Abstract
The adenovirus E1B-55-kDa protein binds and inactivates the tumor suppressor protein p53. However, the role of this interaction during infection is still poorly understood and was therefore examined here. Infection with a virus carrying the E1B-55-kDa mutation R239A, preventing the interaction with p53, led to the accumulation of p53. However, p53 target genes were not activated in the infected cells, although p53 phosphorylation did occur and the p53 antagonists Mdm2 and ΔNp73 did not accumulate. Deletion of E4orf6, alone or in combination with E1B-55-kDa, did not allow the induction of p53-responsive genes either. In transient reporter assays, the viral E1A-13S protein antagonized p53 activity; mutational analysis suggested that this depends partially on p300 binding, but it depends even more strongly on the interaction of E1A with the p400/TRRAP protein complex. However, viruses expressing E1A mutants lacking these binding activities, in combination with E1B-55-kDa R239A, still abolished p53 activity. In contrast, when the mutation of E1B-55-kDa at R239A was combined with a deletion of the apoptosis inhibitor E1B-19-kDa, infected cells showed more extensive apoptosis than after infection with single mutants, suggesting that accumulated p53, albeit transcriptionally inactive, might nonetheless enhance apoptosis. Despite extensive apoptosis of the infected cells, the deletion of E1B-19-kDa, in combination with the E1B-55-kDa mutation or in the presence of the constitutively active p53 mutant p53mt24-28, reduced virus replication less than fivefold. In conclusion, adenovirus does not need direct binding of E1B-55-kDa to inactivate p53, and forced p53 activity with consecutive apoptosis does not severely impair virus replication.
The tumor suppressor protein p53 is the product of the most frequently mutated gene in human cancer. p53, when wild type, acts as a transcription factor to enhance the expression of numerous target genes. The products of these genes mediate the major biological effects of p53, i.e., cell cycle arrest and apoptosis (64-66). In addition, p53 might also induce apoptotic pathways independently of transcription, in particular through its association with mitochondria (13, 37). The activity of p53 is carefully regulated by various mechanisms, including its phosphorylation and the expression of p53 antagonists. These antagonists include the Mdm2 protein that directly binds p53 and mediates its ubiquitination. Another antagonist of p53 was identified more recently; an amino-terminally truncated form of the p53 homologue p73 can be transcriptionally induced by p53 (18, 25, 38) and then inhibits the activity of p53, presumably through the competition for common promoter binding sites (18, 25, 38, 59), thereby creating a negative-feedback loop. Moreover, p53 uses cofactors for transcriptional activation, including the histone acetyltransferase p300 (53) and the protein complex containing p400 and TRRAP (1), suggesting that the availability of such cofactors might also contribute to the regulation of p53.
p53 is antagonized by a variety of tumor viruses, including simian vacuolating virus (simian virus 40), oncogenic human papillomaviruses (e.g., human papillomavirus types 16 and 18), and adenovirus (31). In each case, viral transforming proteins directly interact with p53. Although p53 is not always inactive in virus-transformed cells (33), all viral p53 antagonists can at least temporarily reduce p53 activity in transient reporter assays. Adenovirus and human papillomavirus not only inactivate p53 but also mediate its degradation. In the case of adenovirus, the E1B-55-kDa oncoprotein binds and inactivates p53 (52), whereas E1B-55-kDa and E4 open reading frame 6 (E4orf6, alias E4-34 kDa) together mediate the ubiquitination and degradation of p53 (44, 45, 50, 57). Other adenovirus proteins can indirectly interfere with p53 activity. For instance, the E1A gene products interact with p300, thereby reducing p53-mediated transactivation, at least in transient reporter assays (56). Furthermore, E1A proteins bind the p400/TRRAP complex (16, 30), suggesting another way of diminishing p53 activity. The E1B-19-kDa protein was shown to inhibit the downstream effects of p53 by blocking the induction of apoptosis (7, 68), and the E1B-19-kDa target proteins Bax and Bak, when not antagonized, have been shown to limit adenovirus replication in baby mouse kidney cells (6).
Intuitively, it seems obvious that antagonizing p53 should represent an advantage for virus replication, e.g., by inhibiting premature apoptosis of the infected cell. On the basis of this assumption, it was suggested that an adenovirus lacking the E1B-55-kDa protein might specifically replicate in tumor cells that do not express active p53. Such a virus, termed dl1520 (2) or Onyx015 (3) has been tested as an oncolytic agent in advanced clinical trials, with relatively promising results (26, 29, 39, 40). However, the theoretical basis for using adenovirus deletion mutants in therapy is far from being consolidated, as reviewed recently (8-10). In particular, and despite the extensive studies of p53 using transient-expression or -transformation assays, surprisingly little is known about the activity and impact of p53 in the context of a productive adenovirus infection.
Koch et al. previously analyzed the effect of p53 overexpression on the efficiency of adenovirus replication (27). To do so, they employed a p53 mutant termed p53mt24-28, with the amino acid residues 24 to 28 replaced by the corresponding residues of the p53 homologue TAp73. p53mt24-28 does not show any reduction in transcriptional activity compared with the activity of wild-type p53; however, it lacks the ability to interact with E1B-55-kDa and is therefore resistant to direct inactivation or degradation during adenovirus infection (27, 50). Using an adenovirus vector expressing p53mt24-28 for coinfection with wild-type adenovirus, Koch et al. previously found that p53 activity does not impair virus replication (27). It now remains to be examined whether adenovirus mutants with deletions or substitutions can be rendered sensitive to p53 activity.
While the overexpression of “E1B-resistant” p53mt24-28 represents a useful tool, it does not necessarily reflect the effects of endogenous p53. For analyzing the role of cellular p53 in the context of adenovirus infection, a mutant virus is required that selectively lacks the ability of E1B-55-kDa to interact with p53. Such a mutant recently became available through a systematic screen of E1B-55-kDa mutants. The R239A mutation within a recombinant virus termed Onyx051 eliminates the abilities of E1B-55-kDa to bind and destabilize p53, but it preserves the interaction of E1B-55-kDa with E4orf6 and still induces the host cell shutoff phenomenon (54). However, it is currently unknown whether infection with this type of virus mutant results in p53 activation and whether the p53 status of the infected cell affects the replication of this mutant. Answering this would contribute to understanding the role of p53 in adenovirus infection, and it would help to evaluate the chances of creating a virus with p53-selective replication properties.
We show here that p53 remains transcriptionally inactive in adenovirus-infected cells, independently of the interaction of E1B-55-kDa or E4orf6 with p53, and regardless of E1A binding to p300 or p400/TRRAP. However, even when p53 activity is induced by forced overexpression of p53mt24-28, while deleting the apoptosis inhibitor E1B-19-kDa, virus replication is reduced only fivefold or less. Thus, adenovirus displays resistance to p53 at multiple levels.
MATERIALS AND METHODS
Cells and viruses.
A549, H1299, MRC5 cells (American Type Culture Collection), and human foreskin fibroblasts (generous gift by K. Radsak and coworkers) were maintained in Dulbecco's modified Eagle medium (Life Technologies) with 10% fetal bovine serum (FBS). LS174T cells (American Type Culture Collection) were maintained in RPMI 1640 medium containing 10% FBS. Transfections were done using Lipofectamine 2000 (Life Technologies), and luciferase assays (Promega) were performed 24 h later, as described previously (50). Adenovirus and adenovirus-derived vectors were propagated, and the titer was determined as described previously (27, 67). Briefly, fresh cell monolayers were infected with virus dilutions, followed by immunofluorescence staining of the adenovirus E2A-72-kDa protein and counterstaining with 4,6-diamidino-2-phenylindole (DAPI).
Plasmids and viruses.
Mutant plasmids were created by the QuikChange methodology (Stratagene) for site-directed mutagenesis, using the following primers and their respective reverse complements: E1B-55kDa R239A (GGT TAT TAT GAA TGT AGC GTT TAC GGG CCC CAA TTT TA), ΔE1B-19-kDa (CTT GCT GGA ACA GAG CTC AAG CTT ATC TGG GTC ACC AGG CGC), E1A R2G (CCG GGA CTG AAA ATG GGT CAT ATC ATA TGC CAC GGA GGT GTT), E1A Δ2-36 (GGA CTG AAA ATG CAT TTC GAA CCA CCT ACC CTT CAC G), E1A Δ25-36 (GGA CCA GCT GAT CCA TTT CGA ACC ACC TAC CCT TCA CG), E1A Δ15-25 (GTT ATT ACC GAA GAA GAG GTA CTG GCT GAT), and E1Amut19-24 (GAA ATG GCC GCC AGT TCT AAT GAC CAG AGC GCT GAA GAG GTA CTG GCT).
The expression plasmid pRcCMVp53 (32) was used in reporter assays, along with the reporter plasmid pBP100Gl2 containing the p53-responsive portion of the mdm2 promoter and a luciferase reporter gene (14). Expression plasmids for E1B-55-kDa (11) and for E4orf6 (12) have been described; a cDNA of E1A-13S was cloned into pcDNA3.
For recombinant viruses, the mutations described above were introduced by QuikChange mutagenesis in a plasmid containing the entire E1 region in the background of the vector pShuttle (22). To this end, pShuttle was linearized using PmeI and NotI and filled with an PmeI/NotI-digested PCR product obtained by amplifying the template pXCl (36) using the primers GGA ATT CGT TTA AAC AGG CCT CTC AAG TCT GTA TAC G and GGA ATT CGC GGC CGC GGT TTT AGG CGG ATG TTG TAG. Recombinant viruses were then created by homologous recombination of this plasmid with the vector backbone pAdEasy1 (22) in Escherichia coli strain BJ 5183, followed by transfection of an E1-complementing cell line, subsequent virus amplification and titration, as described previously (27, 67).
