | 41,770 | 19,165 | |
| No. of sires | 1 | 797 | 822 | 804 | 744 | 655 |
| 2 | 672 | 683 | 685 | 653 | 583 | |
| No. of dams | 1 | 112,524 | 116,671 | 71,538 | 38,861 | 18,253 |
| 2 | 63253 | 65,653 | 66,503 | 35,579 | 16,935 | |
| No. of herds | 1 | 3,818 | 3,831 | 3,748 | 3,431 | 2,833 |
| 2 | 3,126 | 3,130 | 3,130 | 3,037 | 2,619 | |
| No. of contemporary groups (herd-year) | 1 | 25,886 | 27,728 | 23,064 | 17,278 | 10,874 |
| 2 | 18,917 | 20,235 | 21,213 | 15,603 | 9,958 | |
| Average age (mo) | 1 | 25.85 | 40.02 | 53.22 | 66.00 | 78.65 |
| 2 | 25.56 | 39.21 | 53.18 | 66.03 | 78.66 |
LSCS, lactation averages of SCS
LSCS1 to 5 are consecutive lactation average somatic cell scores for 1st to 5th lactations.
Table 2.
Descriptive statistics1 on datasets of somatic cell scores for different LSCS traits in Holsteins between 2000 and 2012
| Trait | N | Mean | SD | Min | Max | CV |
|---|---|---|---|---|---|---|
| LSCS1 | 146,738 (77,065) | 3.507 (3.462) | 1.096 (1.075) | 0.165 (0.165) | 8.222 (8.666) | 29.9 (29.9) |
| LSCS2 | 153,680 (80,601) | 3.767 (3.662) | 1.247 (1.182) | 0.121 (0.121) | 9.774 (9.267) | 29.6 (29.7) |
| LSCS3 | 89,135 (81,954) | 4.028 (4.003) | 1.329 (1.319) | 0.365 (0.365) | 10.379 (10.379) | 28.6 (28.7) |
| LSCS4 | 46,299 (41,770) | 4.194 (4.168) | 1.412 (1.401) | 0.448 (0.448) | 10.249 (10.249) | 28.3 (28.4) |
| LSCS5 | 20,976 (19,165) | 4.322 (4.288) | 1.480 (1.473) | 0.189 (0.189) | 10.268 (9.864) | 28.2 (28.3) |
LSCS, lactation averages of somatic cell score; N, number of records; SD, standard deviation of the mean; Min, minimum score; Max, maximum score; CV, coefficient of variation.
Values without and within parentheses indicate dataset 1 and 2, respectively.
Figure 1.
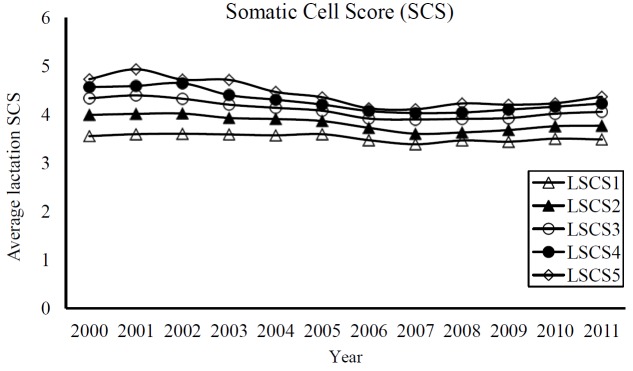
Phenotypic trends for lactation average somatic cell scores (LSCS) in Holstein cows (LSCS1, LSCS2, LSCS3, LSCS4, and LSCS5 are traits of 1st, 2nd, 3rd, 4th, and 5th lactations, respectively).
