Abstract
Improvement for carcass traits related to beef quality is the key concern in beef production. Recent reports found that epigenetics mediates the interaction of individuals with environment and nutrition. The present study was designed to analyze the genetic effect of single nucleotide polymorphisms (SNPs) in seven epigenetic-related genes (DNMT1, DNMT3a, DNMT3b, DNMT3L, Ago1, Ago2, and HDAC5) and two meat quality candidate genes (CAPN1 and PRKAG3) on fourteen carcass traits related to beef quality in a Snow Dragon beef population, and also to identify SNPs in a total of fourteen cattle populations. Sixteen SNPs were identified and genotyped in 383 individuals sampled from the 14 cattle breeds, which included 147 samples from the Snow Dragon beef population. Data analysis showed significant association of 8 SNPs within 4 genes related to carcass and/or meat quality traits in the beef populations. SNP1 (13154420A>G) in exon 17 of DNMT1 was significantly associated with rib-eye width and lean meat color score (p<0.05). A novel SNP (SNP4, 76198537A>G) of DNMT3a was significantly associated with six beef quality traits. Those individuals with the wild-type genotype AA of DNMT3a showed an increase in carcass weight, chilled carcass weight, flank thicknesses, chuck short rib thickness, chuck short rib score and in chuck flap weight in contrast to the GG genotype. Five out of six SNPs in DNMT3b gene were significantly associated with three beef quality traits. SNP15 (45219258C>T) in CAPN1 was significantly associated with chuck short rib thickness and lean meat color score (p<0.05). The significant effect of SNP15 on lean meat color score individually and in combination with each of other 14 SNPs qualify this SNP to be used as potential marker for improving the trait. In addition, the frequencies of most wild-type alleles were higher than those of the mutant alleles in the native and foreign cattle breeds. Seven SNPs were identified in the epigenetic-related genes. The SNP15 in CAPN1 could be used as a powerful genetic marker in selection programs for beef quality improvement in the Snow Dragon Beef population.
Keywords: Association Study, Epigenetic-related Genes, CAPN1, Protein Kinase Subunit Gamma-3 Gene PRKAG3, Beef Quality, Carcass Traits
INTRODUCTION
Carcass traits related to beef quality are normally controlled or regulated by a number of genes and single nucleotide polymorphisms (SNPs) in the genes may be significant markers for improved cattle performance (Dekkers and Hospital, 2002; Shin and Chung, 2007). Beef quality and carcass traits usually have low or moderate heritability and are often recorded post-slaughter (Marshall, 1999; Gill et al., 2009), therefore, SNPs can be used as markers for indirectly improving beef quality traits instead of direct measurements (Ge et al., 2003; Allan et al., 2007).
During the last decade two promising SNPs have been identified and are being used in selection programs of the economically important traits in the livestock industry. One of the SNPs is in the calpain 1 gene (CAPN1, (mu/I) large subunit), which is known to associate with meat tenderness (Smith et al., 2000; White et al., 2005; Casas et al., 2006). The other SNP in the 5′-AMP-activated protein kinase subunit gamma-3 gene (PRKAG3), which affects high glycogen content in porcine skeletal muscle (Li et al., 2012; Ryan et al., 2012).
DNA methylation mediates the interaction of cattle with environment and nutrition (Liu et al., 2012), thus it is important to study the genetic effects of SNPs in epigenetic-related genes in beef quality and carcass traits. Recent studies by our research group fellows (Guo et al., 2012; Liu et al., 2012) suggested that epigenetic related genes (DNA (cytosine-5-)-methyltransferase 3 alpha [DNMT3a], DNA (cytosine-5-)-methyltransferase 3 beta [DNMT3b]) play a role in meat quality and carcass traits. The SNPs in DNMT3b were found significantly associated with meat quality and carcass traits (Liu et al., 2012; McClure et al., 2012). The present study continues our previous study (Liu et al., 2012), which was focused on few SNPs in DNMT3b gene with beef quality traits.
The objectives of this study were to analyze the effects of SNPs on fourteen beef quality and carcass traits in seven epigenetic-related genes (four DNA methyltransferases genes, namely DNMT1, DNMT3a, DNMT3b, DNMT3L; two argonaute genes argonaute RISC catalytic component 1 [Ago1] and Ago2; and histone deacetylase 5 gene [HDAC5]) and the two beef quality candidate genes (CAPN1 and PRKAG3), and to develop potential markers for improvement of meat quality traits in Snow Dragon beef cattle. To provide basic molecular information, we also described the SNPs in other thirteen cattle breeds.