Immunoblotting.
Proteins were separated on sodium dodecyl sulfate (SDS)-polyacrylamide gels and transferred to nitrocellulose membranes, followed by incubation with antibodies in phosphate-buffered saline containing 5% milk powder and 0.1% Tween 20. Peroxidase-coupled secondary antibodies (Fab fragments; Jackson) were then detected by chemiluminescence (Pierce). Antibodies were directed against adenovirus E1A (M73; Calbiochem), adenovirus E2A (B6-8 [47]; obtained from J. Flint), adenovirus hexon protein (goat polyclonal antibody; Biogenesis), p53 (1801 and DO-1; Oncogene Research), phospho-Ser15-p53 (16G8; Cell Signaling), p21/CDKN1A (BD Transduction Laboratories), Mdm2 (2A10 and 3G5 [41]), p73 (rabbit polyclonal antibody; BD Pharmingen), poly(ADP-ribose) polymerase (PARP) (Ab-2; Oncogene Research), Bax (BD Transduction Laboratories), and actin (AC-15; Abcam).
RT-PCR.
Total RNA was isolated from HeLa cells (Trizol reagent; Life Technologies), and mRNA levels were determined by reverse transcription (RT) and semiquantitative PCR, essentially as described previously (5). RT was performed with Superscript II polymerase (Life Technologies) and PCR amplification with Expand HiFi DNA polymerase (Roche). PCR was performed using an initial 3-min denaturation step at 95°C, followed by different numbers of cycles of PCR. One cycle of PCR consisted of 20 s at 95°C, 20 s at 52°C (Mdm2) or 55°C (p21/CDKN1A), and 90 s at 70°C. Specific oligonucleotides were employed for RT and PCR. For p21/CDKN1A, 25 cycles of PCR and RT primer GGA AAA GGA GAA CAC GGG ATG AGG AGG, forward PCR primer CCT GGC ACC TCA CCT GCT CTG CTG, and reverse PCR primer GCA GAA GAT GTA GAG CGG GCC TTT were used. For Mdm2, 25 cycles of PCR and RT primer AAC ATC TGT TGC AAT GTG ATG G, forward PCR primer TCA GGA TTC AGT TTC AGA TCA G, and reverse PCR primer CAT TTC CAA TAG TCA GCT AAG G were used.
Assessment of adenovirus DNA replication.
Infected cells were harvested, and genomic DNA was prepared (QIAGEN) and digested with the restriction enzyme HindIII. DNA was separated by size using a 0.6% agarose gel and visualized by ethidium bromide staining.
RESULTS
The mutation R239A strongly attenuates p53 inhibition by the E1B-55-kDa protein in transient reporter assays.
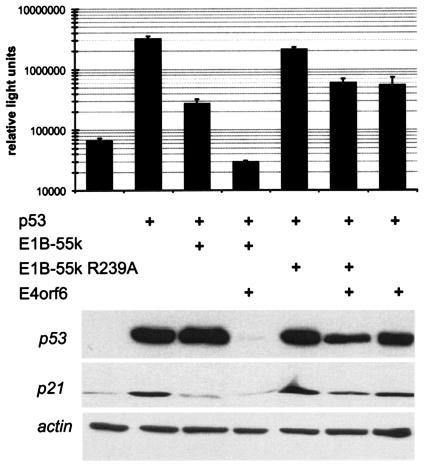
The mutation R239A within the E1B-55-kDa protein of adenovirus type 5 was recently shown to abolish the degradation of p53 during virus infection (54). We first asked whether this mutation also interferes with the ability of E1B-55-kDa to inhibit the transcriptional activity of p53. This was tested by transient transfection of H1299 cells, a p53−/− cell line derived from human lung adenocarcinoma. Expression plasmids for p53 and for E1B-55-kDa were cotransfected together with a luciferase reporter plasmid containing the p53-responsive fragment of the mdm2 promoter (14). Luciferase activity was then determined to assess the extent of p53-induced promoter activation. E1B-55-kDa was found to decrease p53 activity (Fig. 1, top panel), and the addition of E4orf6 virtually abolished any detectable p53-mediated transcription, confirming previous results (49, 50, 57, 69). When E1B-55-kDa and E4orf6 were expressed in combination, the amount of p53, as determined by immunoblot analysis, was drastically reduced (Fig. 1, bottom blot), an effect that was recently shown to be mediated by increased p53 ubiquitination (44, 46). In contrast, when the mutation R239A was introduced into E1B-55-kDa, p53 inhibition was severely impaired, in the presence or absence of E4orf6. Also, p53 degradation was strongly attenuated. The levels of the p53-responsive gene product p21/CDKN1A, as determined by immunoblotting, essentially reflected the luciferase activities, indicating that the effects of E1B-55-kDa and E4orf6 on p53 activity were not limited to a reporter system but pertained to a chromosomally integrated, p53-responsive gene. We conclude that the mutation R239A strongly impairs the ability of E1B-55-kDa to inhibit p53 activity in a transient-transfection assay.
FIG. 1.
Inactivation of p53 by the E1B-55-kDa (E1B-55k) protein in transient reporter assays. H1299 cells (p53−/−) were transiently transfected as indicated with a luciferase reporter plasmid containing a p53-responsive promoter (50 ng), along with expression plasmids for p53 (50 ng), E1B-55-kDa or its mutant R239A (350 ng), and/or E4orf6 (750 ng). When expression plasmids were not used, corresponding empty vector constructs were transfected instead. Luciferase activities were determined after 24 h. The average results from at least three independent experiments are shown on a logarithmic scale, along with standard deviations (error bars). The lysates used for luciferase quantification were then boiled in sample buffer containing SDS and subjected to SDS-polyacrylamide gel electrophoresis and immunoblotting. p53 and its target gene product, p21, were detected by antibodies, along with actin (loading control).
Mutation of E1B-55-kDa allows p53 accumulation but not p53 activity in adenovirus-infected cells.
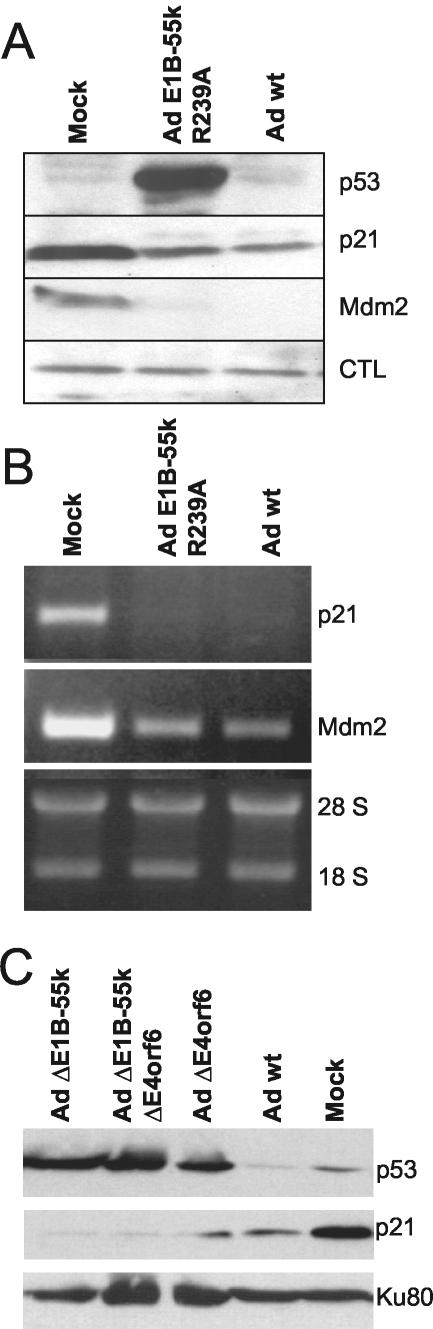
Next, we sought to determine the effects of the mutation R239A on the activity of p53 during an adenovirus infection, as opposed to transient transfections. To test this, the mutation was introduced into a recombinant adenovirus, using a plasmid-based system for virus construction (22). When we infected A549 cells (p53+/+, no p53 mutation) with this virus and subsequently detected p53 and two of its target gene products by immunoblot analysis, it was found that p53 strongly accumulated in cells that were infected with the mutant virus, whereas the levels of p53 were much lower in mock-infected cells or in cells infected with wild-type virus (Fig. 2A), in accordance with previous data (54). Surprisingly, however, the accumulation of p53 did not coincide with increased expression of p53-responsive genes. The levels of the products of such genes, p21/CDKN1A and Mdm2, were decreased, rather than increased, regardless of whether wild-type or mutant virus was used for infection. To rule out the possibility that this decrease in expression occurred only at the level of protein synthesis or stability, we determined the mRNA levels of the same genes by semiquantitative RT-PCR. As shown in Fig. 2B, the decrease in expression of p21/CDKN1A and Mdm2 was also seen at the mRNA level, and again, it was not affected by the R239A mutation in E1B-55-kDa. We conclude that the virus is capable of completely abolishing p53 activity in infected cells by mechanisms that do not require the interaction of E1B-55-kDa and p53.