Despite the difference in use of a constant 4 to obtain a log-transformed SCS where many studies used a value of 3 or that related to differences in measurement of mean, our obtained average LSCSs were somewhat in agreement with earlier reports. Similar averages in Holsteins for logSCC by Ivkić et al. (2012; 3.31), for first parity SCS by Ptak et al. (2009a; 3.51), and for lactation SCS and daily SCS by Ptak et al. (2009b; 3.71, 3.49). Our means were comparable to means found in French Holsteins by Biochard and Rupp (1997), and Rupp and Biochard (1999). However, slightly greater SCS observed in Finnish Holstein by Luttinen and Juga (1997) and Dube et al. (2008). A study of Portuguese Holstein-Friesians by Martins et al. (2011) also showed a similar increasing pattern of SCS as the number of parity increased. This increment of SCS over lactations generally occurs in a dairy breed with faster milk flow, in which the increased milk flow causes an animal more prone to mastitis. The trend of fairly stable and moderate LSCSs found in the study (Figure 2) indicates that mastitis infection in Korean Holstein population was found often; generally in cows with more consecutive parity records.
Figure 2.
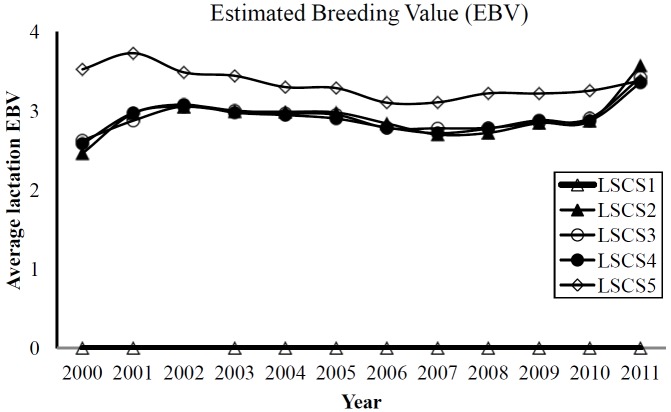
Genetic trends for estimated breeding value of lactation somatic cell scores (LSCS) in Holstein (LSCS1, LSCS2, LSCS3, LSCS4, and LSCS5 are traits of 1st, 2nd, 3rd, 4th, and 5th lactations, respectively).
Heritability estimates
The variance components, heritability estimates of traits derived via multi-trait sire and animal model analyses are shown in Table 3 and 4, respectively. A highest heritability estimate of 0.14 was found in lactation 2 and 4 using sire models. Heritabilities (h2) among parities obtained through sire model analyses were more or less similar among datasets, especially for h2 of LSCS2 and LSCS3. In contrast, the univariate animal model heritabilities were higher than those from sire model estimates and, the observed differences in h2 estimates among datasets were negligible too. However, the analyses using all datasets yielded lowest heritabilities for LSCS5 trait. This is probably because of the bias caused by noticeably fewer records from 5th parity cows as compared to other parities, and resulted underestimated genetic variances for the respective trait; whereas, gradually increased genetic variances were noticed from parity 1 to 4 traits. Another source of bias in the 5th lactation estimates (LSCS5) using animal model might be associated with the presence of noticeably fewer dams than other parities, which was less influential to sire model estimates as expected. With the parity number increases, the proportional increases of variance for herd-year and residual effects deemed relatively higher than the additive genetic variance, indicating more environmental influences on SCS increase. A higher SCS also indicates the consequences of more infection of mastitis in the udder. Environmental variances were found to be relatively lower in the animal model analyses than those found in the sire models, as an opposite of the genetic variances, and those are possibly due to the presence of complete pedigree relationships in the animal model analyses.
Table 3.