MATERIALS AND METHODS
Samples and meat quality traits
Muscle, semen and ear tissue samples were collected from 383 individuals belonging to 14 breeds of cattle. Amongst these 14 breeds, 6 were native Chinese cattle while the other 8 were exotic breeds (Table 1).
Table 1.
Samples used in the study
| Cattle breeds | Numbers | Location of the samples |
|---|---|---|
| Snow Dragon beef | 147 (muscles) | Xuelong Industrial Group(Dalian, China) |
| Holstein | 17* | Beijing Dairy Cattle Centre, China. |
| Simmental | 28 | Beijing Dairy Cattle Centre, Inner Mongolia livestock improvement station, Henan Dingyuan Seedstock Bulls Breeding Ltd. Co, China. |
| Montbeliade | 14 | Coopex, France |
| Norwegian Red Bull | 4 | Geno breeding & AI association, Norway |
| Angus | 9 | Beijing Dairy Cattle Centre, Henan Dingyuan Seedstock Bulls Breeding Ltd. Co, Ningxia Sygen BioEngineering Research Center, China. |
| Charolais | 29 | Beijing Dairy Cattle Centre, Henan Dingyuan Seedstock Bulls Breeding Ltd. Co, Luoyang bull station, Xuchang bull station, Henan, China. |
| Wagyu | 11 (6 ear tissue) | Xuelong Industrial Group and Beijing Dairy Cattle Centre, China |
| Limousin | 24 | Luoyang Baimasi Bull Station, Dingyuan Seedstock Breeding Ltd. Company, Ningxia Sygen BioEngineering Research Center and Beijing Dairy Cattle Centre, China |
| Fuzhou cattle | 7 | National conservation farm of Fuzhou yellow Cattle (Dalian, China) |
| Luxi cattle | 30 | National conservation farm of Shandong Luxi Yellow cattle (Jining, China) |
| Bohai black cattle | 3 | National conservation farm of Shandong Luxi Yellow cattle (Jining, China) |
| Xinjiang brown cattle | 30 | Beijing Dairy Cattle Center, Xinjiang Tianshan Animal Husbandry Bio-engineering Co. Ltd, Xinjiang General Livestock Service, China |
| Sanhe cattle | 30 | Xieertala Breeding Farm, Inner Mongolia, China |
| Total | 383 |
If not noted especially, it means the numbers of semen samples.
The records of 14 meat quality traits were available for 147 individuals from the Snow Dragon beef population (the offspring of Wagyu and Filial 1 hybrid (F1) crossbred cows of Limousin by Fuzhou yellow cattle, supplied by Xuelong Industrial Group, Dalian, China) and were used for association analysis of genotypes with meat quality traits in the present study. The fourteen meat quality traits include dressing percentage (DP), carcass weight (CW), chilled carcass weight (CCW), rib-eye width (RW), rib-eye length (RL), rib-eye area, flank thickness (FT), chuck short rib thickness (CT), chuck short rib score (CS), back-fat thickness (BFT), chuck flap weight (CF), fat color score, lean meat color score (LCS), and marbling score (MS).
Detection and genotyping of the polymorphisms
Genomic DNA was extracted from the muscle and ear tissue samples using the commercial kit of genomic DNA purification Wizard (Promega Corp., Madison, WI, USA) according to the manufacturer’s instructions, while DNA for semen samples were extracted using high salt extraction procedures (Liu, 2007; Qin, 2008). Primer pairs were designed to identify SNPs which covered all coding regions and intron sequences as well as the 5′ regulatory region 2,000 bp upstream of the transcription initiation site using the software Primer 3 (v.0.4.0) and Oligo6.0. A DNA pool was prepared consisting of ten cattle (50 ng/μL per sample) randomly selected from Limousin, Wagyu, Simmental and Angus.