FIG. 2.
p53 levels and activity during infection with the adenovirus mutant E1B-55-kDa R239A. Cells were infected with wild-type adenovirus (Ad wt), adenovirus carrying the E1B-55-kDa R239A mutation (Ad E1B-55k R239A), or mock infected at a multiplicity of infection of 20. The cells were harvested and subjected to immunoblot analysis, and RNA was isolated in parallel experiments at 24 h postinfection. (A) Immunoblot analysis of p53 and its target gene products. p53 and its target gene products p21/CDKN1A and Mdm2 were detected in A549 cell lysates using specific antibodies. A background band served as a loading control (CTL). (B) Levels of p53 target gene transcripts. Total RNA was isolated from infected A549 cells and subjected to agarose gel electrophoresis and ethidium bromide staining. mRNA derived from the p21/CDKN1A and Mdm2 genes was reverse transcribed and amplified by 25 PCR cycles. The PCR products were visualized on agarose gels. (C) Effect of E4orf6 deletion on p53. A549 cells were infected as described above with wild-type adenovirus and/or mutants that lack E1B-55-kDa, E4orf6, or both (4). Subsequently, p53 and p21 levels, along with Ku antigen (loading control), were determined by immunoblot analysis.
It has been reported that the adenovirus E4orf6 protein, at least when transiently expressed, is capable of binding and inactivating p53, even in the absence of E1B-55-kDa (12, 24). Therefore, we tested whether a deletion of E4orf6, alone or in combination with a deletion of E1B-55-kDa, might lead to p53 activity in the context of virus infection. Virus recombinants lacking E1B-55-kDa or E4orf6 or both were previously characterized (4). These viruses were used to infect A549 cells, and then immunoblot analysis of p53 and p21 was performed. As shown in Fig. 2C, deletions of either gene led to the accumulation of p53, consistent with the concept that E1B-55-kDa and E4orf6 must cooperate to ubiquitinate and destabilize p53 (44, 45, 50, 57, 69). Deletion of E4orf6 resulted in somewhat higher p21 levels than those observed in the presence of E4orf6 (Fig. 2C, bottom blot). However, despite the massive accumulation of p53 in all cases, p21 levels remained equal or lower than in mock-infected cells, regardless of the virus mutant employed. We conclude that neither E1B-55-kDa nor E4orf6 is required to block the transcriptional activity of p53 that is not active during infection with adenovirus.
Infection with adenovirus carrying a mutation of E1B-55-kDa induces the accumulation of phosphorylated p53, as well as TAp73, but not Mdm2 or ΔNp73.
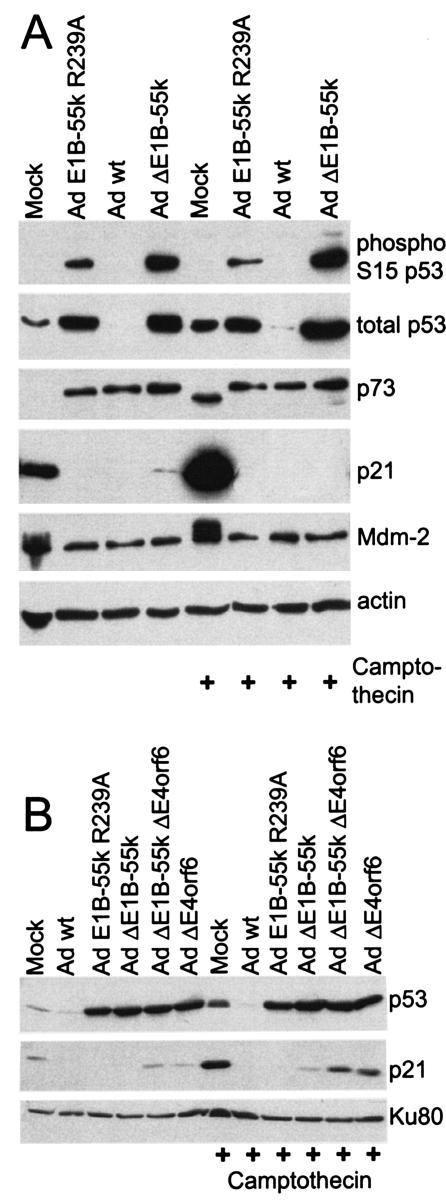
To narrow down the spectrum of possible reasons that might explain the inactivity of p53 during infection, we assessed the degree of p53 phosphorylation. A key residue of p53, serine 15, is known to inhibit Mdm2 binding and to increase the efficiency of transactivation when it becomes phosphorylated (55). Therefore, we infected LS174T cells with adenovirus carrying E1B-55-kDa mutations, and then determined the degree of p53 phosphorylation at this residue, in parallel to the total amounts of p53, using a phosphospecific antibody. A parallel experiment was performed with cells that were treated with the topoisomerase inhibitor camptothecin to analyze the p53 response in the presence of virus-independent DNA damage. As expected, p53 accumulated when the cells were infected with a virus carrying the E1B-55-kDa mutation R239A or lacking E1B-55-kDa entirely (Fig. 3A). Moreover, p53 was found phosphorylated in all these cases, and the degree of phosphorylation even exceeded the amount of phosphorylated p53 in camptothecin-treated and mock-infected cells. At the same time, camptothecin treatment alone led to a considerable increase of p53 activity, as determined by the increased levels of p21/CDKN1A and Mdm2. In contrast, no p53 activity was observed by this criterion in infected cells, even when the cells had been treated with camptothecin. We conclude that a lack of p53 phosphorylation, at least at the key residue S15, is not the reason for missing p53 activity in the infected cells. The expression levels of Mdm2, compared to mock-infected cells, were not increased upon virus infection. This argues against p53 inhibition by Mdm2 during virus infection.
FIG. 3.
p53 phosphorylation and levels of p53 antagonists during adenovirus infection. (A) Effect of adenovirus E1B-55-kDa (Ad E1B-55k) mutation on p53. LS174T cells were mock infected or infected with wild-type adenovirus (Ad wt) or the indicated adenovirus mutants at a multiplicity of infection of 20. The cells were also treated with camptothecin (300 nM) for 3 h at the time of infection. Immunoblot analysis was performed with antibodies against the indicated protein entities. (B) Effect of E4orf6 deletion on p53. LS174T cells were infected and/or treated with camptothecin as described above, using wild-type adenovirus and/or mutants that lack E1B-55-kDa, E4orf6, or both (4). Subsequently, p53 and p21 levels, along with Ku antigen (loading control), were determined by immunoblot analysis.
Further, we determined the levels of p73 in infected cells, since we and others have previously characterized the ΔNp73 isoform to be a negative regulator of p53 (18, 25, 38, 59, 70, 71). It was found that infected cells expressed increased levels of a p73 protein. However, this protein was larger than the ΔNp73α protein that was induced by camptothecin treatment alone in this experiment (Fig. 3A) and the results reported by Kartasheva et al. (25). Since ΔNp73α is the largest isoform of p73 that lacks a functional transactivation domain (70, 71), we conclude that the isoform detected in the infected cells is a transcriptionally active p73 protein. Its size corresponds to the size TAp73α, as determined previously by comparison with a transiently expressed recombinant protein (25; our unpublished observations). The accumulation of TAp73α in infected cells is in accordance with the known activation of E2F-responsive genes during adenovirus infection (28, 48) and the strong E2F responsiveness of the TAp73 promoter (23, 58). Notably, the increased levels of a transcriptionally active isoform of TAp73α do not provide an explanation for p53 inactivity. Rather, it appears that both p53 and its transcriptionally active homologue TAp73 are inactivated during adenovirus infection by as yet unknown mechanisms.
Finally, the impact of E4orf6 on p53 activity was analyzed in LS174T cells in the presence or absence of camptothecin. As found in A549 cells (Fig. 2C), the deletion of E4orf6 strongly increased p53 levels but led to a moderate increase in the level of p21 (Fig. 3B). In any case, the amount of p21 remained equal to (in nontreated cells) or lower than (in camptothecin-treated cells) the level observed in noninfected cells. Thus, the deletion of E4orf6 does not remove the block of p53 activity during adenovirus infection.
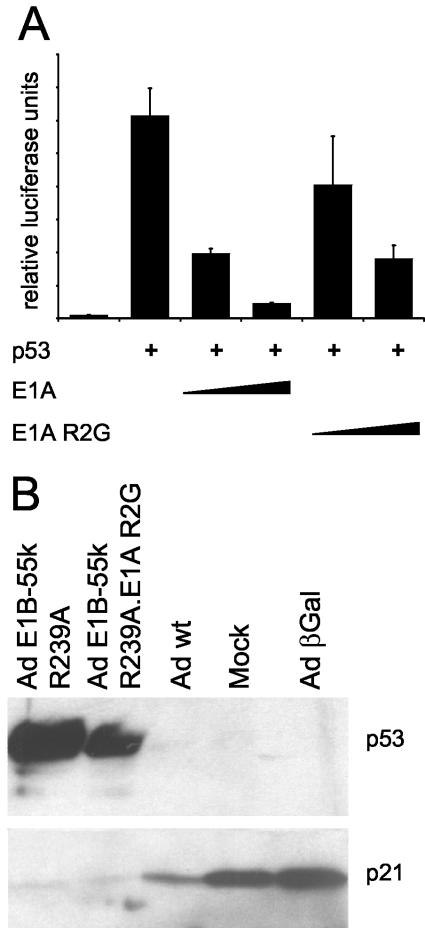
The R2G mutation attenuates p53 inhibition by E1A-13S in transient reporter assays but does not allow p53 activity in adenovirus-infected cells.