Estimates of variance components for LSCS traits from different datasets1
| Trait | Parameter | Sire model | Animal model | ||
|---|---|---|---|---|---|
|
|
|
||||
| Dataset 1 | Dataset 2 | Dataset 1 | Dataset 2 | ||
| LSCS1 | G | 0.037 | 0.040 | 0.235 | 0.258 |
| HY | 0.117 | 0.098 | 0.171 | 0.158 | |
| E | 1.045 | 1.195 | 0.839 | 0.781 | |
| LSCS2 | G | 0.054 | 0.053 | 0.317 | 0.303 |
| HY | 0.181 | 0.116 | 0.331 | 0.302 | |
| E | 1.272 | 1.195 | 0.957 | 0.837 | |
| LSCS3 | G | 0.062 | 0.056 | 0.343 | 0.333 |
| HY | 0.164 | 0.134 | 0.387 | 0.379 | |
| E | 1.524 | 1.510 | 1.085 | 1.081 | |
| LSCS4 | G | 0.074 | 0.070 | 0.348 | 0.327 |
| HY | 0.151 | 0.135 | 0.435 | 0.433 | |
| E | 1.811 | 1.793 | 1.249 | 1.250 | |
| LSCS5 | G | 0.064 | 0.055 | 0.201 | 0.159 |
| HY | 0.214 | 0.204 | 0.538 | 0.533 | |
| E | 2.008 | 2.006 | 1.452 | 1.489 | |
LSCS, lactation averages of somatic cell score; G, additive genetic variance; HY, herd-year class; E, residual variance.
LSCS1 to 5, lactation average somatic cell score traits for 1st to 5th parity.
Variance estimates corresponded to single-trait and multiple-trait analyses using animal and sire models, respectively.
Table 4.
Heritability estimates of somatic cell scores of Holstein through multiple-trait (sire model) and single-trait (animal model) lactation analyses1
| Trait | Sire model | Animal model | ||
|---|---|---|---|---|
|
|
|
|||
| Dataset 1 | Dataset 2 | Dataset 1 | Dataset 2 | |
| LSCS1 | 0.12 | 0.14 | 0.19 | 0.22 |
| LSCS2 | 0.14 | 0.16 | 0.20 | 0.21 |
| LSCS3 | 0.13 | 0.14 | 0.19 | 0.19 |
| LSCS4 | 0.14 | 0.14 | 0.17 | 0.16 |
| LSCS5 | 0.11 | 0.10 | 0.10 | 0.07 |
LSCS1 to 5 are lactation average somatic cell scores for 1st to 5th lactations, respectively.
Generally, a low to medium range of heritability was reviewed in previous reports, such as 0.05 to 0.29 (Schutz et al., 1990) or 0.06 to 0.13 (Ptak et al., 2009a). However, most of the authors have published h2 estimates for LSCS between 0.10 and 0.27 (Monardes et al., 1990; Mrode and Swanson, 1996; Rupp and Boichard, 1999; Mark and Sullivan, 2005). Rupp and Boichard (2003) reviewed that LSCS trait, obtained by averaging the individual test day records, shows consistently higher h2 estimate around 0.15. Estimates of heritabilities in this study deemed very consistent with most of these earlier studies. Further agreement is found in Austrian Fleckvieh cows which showed a mean h2 of 0.09 to 0.13 for SCS traits in first five lactations (Koeck et al., 2010). Heritabilities obtained by Kadarmideen (2004) and Charfeddine et al. (1997) are also comparable (0.13 to 0.14). Our estimates in first three lactations are concordant with Swedish Holstein (Carlen et al., 2004), and Polish Holstein (Rzewuska et al., 2011), even though traits were defined differently. Similar h2 estimates were obtained from the repeated measures test-day SCS model for first three lactations (Reents et al., 1995). However, some disagreements were observed from several reports (Haile-Mariam et al., 2001; Mrode and Swanson, 2003; Heringstad et al., 2008; Dube et al., 2008; Martins et al., 2011; Ivkić et al., 2012), where h2 estimates tend to be higher or lower than those estimates found in this study. Increase of genetic variances and heritability estimates along with the increment in parity numbers have also been reported in earlier studies (Schutz et al., 1990; Samore et al., 2002; Muir et al., 2007; Ptak et al., 2007; Martins et al., 2011).