Polymerase chain reaction (PCR) amplifications for the pooled DNA were performed in a final reaction volume of 25 μL consisting of 50 ng genomic DNA, 0.5 μL each primer, 2.5 μL 10× PCR buffer, 2.5 mM of dNTP and 1 U Taq DNA polymerase (TaKaRa, Dalian, China). The PCR protocol was 5 min at 95°C for initial denaturation followed by 40 cycles at 95°C for 30 s, 59°C for 30 s, 72°C for 30 s and a final extension at 72°C for 10 min for all the primer pairs. Matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS, Squeno MassARRAY, Bioyong Technologeies Inc. HK, China) assay was used for individual genotyping of 383 individuals belonging to 14 breeds of cattle.
Statistical analysis
The genotypic and allelic frequencies of the SNPs were calculated using PopGene 3.2. Association analysis between the 15 SNPs and beef quality traits in the population of Snow Dragon beef was carried out using the general linear model procedure of SAS 9.1 with the following fixed effect model:
| (1) |
Where y represents the phenotype records of carcass and meat quality traits; μ is overall mean; gi is the fixed effect of the genotype; sj is the fixed effect of sex; mk is the fixed effect of month-old; dl is the fixed effects of slaughtering date; e is the random error. The statistics were presented as probability values and least squares means±standard error, and a p value of <0.05 was considered statistically significant.
The combination effects of SNP15 with other 14 SNPs on beef quality traits in the Snow Dragon beef population were analyzed with the following model:
| (2) |
Where y, μ, sj, mk, dl, and e are the same as shown in Equation 1, and ci is the effect of the combination genotypes between two SNPs.
RESULTS
Investigation of the promoter and the exon sequences of pooled DNA revealed 16 SNPs in the 9 genes, amongst these 16 SNPs, 7 were novel (Table 2). A total of 3 SNPs were found in the exon regions including SNP1 (13154420A>G) in exon 17 and SNP2 in exon 18 (13153987T>C) of DNMT1, and SNP15 (45219258C>T) in exon 4 of CAPN1. Whereas, the other 13 SNPs were found in the intron regions (Table 2, Figure 1).
Table 2.
Information of the 16 SNPs found in 8 genes in the study
| SNPs | Gene | Position | SNP | Reference |
|---|---|---|---|---|
| SNP1 | DNMT1 | 7chr 13154420 exon17 | A>G | rs135600607 |
| SNP2 | DNMT1 | 7chr 13153987 exon18 | T>C | rs134240033 |
| SNP3 | DNMT1 | 7chr 13155622 intron33 | G>A | rs110894996 |
| SNP4 | DNMT3a | 11chr 76198537 intron11 | A>G | novel |
| SNP5 | DNMT3b | 13chr 63037313 intron10 | C>T | rs135724333 |
| SNP6 | DNMT3b | 13chr 63037426 intron10 | C>T | rs132930989 |
| SNP7 | DNMT3b | 13chr 63030432 intron4 | A>G | rs134010442 |
| SNP8 | DNMT3b | 13chr 63029349 intron3 | C>T | Novel (Liu et al. 2012) |
| SNP9 | DNMT3b | 13chr 63033215 intron6 | T>C | Novel (Liu et al. 2012) |
| SNP10 | DNMT3b | 13chr 63039420 intron13 | A>G | Novel (Liu et al. 2012) |
| SNP11 | Ago1 | 3chr 117123414 intron3 | C>A | novel |
| SNP12 | Ago2 | 14chr 2398943 intron12 | C>T | rs109412562 |
| SNP13 | Ago2 | 14chr 2390703 intron6 | T>C | novel |
| SNP14 | HDAC5 | 19chr 45363113 intron1 | T>C | novel |
| SNP15 | CAPN1 | 29chr 45219258 exon4 | C>T | rs17872079 |
| SNP16 | PRKAG3 | 2chr 111194388 intron4 | T>C | reference |
SNP, single nucleotide polymorphism; DNMT, DNA (cytosine-5-)-methyltransferase; Ago, argonaute RISC catalytic component; HDAC5, histone deacetylase 5; CAPN1, calpain 1, (mu/I) large subunit; PRKAG3, protein kinase subunit gamma-3.
Figure 1.