Besides E1B-55-kDa, the E1A oncoproteins have previously been implicated in the negative regulation of p53. In particular, it was suggested that the interaction of E1A with the protein p300 might be responsible for attenuation of p53, since p300 can act as a transcriptional cofactor of p53 (53). We first addressed this by transient-transfection and luciferase assays. Indeed, p53-mediated transactivation was negatively regulated by E1A-13S (Fig. 4A). In this experiment, wild-type E1A was compared with the mutant R2G that had previously been shown to abolish p300 binding (reference 15 and references therein). As expected, this mutant inhibited p53 only to a lesser extent. Next, we sought to determine whether p300 binding was the principal mechanism to inactivate p53 during an infection with a virus carrying a E1B-55-kDa mutation. A double mutant virus that expresses E1A with theR2G mutation and E1B-55-kDa with the R239A mutation was constructed. This virus, as well as the single mutant E1B-55-kDa R239A, induced p53 accumulation compared to the p53 levels in mock-infected cells or cells that were infected with wild-type adenovirus. However, in both cases, the p53-responsive gene p21/CDKN1A was not induced, as determined by immunoblot analysis of the encoded protein (Fig. 4B). We conclude that the activity of p53 is abolished in adenovirus-infected cells by mechanisms that function independently of the interactions between E1A and p300 or between E1B-55-kDa and p53.
FIG. 4.
Role of p300 binding in inactivation of p53 by E1A in transient reporter assays. The impact of E1A-p300 interaction on p53 activity was examined by transient reporter assays and immunoblot analysis. (A) Transient reporter assays. H1299 cells (p53−/−) were transiently transfected as indicated with a luciferase reporter plasmid containing a p53-responsive promoter (50 ng), along with expression plasmids for p53 (50 ng) and E1A or its mutant R2G (300 or 1,100 ng). Luciferase activities were determined as described in the legend to Fig. 1. (B) Immunoblot analysis of p53 and p21/CDKN1A in infected cells. A549 cells were mock infected or infected with wild-type adenovirus (Ad wt) or the indicated adenovirus mutants at a multiplicity of infection of 20. Cells were harvested and subjected to immunoblot analysis at 24 h postinfection. p53 and its target gene product p21/CDKN1A were detected using specific antibodies. Ad E1B-55k, adenovirus E1B-55-kDa; Ad βGal, adenovirus expressing β-galactosidase.
E1A mutations that abolish p400/TRRAP binding strongly attenuate p53 inhibition by E1A in transient reporter assays but do not allow p53 activity in adenovirus-infected cells.
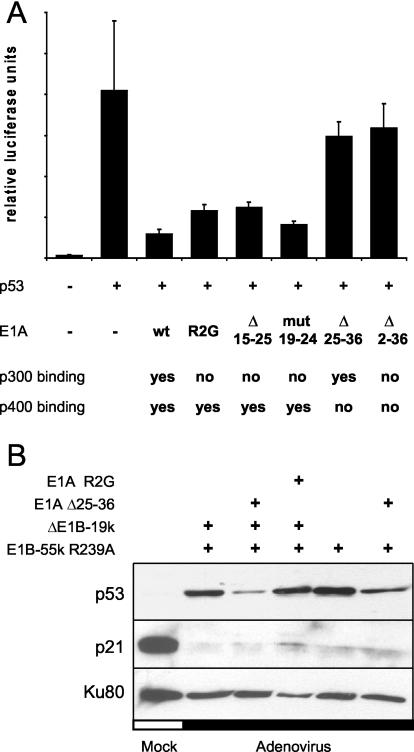
Besides p300, the p400/TRRAP complex of proteins has been found to interact with E1A proteins through a region of E1A that is close to the p300 binding region but can still be distinguished from it (15, 16). Furthermore, p400/TRRAP can act as a cofactor of p53, at least with regard to the activation of the Mdm2 promoter (1). Therefore, we assessed whether E1A might attenuate p53 activity through binding to p400/TRRAP. To test this, E1A mutants that lack the ability to bind p400/TRRAP or that lack p300 binding activity or both (reference 15 and references therein) were constructed, as summarized in Fig. 5A. These E1A mutants were tested for p53 inhibition in transient reporter assays. It was found that the mutants lacking either residues 25 to 36 (selectively lacking p400/TRRAP binding) or residues 2 to 36 (lacking both binding activities) were most compromised for p53 attenuation compared to wild-type E1A or mutants that retain p400/TRRAP binding while lacking p300 binding activity (Fig. 5A). We conclude that, at least in the system under study here, the interaction between E1A and p400/TRRAP might be of even greater impact than the interaction between E1A and p300 for the negative regulation of p53 activity.
FIG. 5.
Role of p400/TRRAP binding in inactivation of p53 by E1A in transient reporter assays. The impact of E1A-p400/TRRAP interaction on p53 activity was examined by transient reporter assays and immunoblot analysis. (A) Transient reporter assays. H1299 cells (p53−/−) were transiently transfected as indicated with a luciferase reporter plasmid containing a p53-responsive promoter (50 ng), along with expression plasmids for p53 (5 ng) and E1A-13S or the indicated mutants of E1A (1,100 ng). The reported abilities of these mutants (15) to bind p300 and/or the p400/TRRAP complex are indicated below the graph. Luciferase activities were determined as described in the legend to Fig. 1. Abbreviations: wt, wild-type; mut, mutant; Δ 15-25, deletion of residues 15 to 25. (B) Immunoblot analysis of p53 and p21/CDKN1A in infected cells. A549 cells were mock infected or infected with wild-type adenovirus or adenoviruses carrying the indicated mutations at a multiplicity of infection of 15. Cells were harvested and subjected to immunoblot analysis at 36 h postinfection. p53 and its target gene product p21/CDKN1A were detected using specific antibodies, along with the Ku antigen (loading control).
Next, we assessed the effect that the deletion of residues 25 to 36 within E1A has on p53 activity in infected cells. A recombinant adenovirus was created; this adenovirus simultaneously carried the deletion of residues 25 to 36 within E1A (thereby abolishing the interaction of E1A with p400/TRRAP) and the R239A mutation within E1B-55-kDa (thereby eliminating the interaction between E1B-55-kDa and p53). In addition, a similar virus in which E1B-19-kDa had been deleted was created to address the impact of apoptosis on virus replication (see below). Subsequently, A549 cells were infected by these recombinant viruses, and the levels of p53 and p53 target gene products were determined by immunoblotting. In all cases, the recombinant viruses caused p53 accumulation, although the mutants lacking the E1A-p400/TRRAP interaction did so only to a somewhat lesser degree (Fig. 5B). It is possible that the p400/TRRAP complex affects or even mediates the ubiquitination and hence the stability of p53, as has been reported for p300 (19). However, as in the previous experiments, the levels of the p53 target gene product p21/CDKN1A were not increased in infected cells. In contrast, a strong decrease in p21/CDKN1A levels was observed in all cases. We conclude that the elimination of the E1A-p400/TRRAP interaction, despite its considerable impact on p53 activity in transient reporter assays, does not restore p53 activity during adenovirus infection. Hence, p53 activity is abolished in adenovirus-infected cells by mechanisms that do not depend on the direct binding of p53, p300, or p400/TRRAP by viral E1 gene products.
The E1B-55-kDa mutation R239A augments apoptosis of infected cells when combined with a deletion of E1B-19-kDa.
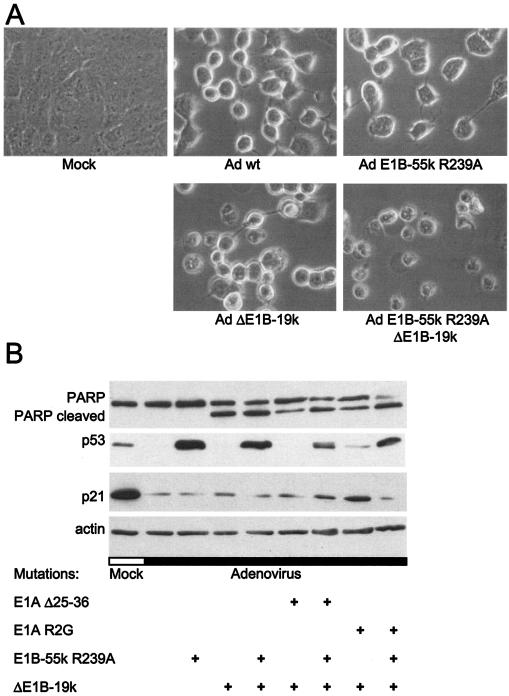
p53 appears to induce apoptosis through transcription-dependent and -independent pathways (reference 42 and references therein). In particular, p53 was found to associate with mitochondria, possibly triggering the initial phase of apoptosis before transcription is induced (13, 37). Therefore, we set up an experiment to determine whether abolishing p53 binding by E1B-55-kDa might lead to increased apoptosis of infected cells, despite the continuously missing transcriptional activity of p53. To test this, the principal adenovirus-expressed inhibitor of apoptosis, E1B-19-kDa, was eliminated by a corresponding deletion within the virus genome. Recombinant viruses that simultaneously carried a deletion of E1B-19-kDa and the R239A mutation within E1B-55-kDa that removes p53 binding were constructed. A549 cells were infected with these viruses, and their morphology was subsequently monitored by phase-contrast microscopy. Forty-eight hours after infection with wild-type adenovirus, the cells appeared enlarged and displayed a round shape, while remaining attached to the culture dish (Fig. 6A). A similar pattern was observed when cells were infected with virus with the single mutation E1B-55-kDa R239A. Also, when cells were infected with a virus carrying the single deletion of E1B-19-kDa, a similar morphology was observed in most cells, with a few smaller and apparently apoptotic cells. In contrast, when cells were infected with a recombinant adenovirus that carried the E1B-55-kDa R239A mutation and deletion of E1B-19-kDa, the cells seemed more damaged; they were largely detached and displayed apoptotic morphology throughout.