Genetic and phenotypic correlations
Genetic correlations among LSCS traits derived via sire models are shown in Table 5. The estimates of genetic correlation deemed strong among lactation SCSs. The highest genetic correlation was observed between LSCS4 and LSCS5 (0.99) whereas, the lowest genetic correlation was found between LSCS1 and LSCS5 (0.62). Genetic correlations were generally strong between successive lactations (0.82 to 0.99), and tended to decrease between distant lactations. Sire model analysis using dataset 2 also derived similar estimates (Table 5). The phenotypic correlations among LSCS traits, however, were expectedly low to moderate. The highest phenotypic association of 0.54 was observed between LSCS4 and LSCS5 in the study. Phenotypic correlations between other lactations ranged between 0.22 and 0.51. Genetic trends of LSCS traits were shown in Figure 2. Even though it was found that heritabilities increased slightly with more parity records per cow (dataset 2), their genetic correlations rather deemed less influenced. In fact, more animals with at least two successive parity records were sufficient to obtain similar correlations as estimated from animals having more than 2 successive parity records.
Table 5.
Genetic (upper diagonal) and phenotypic (lower diagonal) correlations between Holstein lactations using multiple-trait lactation sire models1
| Trait | LSCS1 | LSCS2 | LSCS3 | LSCS4 | LSCS5 |
|---|---|---|---|---|---|
| LSCS1 | - | 0.82 (0.80) | 0.73 (0.72) | 0.68 (0.68) | 0.62 (0.59) |
| LSCS2 | 0.34 (0.36) | - | 0.97 (0.96) | 0.89 (0.87) | 0.83 (0.78) |
| LSCS3 | 0.27 (0.28) | 0.47 (0.48) | - | 0.96 (0.96) | 0.91 (0.89) |
| LSCS4 | 0.25 (0.25) | 0.39 (0.40) | 0.51 (0.51) | - | 0.99 (0.98) |
| LSCS5 | 0.22 (0.22) | 0.34 (0.34) | 0.44 (0.44) | 0.54 (0.54) | - |
LSCS1 to 5, lactation average somatic cell scores for 1st to 5th lactations.
Values without and within parentheses indicate estimates obtained using datasets 1 and 2, respectively.
For correlations between traits, there was a harmony between the present study and previous reports. The strong genetic correlations among traits in the present study are found to be similar with estimates by Dube et al. (2008), Reents et al. (1995), Carlen et al. (2004), Mostert et al. (2004), and Mrode and Swanson (1996). A review by Rupp and Boichard (2003) also depicted similar estimates. A greater agreement for strong genetic correlation between adjacent LSCS was also observed from studies (Haile-Mariam et al., 2001; Koivula et al., 2002, 2004; Odegard et al., 2003; Negussie et al., 2006). These high estimates of genetic correlation (greater than 0.8) indicate that genetic determinism of SCC is close within and across lactations. Phenotypic correlations were also are concordant with earlier studies.
Phenotypically, the yearly trends of LSCS (Figure 1) were consistently lower and higher LSCS at lactation 1 and 5, respectively. Interestingly, the genetic trend of LSCS1 using EBV of animals from dataset 1 (Figure 2) showed fairly consistent and very low EBVs in animals across the years for the same trait. Estimated breeding value of LSCS2, LSCS3 and LSCS4, although higher than EBV of LSCS1, showed somewhat similar trends, indicating less degradations over time. The observed EBVs indicate that genetic merit for high SCS (susceptibility to mastitis) in the first lactation of Holsteins is negligible but increases as lactation number progresses. Evidently, these results seem to be in line with the claim that first lactation trait probably is a different trait from second and later lactations (Reents et al., 1995). They found more incidence of mastitis in later lactations which is also similar to our study. Reents et al. (1995) further suggested that SCS from later lactations might be a better indicator for mastitis susceptibility than only first lactation SCS. Shook and Schutz (1994) also suggested use of later lactation records, ideally in a multiple-trait model.