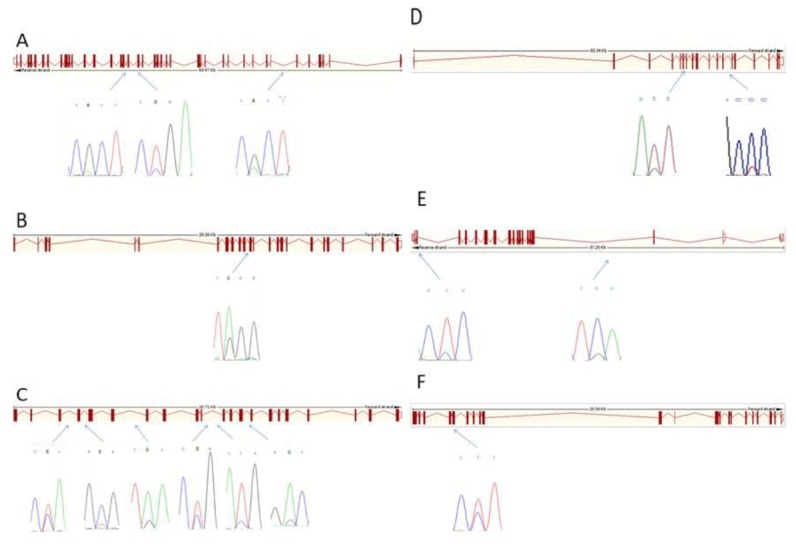
Position of single nucleotide polymorphisms identified in (A) DNMT1, (B) DNMT3a, (C) DNMT3b, (D) Ago2, (E) HDAC5, and (F) CAPN1. DNMT, DNA (cytosine-5-)-methyltransferase; Ago, argonaute RISC catalytic component; HDAC5, histone deacetylase 5; CAPN1, calpain 1, (mu/I) large subunit.
Genotype and allele frequencies
Amongst the 16 SNPs, except SNP14, 15 were successfully genotyped using MassARRAY technique. The Genotype and allele frequencies are shown in Table 3. The wild-type and mutation-type allele frequencies of the 15 SNPs identified in the genes DNMT1, DNMT3a, DNMT3b, Ago2, HDAC5, CAPN1, and PRKAG3 in all 14 breeds of cattle are shown in Figure 2. Generally, the allelic frequencies of wild-type allele were higher than those of the mutation-type allele in each breed, and also there was a plain tendency in the allelic frequency of the native and foreign breeds i.e. the allele T frequencies of SNP2 (13153987T>C) in native breeds (p<0.5 except for Sanhe cattle) were lower (>0.5), whereas, the allele C frequencies of SNP5 (63037313C>T) in native Chinese breeds were higher than those in exotic breeds. The allele and genotype frequencies of the SNPs were very similar between Snow Dragon beef and Wagyu cattle.
Table 3.
Frequencies of alleles and genotypes of the 16 SNPs in the 14 cattle breeds
| SNPs | SNP frequency* | Genotype frequency | |||
|---|---|---|---|---|---|
|
|
|
||||
| SNP1 | A | G | AA | AG | GG |
| 0.52 | 0.48 | 0.29 (111) | 0.45 (168) | 0.25 (94) | |
| SNP2 | T | C | TT | CT | CC |
| 0.52 | 0.48 | 0.30 (112) | 0.44 (164) | 0.25 (94) | |
| SNP3 | G | A | GG | GA | AA |
| 0.57 | 0.43 | 0.35 (132) | 0.43 (158) | 0.22(80) | |
| SNP4 | A | G | AA | AG | GG |
| 0.72 | 0.28 | 0.52 (190) | 0.41 (151) | 0.07 (27) | |
| SNP5 | C | T | CC | CT | TT |
| 0.73 | 0.27 | 0.54 (187) | 0.37(127) | 0.09(31) | |
| SNP6 | C | T | CC | CT | TT |
| 0.79 | 0.21 | 0.63 (227) | 0.33 (117) | 0.04 (16) | |
| SNP7 | A | G | AA | AG | GG |
| 0.72 | 0.28 | 0.54(201) | 0.36 (135) | 0.10 (38) | |
| SNP8 | C | T | CC | CT | TT |
| 0.81 | 0.19 | 0.69 (242) | 0.28(79) | 0.07 (26) | |
| SNP9 | T | C | TT | CT | CC |
| 0.80 | 0.20 | 0.64 (241) | 0.31 (117) | 0.05(17) | |
| SNP10 | A | G | AA | AG | GG |
| 0.79 | 0.21 | 0.65(232) | 0.29 (107) | 0.06 (23) | |
| SNP11 | C | T | CC | CT | TT |
| 0.96 | 0.04 | 0.93(347) | 0.07 (28) | 0(0) | |
| SNP12 | T | C | TT | CT | CC |
| 0.92 | 0.08 | 0.86(323) | 0.13 (50) | 0.01 (4) | |
| SNP13 | T | C | TT | TC | CC |
| 0.47 | 0.53 | 0.25(92) | 0.45 (167) | 0.30 (112) | |
| SNP15 | C | T | CC | TC | TT |
| 0.73 | 0.27 | 0.56(202) | 0.35 (127) | 0.09 (33) | |
| SNP16 | T | C | TT | TC | CC |
| 0.59 | 0.41 | 0.36(132) | 0.46 (170) | 0.18 (67) | |
The wild-type allele was listed in the left.