FIG. 6.
Apoptosis of cells upon infection with adenovirus carrying multiple E1 mutations. The impact of E1B mutations on the apoptotic response of infected cells was studied by examining cell morphology and PARP cleavage. (A) Morphology of infected cells. A549 cells were mock infected or infected with wild-type adenovirus (Ad wt) or the indicated adenovirus mutants at a multiplicity of infection of 10 for 48 h. Cell morphology was then examined by light microscopy. (B) PARP cleavage in infected cells. LS174T cells were mock infected or infected with the indicated adenovirus mutants at a multiplicity of infection of 20 for 30 h. The cells were then harvested and subjected to immunoblot analysis of PARP, as well as p53 and its target genes and actin (loading control). The top band in the PARP detection panel represents full-length PARP; the lower band (PARP cleaved) corresponds to the characteristic cleavage fragment that caspase 3 leaves after proteolysis.
A similar experiment was performed with LS174T cells that allowed the detection of PARP. Cleavage of PARP is a well-established hallmark of apoptosis. Cells were infected with wild-type adenovirus and with a virus carrying the R239A mutation of E1B-55-kDa with a deletion of E1B-19-kDa. On top of this, these mutations were also combined with E1A mutations affecting either p300 binding or p400/TRRAP binding. As shown in Fig. 6B, increased PARP cleavage was observed in those cells that were infected with viruses carrying the E1B-55-kDa mutation on top of the E1B-19-kDa deletion. The disappearance of the upper band (full-length PARP) and increase of the lower band (caspase-cleaved PARP) was particularly enforced by the E1B-55-kDa mutation in the background of an additional E1A mutation. This was observed despite the absence of any detectable transcriptional activity of p53, as determined by immunoblot detection of p21 (Fig. 6B). Thus, accumulating p53 in adenovirus-infected cells still appears to increase the apoptotic response of infected cells. This is compatible with the assumption that p53 might contribute to programmed cell death through transcription-independent pathways.
In general, the E1B-55-kDa mutation R239A, alone or in combination with a deletion of E1B-19-kDa, does not decrease virus replication.
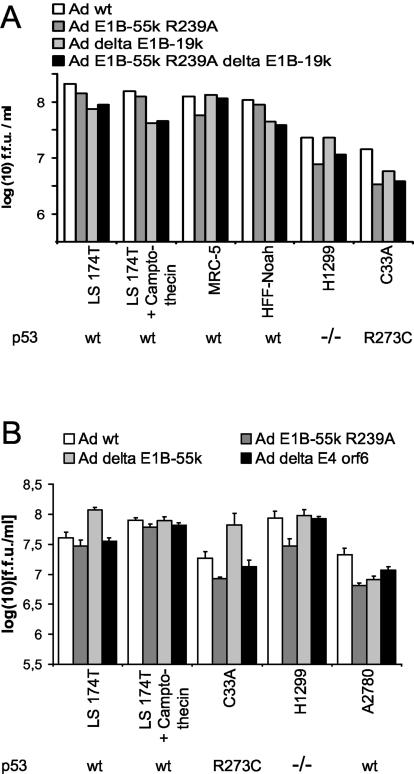
Next, we determined whether the increased apoptosis of infected cells upon mutation of E1B-55-kDa and/or deletion of E1B-19-kDa leads to a decrease in virus replication. A number of different cell lines with different p53 status were infected with the four viruses that either did or did not contain the corresponding E1B mutations. Forty-eight hours postinfection, the cells were harvested, and virus yield (in fluorescence-forming units) was determined by titration on H1299 cells. In most cases, there were only subtle changes in virus yield, differences of less than 1 log unit when the different virus mutants for each cell line were compared (Fig. 7A). The differences between the mutant lacking E1B-19-kDa and the double mutant were even less pronounced. This was observed in tumor cell-derived cell lines in the presence or absence of the DNA-damaging (and p53-activating) agent camptothecin, as well as in human fibroblasts. We conclude that despite the enhanced apoptosis of cells infected with virus carrying both E1B mutations, the efficiency of the replication cycle is not grossly compromised by these mutations. Hence, the viruses can apparently finish most of their replication cycles before the infected cells become too distorted by apoptosis to support virus replication.
FIG. 7.
Replication of adenovirus mutants carrying E1B mutations. The replication of adenovirus mutants was assessed in the indicated cell lines. The p53 status of each cell line is indicated below the graph. wt, wild type. (A) Mutation of adenovirus E1B-55-kDa (Ad E1B-55k) and deletion of E1B-19-kDa (Ad delta E1B-19k). Cells of the indicated lines were infected with wild-type adenovirus (Ad wt) or adenoviruses carrying the indicated mutations of E1B-55-kDa and/or E1B-19-kDa at a multiplicity of infection that leads to infection of less than 50% of the cells for 48 h. Where indicated, the cells were treated with camptothecin (300 nM) for 3 h at the time of infection. The cells were harvested and lysed by freeze-thawing, and the titer of newly synthesized virus was determined by counting the fluorescence-forming units (f.f.u.) in the cell lysate. Note that the results are displayed on a logarithmic scale. (B) Deletion of E1B-55-kDa. The replication of a virus that lacks E1B-55-kDa, dl338 (43) was compared to a corresponding wild-type virus, dl309 (61), as in panel A.
To compare the effects of an E1B-55-kDa mutation with the complete deletion of E1B-55-kDa, virus lacking E1B-55-kDa entirely was used in parallel with wild-type virus to infect a subset of the cells under study, and virus yield was determined. As shown in Fig. 7B, even the deletion of E1B-55-kDa did not grossly reduce the efficiency of virus replication. This is consistent with previous results reporting that a severe effect of E1B-55-kDa deletion on virus yield is observed in relatively few cell lines, such as HeLa cells (17, 21).
Overexpression of active p53 strongly augments apoptosis of infected cells when combined with a deletion of E1B-19-kDa.
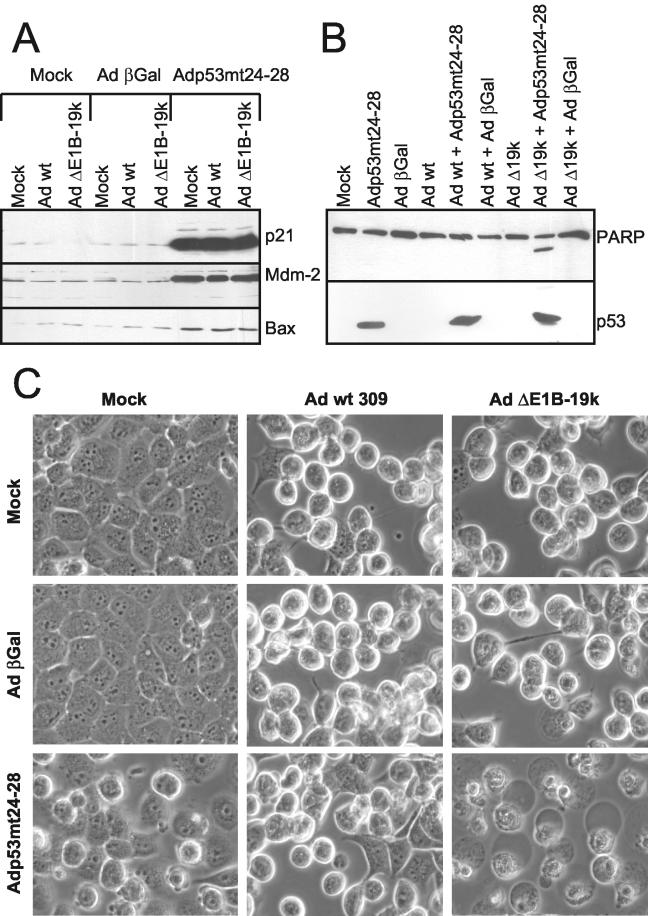
To corroborate the concept that p53 can increase the extent of apoptosis of infected cells when E1B-19-kDa is absent, we took advantage of a previously developed system to ensure p53 activity in cells that are infected with wild-type adenovirus (27). H1299 cells (p53−/−) had previously been infected with replication-competent adenoviruses, in combination with an adenovirus virus vector to express p53mt24-28, a p53 mutant that fully retains transcriptional activity but does not detectably interact with E1B-55-kDa (27, 50, 69). In addition to wild-type adenovirus, in this study we used a recombinant adenovirus in which E1B-19-kDa had been deleted for coinfection with the expression vector. p53mt24-28 was capable of inducing the p53-responsive genes p21/CDKN1A, Mdm2, and Bax, regardless of the coinfection with replicative viruses, as verified by immunodetection of the corresponding gene products (Fig. 8A). Also, p53 expression levels were not compromised by coinfection with replicative viruses (Fig. 8B). Since the p53 expression vector and a control vector expressing β-galactosidase have the E1 region entirely deleted, the coinfected cells did not contain the apoptosis inhibitor E1B-19-kDa. Thus, we determined the extent of apoptosis in the infected cells by monitoring PARP cleavage and cell morphology. At 30 h postinfection, neither transduction with an expression vector for p53mt24-28 nor infection with adenovirus lacking E1B-19-kDa alone had induced detectable PARP cleavage, whereas the combination of both had already led to the appearance of a visible PARP fragment and corresponding reduction of full-length PARP (Fig. 8B). Similarly, only the combination of adenovirus lacking E1B-19-kDa and the p53mt24-28 expression vector induced the extensive morphological changes typical of apoptosis in the infected cells (Fig. 8C). In conclusion, forced p53 activity clearly enhances apoptosis in cells upon infection with an adenovirus lacking E1B-19-kDa.