Mastitis resistance is a complex trait that depends on genetic component as well as physiological and environmental factors including infection pressure (Rupp and Biochard, 2003). It is difficult to improve this trait by udder health management only when its genetic trend shows an increment. So, a selection for reducing milk SCC through breeding approaches should be considered. Otherwise, if SCC is kept unnoticed the SCC could increase due to correlated response when intense selection for high milk yield is performed. The selection of rear udder height and udder cleft type traits can also result the same. Contrarily, if SCS is the only criteria for resistance against mastitis, the response will be slow because of its lower heritability, which perhaps will even be slowed down by the lack of perfect genetic correlation between SCS and mastitis (Koivula et al., 2005). So, the best way to SCS evaluations might be through an economic index with a lesser emphasis on SCS relative to the most desired high milk yield traits; otherwise, a greater emphasis would decrease genetic gain in yield traits which are economically more important (Schutz, 1994).
Statistical methods for SCS evaluation vary widely among reports such as, lactation average model, test-day model, or random regression models. Each of them provides particular advantages than the other. Lactation average model is the simplest model of evaluation where a LSCS, the mean (or a weighted mean) of the test-day SCS are used. Considering high correlations among lactations test-day models using monthly repeated records of SCC were also developed (Reents et al., 1995). Random regression model, which allows more accurate modelling of the each test-day records, supports more complex statistical methods, but are computationally limited in many countries computation infrastructure. As a result, the preferred statistical model in earlier reports commonly reflected their available data structure, and proper computational tools to analyze them. In this study, we considered the lactation average model, based on the current limitation with the data structure (i.e. missing lactation records/animal), and computation platform. The choice of LSCS is justified by the common genetic determinism of SCS all along the lactation (Rupp and Biochard, 2003). In addition to data pruning and transformation, we fitted SCS with test-day (DIM) via Wilmink’s function, which performed an adjustment for the test-days as well as for the time at minimum SCS occurs (50 days post-partum approximately), to better fit the lactation curve of animals and to obtain less biased estimates. Further, the consideration of multi-trait lactation model, due to high correlation between lactation data, also exploited the existing (co)variance between lactation records.
To conclude, it can be noted that genetic parameters in this study are consistent with many previous reports. Heritability estimates and (co)variances components indicate the possibility of selection for SCS or mastitis resistance. Genetic correlations are favorable for improvement of one lactation SCS trait by selection of another LSCS trait. Although the evidence for genetic merit degradation of SCS traits is not strong, it is not to be ignored either. And, the EBV estimate indicates that it will be better if later lactation records are given priority for selection than only first lactation SCS. Strongly correlated successive LSCS traits in this study also suggest that multiple-trait evaluation may yield better result against selection of mastitis. It is expected that these results will be useful in setting up breeding targets, and selection criteria for dairy development in Korea and, as a valuable resource while working alongside the research goals of INTERBULL. We hope that the estimated variances and covariances will provide a better step to start with and compare estimates when random regression models will be implemented for the genomic evaluation of Korean Holsteins.
ACKNOWLEDGMENTS
This research was supported by the funding from the project “Development of economically important traits and technologies for sustainable dairy production (Project No: PJ907160)” and the “Postdoctoral Fellowship Program” of the National Institute of Animal Science, RDA, Cheonan, Korea.