Figure 2.
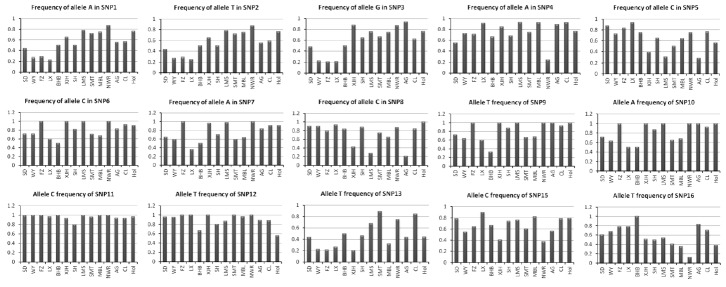
The wild-type allele frequencies of the 15 single nucleotide polymorphisms in the 14 breeds of cattle. SD, Snow Dragon beef; WY, Wagyu; FZ, Fuzhou cattle; LX, Luxi cattle; BHB, Bohai Black cattle; XJH, Xinjiang Brown cattle; SH, Sanhe cattle, LMS, Limousin; SMT, Simmental; MBL, Montbeliade; NWR, Norwegian Red Bull; AG, Angus; CL, Charolais; Hol, Holstein.
Association studies
Out of the 15 SNPs, 8 SNPs in 4 genes (DNMT1, DNMT3a, DNMT3b, and CAPN1) were found significantly associated with carcass and meat quality traits (Table 4). SNP1 (13154420A>G, National Center for Biotechnology Information [NCBI] reference no rs135600607) of DNMT1 was a silent mutation located in exon 17 of chromosome 7 and was found significantly associated with RW and LCS (p<0.05). Animals with mutation genotype GG in SNP1 performed better than the wild type genotype AA for the two traits.
Table 4.
Significant association analysis between SNP effects and meat quality traits in Snow Dragon beef population (LSM±SE)*
| SNP | Genotype | CW (kg) | CCW (kg) | DP (%) | RA (cm2) | RW (cm) | RL (cm) | FT (cm) | CT (cm) | CS | BFT (cm) | CF (kg) | FCS | LCS | MS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP1 | AA | 344.39±10.71 | 268.85±21.93 | 0.46±0.04 | 33.99±1.10 | 5.44±0.11a | 8.91±0.16 | 5.58±0.15 | 2.29±0.13 | 3.04±0.27 | 2.27±0.13 | 2.34±0.09 | 4.00±0.06 | 2.59±0.14 | 2.19±0.22 |
| AG | 344.92±6.70 | 311.85±13.72 | 0.53±0.02 | 36.60±0.69 | 5.65±0.07ab | 9.21±0.10 | 5.62±0.09 | 2.43±0.08 | 3.23±0.17 | 2.16±0.08 | 2.47±0.06 | 4.03±0.04 | 2.93±0.09 | 2.77±0.14 | |
| GG | 355.35±8.39 | 315.35±17.18 | 0.54±0.03 | 37.19±0.87 | 5.79±0.08b | 9.15±0.13 | 5.77±0.11 | 2.27±0.10 | 3.25±0.21 | 1.94±0.10 | 2.44±0.07 | 3.95±0.05 | 2.64±0.11 | 2.55±0.17 | |
| p value | >0.05 | >0.05 | >0.05 | >0.05 | <0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.05 | >0.05 | |
| SNP4 | AA | 376.08±9.19A | 349.01±19.10a | 0.58±0.03 | 37.97±0.99 | 5.72±0.10 | 9.44±0.15 | 6.06±0.12A | 2.66±0.11A | 3.69±0.24 | 2.06±0.12 | 2.61±0.08a | 4.03±0.05 | 2.71±0.12 | 2.74±0.20 |
| AG | 340.51 ±5.83B | 290.81 ±12.11b | 0.49±0.02 | 35.83±0.63 | 5.64±0.06 | 9.04±0.09 | 5.52±0.08B | 2.24±0.07B | 3.05±0.15 | 2.17±0.07 | 2.37±0.05b | 3.99±0.03 | 2.82±0.08 | 2.51±0.13 | |