FIG. 8.
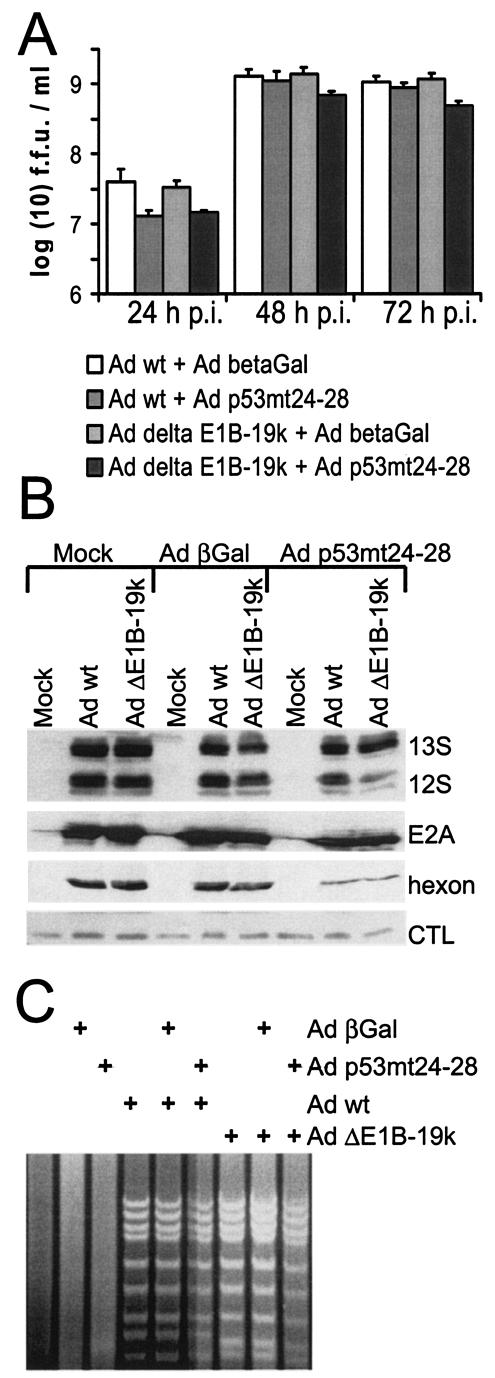
Apoptosis of cells upon infection with adenovirus lacking E1B-19-kDa, in combination with an expression vector for active p53. The effect that a deletion of E1B-19-kDa and the simultaneous overexpression of active p53 have on apoptosis was assessed as follows. In each case, H1299 cells were mock infected or infected with a combination of replication-competent adenovirus (wild-type adenovirus [Ad wt]or virus lacking E1B-19-kDa [Ad ΔE1B-19k] at a multiplicity of infection of 5) and adenovirus-based expression vectors expressing β-galactosidase (βGal) or p53mt24-28, a mutant of p53 that retains transcriptional activity but lost the interaction with E1B-55-kDa (multiplicity of infection of 30) as described previously (27, 50). (A) Expression levels of p53-responsive gene products. The cells were harvested at 30 h postinfection, followed by immunoblot analysis of the indicated p53-responsive gene products. (B) PARP cleavage in infected cells. Similarly, PARP and its cleavage product (the lower, unlabeled band), as well as p53, were detected. (C) Morphology of infected cells. The extent of morphological changes was assessed by phase-contrast microscopy.
Overexpression of active p53 only marginally interferes with virus replication, even when combined with a deletion of E1B-19-kDa.
Finally, we asked whether forced p53 activity in infected cells can affect virus yield. Previously, Koch et al. had found that active p53 does not interfere with the replication of wild-type adenovirus (27), raising the question whether p53 inhibition by adenovirus might represent a redundant function for virus replication, which is of importance only when other functions are missing. Given the enhanced apoptosis induced by p53mt24-28 when E1B-19-kDa was absent, we sought to determine whether the combination of p53 activity and E1B-19-kDa deletion might reduce virus yield. To test this, H1299 cells were again infected with combinations of adenovirus (wild type or lacking E1B-19-kDa) and virus-derived expression vectors for β-galactosidase (control) or p53mt24-28. At different time points, the cells were harvested, and virus yield (in fluorescence-forming units) was quantified by titration on H1299 cells.
As shown in Fig. 9, the differences in virus yield observed between the four different combinations were small. The combination of p53mt24-28 with E1B-19-kDa deletion led to a decrease in virus yield of less than fivefold compared to the other pairs of viruses (Fig. 9A), despite extensive apoptosis occurring with this combination. The expression levels of early virus gene products (E1A proteins and E2A-72-kDa DNA binding protein) and the late gene product hexon were reduced even less under these circumstances (Fig. 9B), and a similarly moderate reduction was found when the levels of virus DNA were analyzed (Fig. 9C). We conclude that, despite extensive apoptosis in the absence of E1B-19-kDa, forced p53 activity does not severely compromise virus replication.
FIG. 9.
Replication of adenovirus lacking E1B-19-kDa (Ad delta E1B-19k) in the presence of active p53. The impact of p53 activity on the replication of an adenovirus lacking E1B-19-kDa was assessed by examining virus yields, expression levels of virus proteins, and viral DNA replication in infected cells. (A) Virus yield. H1299 cells were infected with a wild-type-like adenovirus, dl309, or a virus mutant lacking E1B-19-kDa, dl337, at a multiplicity of infection of 5. Simultaneously, the cells were transduced with expression vectors for p53mt24-28 or β-galactosidase (betaGal) (multiplicity of infection of 50). After the indicated time points, the cells were harvested and lysed by freeze-thawing, and the titer of newly synthesized virus was determined by counting the fluorescence-forming units (f.f.u.) in the cell lysate. Note that the results are displayed on a logarithmic scale. p.i., postinfection; Ad wt, wild-type adenovirus. (B) Expression levels of virus proteins. H1299 cells were infected as described above for panel A. After 30 h, the cells were harvested, and the following viral gene products were detected by immunoblot analysis, using corresponding antibodies: E1A (13S and 12S isoforms); E2A-72-kDa DNA binding protein; and the hexon structural protein. A background band served as a loading control (CTL). (C) Extent of viral DNA replication in infected cells. H1299 cells were infected as described above for panel A. After 30 h, the cells were harvested, and genomic DNA was prepared. After restriction with HindIII, the fragments were separated on an agarose gel and visualized by ethidium bromide staining.
DISCUSSION
Our results show that p53 remains inactive in adenovirus-infected cells when the interactions between the E1B-55-kDa protein and p53 and/or the interactions between E1A and p300 or the p400/TRRAP complex are eliminated. However, even in response to forced p53 activity and upon deletion of the apoptosis inhibitor E1B-19-kDa, the virus still replicated to yield more than 20% of the wild-type levels. Thus, the virus not only inactivates p53 by multiple mechanisms, it is also remarkably resistant to any p53 activity that occurs despite these antagonisms and even replicates relatively efficiently when p53-induced apoptosis is permitted. Thus, despite the fact that p53 is one of the most uniform targets of small DNA tumor viruses, its elimination appears to be of little obvious relevance during adenovirus infection of cultured cells.
What are the mechanisms that lead to p53 inactivation despite its accumulation when cells are infected with an E1B-55-kDa mutant virus? We have ruled out a number of possibilities. Neither E4orf6 nor the interaction of E1A with p53 cofactors appears to be required for p53 inactivation. We did not find evidence for the induction of known p53 antagonists. Another possibility is the relocalization of p53 to intracellular sites where it is no longer available for the induction of cellular genes. However, immunofluorescence studies revealed that p53 remained in its usual, diffusely nuclear localization during virus replication and did not detectably relocalize to the cytoplasm or any particular nuclear structure (data not shown). At this time, we do not know the mechanism(s) that inactivates p53 during infection with the mutant viruses. Possibilities include the direct inactivation of p53 by an as yet unknown viral gene product or the recruitment of a limiting transcription factor to the viral replication centers. We have recently reported that only a small number of cellular genes are affected in their expression by an adenovirus infection, and all these genes were downregulated (34). This raises the possibility that adenovirus might be generally capable of preventing increases in the transcription of cellular genes. We speculate that adenovirus might inactivate factors that are required for augmenting gene expression, e.g., chromatin modifiers. Such factors might for instance be tethered to the virus replication centers, making them unavailable for cellular genes.
Despite the lack of transcriptional activity, accumulated p53 appears to increase the susceptibility of infected cells for apoptosis. Thus, transcriptional activation and death induction by p53 was separated in this system. This can be taken as a novel piece of evidence supporting a transcription-independent role of p53 in the induction of programmed cell death. It is tempting to speculate that p53 might enhance the death of infected cells by associating with mitochondria (13, 37). In the future, this system might be suitable to assess the domains, mechanisms, and antagonists that regulate the mitochondrial pathways employed by p53 to start apoptosis. For instance, it is possible that the recently suggested role of gain-of-function mutants of p53 in the replication of adenovirus (20) might be related to the association of such mutants with mitochondria.
Another open question concerns the fact that overexpressed p53mt24-28 activates transcription of p53 target genes in productively infected cells, whereas accumulated endogenous p53 does not. One might hypothesize that this is an inherent property conferred by the mutation of residues 24 to 28. However, earlier studies had revealed that even overexpressed wild-type p53 retains the ability to induce p21/CDKN1A expression in adenovirus-infected cells (27). We therefore assume that the sheer amount of overexpressed p53 overcomes any mechanism that would normally antagonize p53 in infected cells.