REFERENCES
- Ali AKA, Shook GE. An optimum transformation for somatic cell concentration in milk. J Dairy Sci. 1980;63:487–490. [Google Scholar]
- Bloemhof S, de Jong G, de Haas Y. Genetic parameters for clinical mastitis in the first three lactations of Dutch Holstein cattle. Vet Microbiol. 2009;134:165–171. doi: 10.1016/j.vetmic.2008.09.024. [DOI] [PubMed] [Google Scholar]
- Boettcher P, Samore A, Pagnacco G. Relationships between normal levels of somatic cells and the duration of mastitis infections. Proceedings of 7th World Congress on Genetics Applied to Livestock Production (WCGALP); August 2002; Montpellier, France. 2002. pp. 9–23. [Google Scholar]
- Boichard D, Rupp R. INTERBULL Bull No. 15. Int Bull Eval Serv; Uppsala, Sweden: 1997. Genetic analysis and genetic evaluation for somatic cell score in French dairy cattle; pp. 54–60. [Google Scholar]
- Carlen E, Strandberg E, Roth A. Genetic parameters for clinical mastitis, somatic cell score, and production in the first three lactations of Swedish Holstein cows. J Dairy Sci. 2004;87:3062–3070. doi: 10.3168/jds.S0022-0302(04)73439-6. [DOI] [PubMed] [Google Scholar]
- Charfeddine N, Alenda R, Carabaño MJ. Relationships between somatic cell score and longevity, production and type traits in Spanish Holstein-Friesian cows. Proceedings of the 48th Annual Meeting of the EAAP; August 25–28 1997; Vienna, Austria. 1997. p. 7. [Google Scholar]
- Dematawewa CMB, Berger PJ. Genetic and phenotypic parameters for 305-day yield, fertility, and survival in Holsteins. J Dairy Sci. 1998;81:2700–2709. doi: 10.3168/jds.S0022-0302(98)75827-8. [DOI] [PubMed] [Google Scholar]
- Dube B, Dzama K, Banga CB. Genetic analysis of somatic cell score and udder type traits in South African Holstein cows. S Afr J Anim Sci. 2008;38:1–11. [Google Scholar]
- Groeneveld E, Kovac M, Wang T. PEST, a general purpose BLUP package for multivariate prediction and estimation. Proceedings of the 4th World Congress on Genetics applied to Livestock Production. XIII. Plenary lectures, molecular genetics and mapping, selection, prediction and estimation; July 23–27 1990; Edinburgh, UK. 1990. pp. 488–491. [Google Scholar]
- Haile-Mariam M, Goddard ME, Bowman PJ. Estimates of genetic parameters for daily somatic cell count of Australian dairy cattle. J Dairy Sci. 2001;84:1255–1264. doi: 10.3168/jds.S0022-0302(01)74587-0. [DOI] [PubMed] [Google Scholar]
- Heringstad B, Sehested E, Steine T. Correlated selection responses in somatic cell count from selection against clinical mastitis. J Dairy Sci. 2008;91:4437–4439. doi: 10.3168/jds.2008-1330. [DOI] [PubMed] [Google Scholar]
- Ivkić Z, Špehar M, Bulić V, Mijić P, Ivanković A, Solić D. Estimation of genetic parameters and environmental effects on somatic cell count in Simmental and Holstein breeds. Mljekarstvo. 2012;62:143–150. [Google Scholar]
- Kadarmideen H. Genetic correlations among body condition score, somatic cell score, milk production, fertility and conformation traits in dairy cows. Anim Sci. 79(2004):191–201. [Google Scholar]
- Koeck A, Heringstad B, Egger-Danner C, Fuerst C, Winter P, Fuerst-Walt B. Genetic analysis of clinical mastitis and somatic cell count traits in Austrian Fleckvieh cows. J Dairy Sci. 2010;93:5987–5995. doi: 10.3168/jds.2010-3451. [DOI] [PubMed] [Google Scholar]
- Koivula M, Mantysaari EA, Negussie E, Serenius T. Genetic and phenotypic relationships among milk yield and somatic cell count before and after clinical mastitis. J Dairy Sci. 2005;88:827–833. doi: 10.3168/jds.S0022-0302(05)72747-8. [DOI] [PubMed] [Google Scholar]