| GG | 340.04±12.47AB | 296.95±25.92ab | 0.51±0.04 | 35.72±1.35 | 5.57±0.13 | 9.12±0.20 | 5.63±0.17AB | 2.31±0.15AB | 2.89±0.32 | 1.84±0.16 | 2.46±0.11ab | 4.00±0.07 | 2.58±0.17 | 2.58±0.27 | |
| p value | <0.01 | <0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.01 | <0.01 | <0.05 | >0.05 | <0.05 | >0.05 | >0.05 | >0.05 | |
| SNP5 | CC | 341.68±5.16A | 298.82±10.78 | 0.51±0.02 | 35.88±0.56 | 5.64±0.05 | 9.07±0.08A | 5.55±0.07A | 2.31±0.06 | 3.18±0.13 | 2.15±0.07a | 2.39±0.04 | 3.99±0.03 | 2.76±0.07 | 2.58±0.11 |
| CT | 370.05±9.93B | 325.69±20.74 | 0.54±0.03 | 37.44±1.08 | 5.71±0.10 | 9.31±0.16A | 6.02±0.14B | 2.48±0.12 | 3.27±0.26 | 2.00±0.13ab | 2.58±0.08 | 4.03±0.06 | 2.80±0.13 | 2.63±0.21 | |
| TT | 425.80±38.44AB | 418.60±80.33 | 0.61±0.13 | 42.70±4.16 | 5.50±0.40 | 11.00±0.60B | 6.25±0.53AB | 2.25±0.47 | 1.00±0.99 | 1.00±0.49b | 3.00±0.32 | 4.00±0.22 | 2.00±0.51 | 1.00±0.82 | |
| p value | <0.01 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.01 | >0.05 | >0.05 | <0.05 | <0.05 | >0.05 | >0.05 | >0.05 | |
| SNP6 | CC | 351.15±6.69 | 317.36±13.65 | 0.54±0.02 | 36.09±0.71 | 5.61±0.07 | 9.16±0.10 | 5.60±0.09 | 2.28±0.08 | 3.28±0.17 | 2.13±0.08 | 2.39±0.05A | 4.00±0.04 | 2.79±0.09 | 2.65±0.14 |
| CT | 346.36±7.22 | 292.45±14.72 | 0.50±0.02 | 36.59±0.77 | 5.68±0.07 | 9.16±0.11 | 5.72±0.10 | 2.45±0.09 | 3.08±0.18 | 2.03±0.09 | 2.43±0.06A | 4.00±0.04 | 2.79±0.09 | 2.52±0.15 | |
| TT | 352.87±18.79 | 308.16±38.33 | 0.51±0.06 | 36.77±2.00 | 5.80±0.19 | 9.00±0.29 | 5.83±0.26 | 2.24±0.22 | 2.78±0.48 | 2.38±0.23 | 2.89±0.15AB | 4.00±0.11 | 2.33±0.24 | 2.11±0.39 | |
| p value | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.01 | >0.05 | >0.05 | >0.05 | |
| SNP7 | AA | 348.35±7.38 | 314.66±15.11 | 0.54±0.02 | 35.88±0.78 | 5.61±0.08 | 9.12±0.12 | 5.61±0.10 | 2.29±0.09 | 3.36±0.19 | 2.12±0.08 | 2.38±0.05A | 4.00±0.04 | 2.78±0.09 | 2.72±0.15 |
| AG | 349.19±6.77 | 301.62±13.86 | 0.51±0.02 | 36.79±0.72 | 5.68±0.07 | 9.22±0.11 | 5.69±0.09 | 2.42±0.08 | 3.13±0.17 | 2.03±0.09 | 2.43±0.06B | 4.00±0.04 | 2.79±0.09 | 2.55±0.14 | |
| GG | 353.53±14.52 | 296.24±29.72 | 0.49±0.05 | 36.01±1.54 | 5.71±0.15 | 8.97±0.23 | 5.73±0.20 | 2.25±0.17 | 2.60±0.36 | 2.49±0.25 | 2.96±0.16B | 4.00±0.11 | 2.38±0.26 | 2.11±0.39 | |
| p value | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.01 | >0.05 | >0.05 | >0.05 | |