The relatively efficient replication of adenovirus despite the massive apoptosis of infected cells came as a surprise. It appears that the essential steps of virus replication are taken before inhibition by cell death. Our results also suggest that the late steps of the virus replication cycle, such as virus assembly, can be carried out in cells despite advanced apoptosis. Indeed, even wild-type adenovirus actively induces apoptosis at the end of its replication cycle, through expression of the adenovirus death protein encoded by the E3 region. This may help to release viral progeny (62, 63). Apparently, the virus is quite robust with regard to the timing of death induction, largely obviating the need for early inhibition of apoptosis.
Our results argue against the possibility of using p53-selective viruses for therapeutic purposes. p53 remains inactive despite the elimination of several known p53-antagonizing mechanisms. Even if it were possible to restore p53 activity by additional modifications of the virus, our results obtained with p53mt24-28 strongly suggest that p53 activity would still not severely reduce the replication of such a virus mutant. However, it might still be possible to create a virus that is suppressed by wild-type p53 and/or activated by mutant p53. For instance, Roth et al. recently reported an adaptor system for reactivating mutant p53 to activate cellular p53-responsive genes (51). A similar system might be used to activate early virus genes, using an adaptor that recruits mutant p53 to viral promoters. An obvious alternative would consist in the integration of p53-repressible promoters into the virus genome. However, since p53-mediated repression of transcription requires the induction of p21/CDKN1A (35) and since the expression of this gene is impaired by adenovirus regardless of p53 accumulation (this study), we do not favor this approach.
Finally, the question remains why adenovirus has evolved specific and direct mechanisms to eliminate p53, if additional ways of p53 inactivation exist. Forced p53 activity in the absence of E1B-19-kDa moderately reduces virus replication, and such a reduction might provide enough selective pressure to evolve p53 antagonisms. However, this occurs only in the presence of highly overexpressed p53 (Fig. 9) and much less with endogenous p53 (Fig. 7). The question of what advantage has been gained by the evolution of p53 antagonisms pertains to viruses other than adenovirus. Human papillomaviruses also evolved in numerous types, and only a fraction have developed efficient mechanisms to antagonize p53. Thus, such mechanisms appear to be selected for only under special and currently unknown circumstances. It should be kept in mind, however, that all experiments shown here were performed in cell culture systems. This raises the possibility that specific p53 antagonisms are more important for the interaction of the virus with an infected organism. Accordingly, recent observations regarding the induction of p53 expression by interferons (60) emphasize the existence of links between p53 and the immune response. We therefore propose that a complete picture reflecting the role of p53 in virus infection can be revealed only in immunocompetent animal models and that its final assessment will require the analysis of virus-induced pathogenesis.
Acknowledgments
We thank H.-D. Klenk for continuous support. We are indebted to J. Flint, G. Ketner, A. J. Levine, T. Shenk, and B. Vogelstein for the generous gift of plasmids, antibodies, viruses, and cells.
This work was supported in part by the German Research Foundation (DFG), the Deutsche Krebshilfe/Mildred Scheel Stiftung, and the P. E. Kempkes Stiftung.
REFERENCES
- 1.Ard, P. G., C. Chatterjee, S. Kunjibettu, L. R. Adside, L. E. Gralinski, and S. B. McMahon. 2002. Transcriptional regulation of the mdm2 oncogene by p53 requires TRRAP acetyltransferase complexes. Mol. Cell. Biol. 22:5650-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker, D. D., and A. J. Berk. 1987. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 156:107-121. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff, J. R., D. H. Kirn, A. Williams, C. Heise, S. Horn, M. Muna, L. Ng, J. A. Nye, A. Sampson-Johannes, A. Fattaey, and F. McCormick. 1996. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science 274:373-376. [DOI] [PubMed] [Google Scholar]
- 4.Bridge, E., and G. Ketner. 1990. Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174:345-353. [DOI] [PubMed] [Google Scholar]
- 5.Contente, A., A. Dittmer, M. C. Koch, J. Roth, and M. Dobbelstein. 2002. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat. Genet. 30:315-320. [DOI] [PubMed] [Google Scholar]
- 6.Cuconati, A., K. Degenhardt, R. Sundararajan, A. Anschel, and E. White. 2002. Bak and Bax function to limit adenovirus replication through apoptosis induction. J. Virol. 76:4547-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debbas, M., and E. White. 1993. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 7:546-554. [DOI] [PubMed] [Google Scholar]
- 8.Dix, B. R., S. J. Edwards, and A. W. Braithwaite. 2001. Does the antitumor adenovirus ONYX-015/dl1520 selectively target cells defective in the p53 pathway? J. Virol. 75:5443-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobbelstein, M. 2003. Replicating adenoviruses in tumor therapy. Curr. Top. Microbiol. Immunol. 273:291-334. [DOI] [PubMed] [Google Scholar]
- 10.Dobbelstein, M. 2003. Viruses in therapy—royal road or dead end? Virus Res. 92:219-221. [DOI] [PubMed] [Google Scholar]
- 11.Dobbelstein, M., J. Roth, W. T. Kimberly, A. J. Levine, and T. Shenk. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 16:4276-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobner, T., N. Horikoshi, S. Rubenwolf, and T. Shenk. 1996. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science 272:1470-1473. [DOI] [PubMed] [Google Scholar]
- 13.Dumont, P., J. I. Leu, A. C. Della Pietra III, D. L. George, and M. Murphy. 2003. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 33:357-365. [DOI] [PubMed] [Google Scholar]
- 14.Freedman, D. A., C. B. Epstein, J. C. Roth, and A. J. Levine. 1997. A genetic approach to mapping the p53 binding site in the MDM2 protein. Mol. Med. 3:248-259. [PMC free article] [PubMed] [Google Scholar]
- 15.Frisch, S. M., and J. S. Mymryk. 2002. Adenovirus-5 E1A: paradox and paradigm. Nat. Rev. Mol. Cell Biol. 3:441-452. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297-307. [DOI] [PubMed] [Google Scholar]
- 17.Goodrum, F. D., and D. A. Ornelles. 1998. p53 status does not determine outcome of E1B 55-kilodalton mutant adenovirus lytic infection. J. Virol. 72:9479-9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grob, T. J., U. Novak, C. Maisse, D. Barcaroli, A. U. Luthi, F. Pirnia, B. Hugli, H. U. Graber, V. De Laurenzi, M. F. Fey, G. Melino, and A. Tobler. 2001. Human ΔNp73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ. 8:1213-1223. [DOI] [PubMed] [Google Scholar]
- 19.Grossman, S. R., M. E. Deato, C. Brignone, H. M. Chan, A. L. Kung, H. Tagami, Y. Nakatani, and D. M. Livingston. 2003. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 300:342-344. [DOI] [PubMed] [Google Scholar]
- 20.Hann, B., and A. Balmain. 2003. Replication of an E1B 55-kilodalton protein-deficient adenovirus (ONYX-015) is restored by gain-of-function rather than loss-of-function p53 mutants. J. Virol. 77:11588-11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harada, J. N., and A. J. Berk. 1999. p53-independent and -dependent requirements for E1B-55K in adenovirus type 5 replication. J. Virol. 73:5333-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irwin, M., M. C. Marin, A. C. Phillips, R. S. Seelan, D. I. Smith, W. Liu, E. R. Flores, K. Y. Tsai, T. Jacks, K. H. Vousden, and W. G. Kaelin, Jr. 2000. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407:645-648. [DOI] [PubMed] [Google Scholar]
- 24.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683-689. [DOI] [PubMed] [Google Scholar]
- 25.Kartasheva, N. N., A. Contente, C. Lenz-Stoppler, J. Roth, and M. Dobbelstein. 2002. p53 induces the expression of its antagonist p73ΔN, establishing an autoregulatory feedback loop. Oncogene 21:4715-4727. [DOI] [PubMed] [Google Scholar]
- 26.Khuri, F. R., J. Nemunaitis, I. Ganly, J. Arseneau, I. F. Tannock, L. Romel, M. Gore, J. Ironside, R. H. MacDougall, C. Heise, B. Randlev, A. M. Gillenwater, P. Bruso, S. B. Kaye, W. K. Hong, and D. H. Kirn. 2000. A controlled trial of intratumoral ONYX-015, a selectively-replicating adenovirus, in combination with cisplatin and 5-fluorouracil in patients with recurrent head and neck cancer. Nat. Med. 6:879-885. [DOI] [PubMed] [Google Scholar]
- 27.Koch, P., J. Gatfield, C. Löber, U. Hobom, C. Lenz-Stöppler, J. Roth, and M. Dobbelstein. 2001. Efficient replication of adenovirus despite the overexpression of active and non-degradable p53. Cancer Res. 61:5941-5947. [PubMed] [Google Scholar]