- Koivula M, Negussie E, Mantysaari EA. Genetic parameters for test-day somatic cell count at different stages of lactation in Finnish Ayrshire cattle. Proceedings of 7th World Congress on Genetics Applied to Livestock Production (WCGALP); August 2002; Montpellier, France. 2002. pp. 9–20. [Google Scholar]
- Koivula M, Negussie E, Mantysaari EA. Genetic parameters for test-day somatic cell count at different lactation stages of Finnish dairy cattle. Livest Prod Sci. 2004;90:145–157. [Google Scholar]
- Luttinen A, Juga J. INTERBULL Bull No. 15. Int Bull Eval Serv; Uppsala, Sweden: 1997. Genetic relationship between milk yield, somatic cell count, mastitis, milkability and leakage in Finnish dairy cattle; pp. 78–83. [Google Scholar]
- Madsen P, Jensen J. A Package for Analysing Multivariate Mixed Models, Version 6, Release 4.7. Danish Institute of Agricultural Sciences; Tjele, Denmark: 2008. An User’s Guide to DMU. [Google Scholar]
- Mark T, Sullivan PG. Multiple-trait-multiple-country genetic evaluations for udder health. Interbull Bull. 2005;33:8–15. doi: 10.3168/jds.S0022-0302(06)72536-X. [DOI] [PubMed] [Google Scholar]
- Martins AM, Silvestre AM, Petim-Batista MF, Colaço JA. Somatic cell score genetic parameter estimates of dairy cattle in Portugal using fractional polynomials. J Anim Sci. 2011;89:1281–1285. doi: 10.2527/jas.2010-3211. [DOI] [PubMed] [Google Scholar]
- Monardes HG, Cue RI, Hayes JF. Correlations between udder conformation traits and somatic cell count in Canadian Holstein cows. J Dairy Sci. 1990;73:1337–1342. doi: 10.3168/jds.S0022-0302(90)78800-5. [DOI] [PubMed] [Google Scholar]
- Mostert BE, Banga C, Groeneveld E, Kanfer FHJ. Breeding value estimation for somatic cell score in South African dairy cattle. S Afr J Anim Sci. 2004;34(Suppl 2):32–34. [Google Scholar]
- Mrode RA, Swanson GJT. Genetic and statistical properties of somatic cell count and its suitability as an indirect means of reducing the incidence of mastitis in dairy cattle. Anim Breed Abstr. 1996;64:847–857. [Google Scholar]
- Mrode RA, Swanson GJT. Estimation of genetic parameters for somatic cell count in the first three lactations using random regression. Livest Prod Sci. 2003;79:239–247. [Google Scholar]
- Muir BL, Kistemaker G, Jamrozik J, Canavesi F. Genetic parameters for a multiple-trait multiple-lactation random regression test-day model in Italian Holsteins. J Dairy Sci. 2007;90:1564–1574. doi: 10.3168/jds.S0022-0302(07)71642-9. [DOI] [PubMed] [Google Scholar]
- Nash DL, Rogers GW, Cooper JB, Hargrove GI, Keown JF, Hansen LB. Heritability of clinical mastitis incidence and relationships with sire transmitting abilities for somatic cell score, udder type traits, productive life, and protein yield. J Dairy Sci. 2000;83:2350–2360. doi: 10.3168/jds.S0022-0302(00)75123-X. [DOI] [PubMed] [Google Scholar]
- Negussie E, Stradén I, Mäntysaari EA. Genetic association of clinical mastitis with test-day somatic cell score and milk yield during first lactation of Finnish Ayrshire cows. J Dairy Sci. 2008;91:1189–1197. doi: 10.3168/jds.2007-0510. [DOI] [PubMed] [Google Scholar]
- Negussie E, Koivula M, Mantysaari EA, Lidauer M. Genetic evaluation of somatic cell score in dairy cattle considering first and later lactations as two different but correlated traits. J Anim Breed Genet. 2006;123:224–238. doi: 10.1111/j.1439-0388.2006.00594.x. [DOI] [PubMed] [Google Scholar]
- Neumaier A, Groeneveld E. Restricted maximum likelihood estimation of covariances in sparse linear models. Genet Sel Evol. 1998;30:3–26. [Google Scholar]