| SNP9 | TT | 350.18±6.65 | 316.78±13.56 | 0.54±0.02 | 35.96±0.71 | 5.59±0.07 | 9.16±0.10 | 5.59±0.09 | 2.27±0.08 | 3.28±0.17 | 2.15±0.07a | 2.39±0.04 | 3.99±0.03 | 2.76±0.07 | 2.65±0.14 |
| CT | 347.25±7.22 | 293.33±14.73 | 0.50±0.02 | 36.81±0.77 | 5.71±0.07 | 9.18±0.11 | 5.73±0.10 | 2.45±0.09 | 3.08±0.18 | 2.00±0.13ab | 2.58±0.08 | 4.03±0.06 | 2.80±0.13 | 2.51±0.15 | |
| CC | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | |
| p value | 0.9133 | 0.5055 | 0.5052 | 0.7190 | 0.4342 | 0.7683 | 0.4121 | 0.2867 | 0.5147 | <0.05 | <0.05 | 0.8089 | 0.3202 | 0.4334 | |
| SNP10 | AA | 344.07 ±8.38 | 302.29 ±17.40 | 0.59 ±0.14a | 36.59 ±0.89 | 5.75 ±0.09 | 9.06 ±0.13 | 5.63 ±0.12 | 2.38 ±0.10 | 2.93±0.44 | 1.92 ±0.32a | 2.39 ±0.07 | 4.05 ±0.05 | 2.63 ±0.23 | 2.42±0.61 |
| AG | 347.08 ±6.55 | 306.81 ±13.60 | 0.76 ±0.13b | 35.98 ±0.69 | 5.58 ±0.07 | 9.17 ±0.10 | 5.63 ±0.09 | 2.31 ±0.08 | 2.74 ±0.41 | 2.48 ±0.30b | 2.43 ±0.05 | 3.97 ±0.04 | 2.55 ±0.22 | 2.99±0.58 | |
| GG | 360.46 ±11.35 | 304.43 ±23.56 | 0.56 ±0.19ab | 36.43 ±1.20 | 5.68 ±0.12 | 9.17 ±0.17 | 5.80 ±0.16 | 2.41 ±0.14 | 2.22 ±0.59 | 2.00±0.43ab | 2.53 ±0.09 | 4.00 ±0.06 | 2.52 ±0.32 | 1.74±0.83 | |
| p value | >0.05 | >0.05 | <0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.05 | >0.05 | >0.05 | >0.05 | <0.05 | |
| SNP15 | CC | 351.85±5.90 | 294.37±12.14 | 0.49±0.02 | 35.99±0.62 | 5.63±0.06 | 9.11±0.09 | 5.63±0.08 | 2.28±0.07 | 3.20±0.15 | 2.24±0.07 | 2.40±0.05 | 4.03±0.03 | 2.88±0.07A | 2.60±0.12 |
| CT | 341.48±8.48 | 327.20±17.46 | 0.56±0.03 | 36.28±0.89 | 5.63±0.09 | 9.18±0.13 | 5.69±0.12 | 2.56±0.10 | 3.35±0.21 | 1.93±0.10 | 2.46±0.07 | 3.93±0.05 | 2.72±0.11A | 2.65±0.18 | |
| TT | 353.98±19.67 | 309.73±40.49 | 0.54±0.07 | 39.39±2.06 | 5.96±0.20 | 9.31±0.30 | 5.81±0.27 | 2.08±0.23 | 2.38±0.49 | 1.69±0.24 | 2.63±0.16 | 4.00±0.11 | 1.88±0.24B | 2.13±0.41 | |
| p value | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.01 | >0.05 |
SNP, single nucleotide polymorphism; LSM, least squares means; SE, standard error; CW, Carcass weight; CCW, chilled carcass weight; DP, dressing percentage; RA, rib-eye area; RW, rib-eye width; RL, rib-eye length; FT, flank thickness; CT, chuck short rib thickness; CS, chuck short rib score; BFT, back-fat thickness; CF, chuck flap weight; FCS, fat color score; LCS, lean meat color score; MS, marbling score.