- 28.Kovesdi, I., R. Reichel, and J. R. Nevins. 1986. Identification of a cellular transcription factor involved in E1A trans-activation. Cell 45:219-228. [DOI] [PubMed] [Google Scholar]
- 29.Lamont, J. P., J. Nemunaitis, J. A. Kuhn, S. A. Landers, and T. M. McCarty. 2000. A prospective phase II trial of ONYX-015 adenovirus and chemotherapy in recurrent squamous cell carcinoma of the head and neck (the Baylor experience). Ann. Surg. Oncol. 7:588-592. [DOI] [PubMed] [Google Scholar]
- 30.Lang, S. E., and P. Hearing. 2003. The adenovirus E1A oncoprotein recruits the cellular TRRAP/GCN5 histone acetyltransferase complex. Oncogene 22:2836-2841. [DOI] [PubMed] [Google Scholar]
- 31.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 32.Lin, J., J. Chen, B. Elenbaas, and A. J. Levine. 1994. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 8:1235-1246. [DOI] [PubMed] [Google Scholar]
- 33.Lober, C., C. Lenz-Stoppler, and M. Dobbelstein. 2002. Adenovirus E1-transformed cells grow despite the continuous presence of transcriptionally active p53. J. Gen. Virol. 83:2047-2057. [DOI] [PubMed] [Google Scholar]
- 34.Lohr, K., O. Hartmann, H. Schafer, and M. Dobbelstein. 2003. Mutual interference of adenovirus infection and myc expression. J. Virol. 77:7936-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lohr, K., C. Moritz, A. Contente, and M. Dobbelstein. 2003. p21/CDKN1A mediates negative regulation of transcription by p53. J. Biol. Chem. 278:32507-32516. [DOI] [PubMed] [Google Scholar]
- 36.McKinnon, R. D., S. Bacchetti, and F. L. Graham. 1982. Tn5 mutagenesis of the transforming genes of human adenovirus type 5. Gene 19:33-42. [DOI] [PubMed] [Google Scholar]
- 37.Mihara, M., S. Erster, A. Zaika, O. Petrenko, T. Chittenden, P. Pancoska, and U. M. Moll. 2003. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell 11:577-590. [DOI] [PubMed] [Google Scholar]
- 38.Nakagawa, T., M. Takahashi, T. Ozaki, K. Watanabe Ki, S. Todo, H. Mizuguchi, T. Hayakawa, and A. Nakagawara. 2002. Autoinhibitory regulation of p73 by ΔNp73 to modulate cell survival and death through a p73-specific target element within the ΔNp73 promoter. Mol. Cell. Biol. 22:2575-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nemunaitis, J., I. Ganly, F. Khuri, J. Arseneau, J. Kuhn, T. McCarty, S. Landers, P. Maples, L. Romel, B. Randlev, T. Reid, S. Kaye, and D. Kirn. 2000. Selective replication and oncolysis in p53 mutant tumors with ONYX-015, an E1B-55kD gene-deleted adenovirus, in patients with advanced head and neck cancer: a phase II trial. Cancer Res. 60:6359-6366. [PubMed] [Google Scholar]
- 40.Nemunaitis, J., F. Khuri, I. Ganly, J. Arseneau, M. Posner, E. Vokes, J. Kuhn, T. McCarty, S. Landers, A. Blackburn, L. Romel, B. Randlev, S. Kaye, and D. Kirn. 2001. Phase II trial of intratumoral administration of ONYX-015, a replication-selective adenovirus, in patients with refractory head and neck cancer. J. Clin. Oncol. 19:289-298. [DOI] [PubMed] [Google Scholar]
- 41.Olson, D. C., V. Marechal, J. Momand, J. Chen, C. Romocki, and A. J. Levine. 1993. Identification and characterization of multiple mdm-2 proteins and mdm-2-p53 protein complexes. Oncogene 8:2353-2360. [PubMed] [Google Scholar]
- 42.Oren, M. 2003. Decision making by p53: life, death and cancer. Cell Death Differ. 10:431-442. [DOI] [PubMed] [Google Scholar]
- 43.Pilder, S., M. Moore, J. Logan, and T. Shenk. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Querido, E., R. C. Marcellus, A. Lai, R. Charbonneau, J. G. Teodoro, G. Ketner, and P. E. Branton. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 71:3788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Querido, E., M. R. Morrison, H. Chu-Pham-Dang, S. W. Thirlwell, D. Boivin, P. E. Branton, and M. R. Morisson. 2001. Identification of three functions of the adenovirus E4orf6 protein that mediate p53 degradation by the E4orf6-E1B55K complex. J. Virol. 75:699-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reich, N. C., P. Sarnow, E. Duprey, and A. J. Levine. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128:480-484. [DOI] [PubMed] [Google Scholar]
- 48.Reichel, R., I. Kovesdi, and J. R. Nevins. 1987. Developmental control of a promoter-specific factor that is also regulated by the E1A gene product. Cell 48:501-506. [DOI] [PubMed] [Google Scholar]
- 49.Roth, J., and M. Dobbelstein. 2003. Interaction of p53 with the adenovirus E1B-55 kDa protein. Methods Mol. Biol. 234:135-150. [DOI] [PubMed] [Google Scholar]
- 50.Roth, J., C. König, S. Wienzek, S. Weigel, S. Ristea, and M. Dobbelstein. 1998. Inactivation of p53 but not p73 by adenovirus type 5 E1B 55-kilodalton and E4 34-kilodalton oncoproteins. J. Virol. 72:8510-8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roth, J., C. Lenz-Bauer, A. Contente, K. Lohr, P. Koch, S. Bernard, and M. Dobbelstein. 2003. Reactivation of mutant p53 by a one-hybrid adaptor protein. Cancer Res. 63:3904-3908. [PubMed] [Google Scholar]
- 52.Sarnow, P., Y. S. Ho, J. Williams, and A. J. Levine. 1982. Adenovirus E1b-58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell 28:387-394. [DOI] [PubMed] [Google Scholar]
- 53.Scolnick, D. M., N. H. Chehab, E. S. Stavridi, M. C. Lien, L. Caruso, E. Moran, S. L. Berger, and T. D. Halazonetis. 1997. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 57:3693-3696. [PubMed] [Google Scholar]
- 54.Shen, Y., G. Kitzes, J. A. Nye, A. Fattaey, and T. Hermiston. 2001. Analyses of single-amino-acid substitution mutants of adenovirus type 5 E1b-55K protein. J. Virol. 75:4297-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shieh, S. Y., M. Ikeda, Y. Taya, and C. Prives. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325-334. [DOI] [PubMed] [Google Scholar]
- 56.Somasundaram, K., and W. S. El-Deiry. 1997. Inhibition of p53-mediated transactivation and cell cycle arrest by E1A through its p300/CBP-interacting region. Oncogene 14:1047-1057. [DOI] [PubMed] [Google Scholar]
- 57.Steegenga, W. T., N. Riteco, A. G. Jochemsen, F. J. Fallaux, and J. L. Bos. 1998. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene 16:349-357. [DOI] [PubMed] [Google Scholar]
- 58.Stiewe, T., and B. M. Putzer. 2000. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat. Genet. 26:464-469. [DOI] [PubMed] [Google Scholar]
- 59.Stiewe, T., C. C. Theseling, and B. M. Putzer. 2002. Transactivation-deficient ΔTA-p73 inhibits p53 by direct competition for DNA binding: implications for tumorigenesis. J. Biol. Chem. 277:14177-14185. [DOI] [PubMed] [Google Scholar]
- 60.Takaoka, A., S. Hayakawa, H. Yanai, D. Stoiber, H. Negishi, H. Kikuchi, S. Sasaki, K. Imai, T. Shibue, K. Honda, and T. Taniguchi. 2003. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature 424:516-523. [DOI] [PubMed] [Google Scholar]
- 61.Thimmappaya, B., C. Weinberger, R. J. Schneider, and T. Shenk. 1982. Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell 31:543-551. [DOI] [PubMed] [Google Scholar]
- 62.Tollefson, A. E., J. S. Ryerse, A. Scaria, T. W. Hermiston, and W. S. Wold. 1996. The E3-11.6-kDa adenovirus death protein (ADP) is required for efficient cell death: characterization of cells infected with adp mutants. Virology 220:152-162. [DOI] [PubMed] [Google Scholar]
- 63.Tollefson, A. E., A. Scaria, T. W. Hermiston, J. S. Ryerse, L. J. Wold, and W. S. Wold. 1996. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J. Virol. 70:2296-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 65.Vousden, K. H. 2000. p53: death star. Cell 103:691-694. [DOI] [PubMed] [Google Scholar]
- 66.Vousden, K. H., and X. Lu. 2002. Live or let die: the cell's response to p53. Nat. Rev. Cancer 2:594-604. [DOI] [PubMed] [Google Scholar]
- 67.Weigel, S., and M. Dobbelstein. 2000. The nuclear export signal within the E4orf6 protein of adenovirus type 5 supports virus replication and cytoplasmic accumulation of viral mRNA. J. Virol. 74:764-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.White, E., P. Sabbatini, M. Debbas, W. S. Wold, D. I. Kusher, and L. R. Gooding. 1992. The 19-kilodalton adenovirus E1B transforming protein inhibits programmed cell death and prevents cytolysis by tumor necrosis factor alpha. Mol. Cell. Biol. 12:2570-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wienzek, S., J. Roth, and M. Dobbelstein. 2000. E1B 55-kilodalton oncoproteins of adenovirus types 5 and 12 inactivate and relocalize p53, but not p51 or p73, and cooperate with E4orf6 proteins to destabilize p53. J. Virol. 74:193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang, A., and F. McKeon. 2000. p63 and p73: p53 mimics, menaces and more. Nat. Rev. Mol. Cell Biol. 1:199-207. [DOI] [PubMed] [Google Scholar]
- 71.Yang, A., N. Walker, R. Bronson, M. Kaghad, M. Oosterwegel, J. Bonnin, C. Vagner, H. Bonnet, P. Dikkes, A. Sharpe, F. McKeon, and D. Caput. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404:99-103. [DOI] [PubMed] [Google Scholar]