- Odegard J, Jensen J, Klemetsdal G, Madsen P, Heringstad B. Genetic analysis of somatic cell score in Norwegian cattle using random regression test-day models. J Dairy Sci. 2003;86:4103–4114. doi: 10.3168/jds.S0022-0302(03)74024-7. [DOI] [PubMed] [Google Scholar]
- Poso J, Mantysaari EA. Relationships between clinical mastitis, somatic cell score, and production for the first three lactations of Finnish Ayshire. J Dairy Sci. 1996;79:1284–1291. doi: 10.3168/jds.S0022-0302(96)76483-4. [DOI] [PubMed] [Google Scholar]
- Ptak E, Brzozowski P, Jagusiak W, Zdziarski K. Genetic parameters for somatic cell score for Polish Black-and-White cattle estimated with a random regression model. J Anim Feed Sci. 2007;16:357–369. [Google Scholar]
- Ptak E, Jagusiak W, Zarnecki A. Heritabilities and genetic correlations of lactational and daily somatic cell score with conformation traits in Polish Holstein cattle. 60th Annual Meeting of the European Association for Animal Production; August 24–27 2009; Barcelona, Spain. 2009b. Session S.15. Free communications in dairy genetics. [Google Scholar]
- Ptak E, Jagusiak W, Zarnecki A, Otwinowska-Mindur A. Genetic parameters of daily somatic cell score and some conformation traits in Polish Holstein cattle. Ann Anim Sci. 2009a;9:355–361. [Google Scholar]
- Reents R, Jamrozik J, Schaeffer LR, Dekkers JCM. Estimation of genetic parameters for test day records of somatic cell score. J Dairy Sci. 1995;78:2847–2857. doi: 10.3168/jds.S0022-0302(95)76915-6. [DOI] [PubMed] [Google Scholar]
- Rupp R, Boichard D. Genetic resistance to mastitis in dairy cattle. Vet Res. 2003;34:671–688. doi: 10.1051/vetres:2003020. [DOI] [PubMed] [Google Scholar]
- Rupp R, Boichard D. Genetic parameters for clinical mastitis, somatic cell score, production, udder type traits, and milking ease in first lactation Holsteins. J Dairy Sci. 1999;82:2198–2204. doi: 10.3168/jds.S0022-0302(99)75465-2. [DOI] [PubMed] [Google Scholar]
- Rupp R, Beaudeau F, Boichard D. Relationship between milk somatic-cell counts in the first lactation and clinical mastitis occurrence in the second lactation of French Holstein cows. Prev Vet Med. 2000;46:99–111. doi: 10.1016/s0167-5877(00)00142-2. [DOI] [PubMed] [Google Scholar]
- Rzewuska K, Jamrozik J, Żarnecki A, Strabel T. Genetic parameters of test-day somatic cell scores for the first three lactations of Polish Holstein-Friesian cattle. Czech J Anim Sci. 2011;56:381–389. [Google Scholar]
- Samore AB, Boettcher P, Jamrozik J, Bagnato A, Groen AF. Genetic parameters for production traits and somatic cell scores estimated with a multiple trait random regression model in Italian Holsteins. Proceedings of 7th World Congress on Genetics Applied to Livestock Production (WCGALP); Montpellier, France. 2002. pp. 1–7. [Google Scholar]
- Schutz MM. Genetic evaluation of somatic cell scores for United States dairy cattle. J Dairy Sci. 1994;77:2113–2129. doi: 10.3168/jds.S0022-0302(94)77154-X. [DOI] [PubMed] [Google Scholar]
- Schutz MM, Hansen LB, Steuernagel GR, Reneau JK. Genetic parameters for somatic cells, protein, and fat in milk of Holsteins. J Dairy Sci. 1990;73:494–502. [Google Scholar]
- Shook GE, Schutz MM. Selection on somatic cell score to improve resistance to mastitis in the United States. J Dairy Sci. 1994;77:648–658. doi: 10.3168/jds.S0022-0302(94)76995-2. [DOI] [PubMed] [Google Scholar]
- Shook GE. Major advances in determining appropriate selection goals. J Dairy Sci. 2006;89:1349–1361. doi: 10.3168/jds.S0022-0302(06)72202-0. [DOI] [PubMed] [Google Scholar]
- Wilmink JBM. Adjustment of test-day milk, fat and protein yield for age, season and stage of lactation. Livest Prod Sci. 1987;16:335–348. [Google Scholar]