The Bonferroni t test was used for pair comparison in the study.
Means within a column with no common superscript letters differ at p<0.05.
Means within a column with no common superscript letters differ at p<0.01.
SNP4 (76198537A>G) was found to be a novel SNP and located at intron 11 of DNMT3a gene. Allele A of SNP5 was significantly associated with CF (p<0.05) and highly significantly (p<0.01) associated with CW, CCW, FT, and CT. Compared with the mutation genotype GG, individuals with the wild-type genotype AA showed remarkable increase in CW, CCW, FT, CT, CS, and in CF.
Six SNPs were uncovered in the DNMT3b gene. Of which, five SNPs i.e. SNP5 (63037313C>T), SNP6 (63037426C>T), SNP7 (63030432A>G), SNP9 (63033215T>C) and SNP10 (63039420A>G) were significantly associated with beef quality traits (p<0.05). Individuals with the mutation genotype TT of SNP5 (63037313C>T), compared to wild-type genotype CC, showed an increase of 84.12 kg in CW, 1.93 cm in RL, 0.70 cm in FT, 0.61 kg in CF and a decrease of 1.15 cm in BFT. In SNP6 (63037426C>T) the difference between the wild type CC and mutation type TT genotype was 0.50 kg for CF. The mutation genotype GG of SNP8 showed an increase of 0.58 in CF, whereas, for SNP9 the differences between the wild type genotype TT and the mutation genotype CC for CF and BFT were 1.15 kg and 0.61 cm, respectively. Individuals with the heterozygous genotype (AG) in SNP10 showed an increase in DP, BFT, and MS in contrast with the homozygotes. The rest of the SNPs were significantly (p<0.05) associated with CF except SNP11 (63039420A>G).
The SNP15 (45219258C>T, NCBI reference No rs17872079) of CAPN1 gene is a silent mutation in exon 4 on chromosome 29, which was significantly associated with CT and LCS (p<0.05). Individuals with the wild type CC genotype showed increases of 0.20 cm in CT and 1.00 in LCS.
The effects of combined genotypes on meat quality
The SNP15 (45219258C>T) of CAPN1 was significantly associated with CT and LCS. Therefore, SNP15 (45219258C>T) was selected for analysis in combination with 14 other SNPs to investigate the effects on beef quality traits in the Snow Dragon beef population. Interestingly, all of the 14 combination genotypes were associated with LCS, and the lowest score was showed by combination genotype of NNTT, probably because of the strong association of TT genotype of SNP15 with lower LCS. The results of combination genotypes are mentioned in Table 5.
Table 5.
p Values of the effects of the combination genotypes on beef quality traits in Snow Dragon beef population
| Combination SNPs | CW | CCW | DP | RA | RW | RL | FT | CT | CS | BFT | CF | FCS | LCS | MS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||
| (kg) | (kg) | (%) | (cm2) | (cm) | (cm) | (cm) | (cm) | (cm) | (kg) | |||||
| SNP1-15 | >0.05 | >0.05 | <0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.01 | >0.05 |
| SNP2-15 | >0.05 | >0.05 | <0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.01 | >0.05 |
| SNP3-15 | >0.05 | >0.05 | <0.05 | >0.05 | >0.05 | >0.05 | <0.05 | <0.05 | <0.05 | <0.05 | >0.05 | >0.05 | <0.01 | >0.05 |
| SNP4-15 | <0.01 | >0.05 | >0.05 | >0.05 | >0.05 | <0.05 | <0.01 | <0.05 | >0.05 | >0.05 | <0.01 | >0.05 | <0.01 | >0.05 |
| SNP5-15 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.05 | >0.05 | >0.05 | <0.01 | >0.05 |
| SNP6-15 | >0.05 | >0.05 | >0.05 | <0.05 | >0.05 | <0.05 | >0.05 | >0.05 | >0.05 | <0.05 | >0.05 | >0.05 | <0.01 | >0.05 |
| SNP7-15 | >0.05 | >0.05 | <0.05 | <0.01 | <0.05 | <0.05 | <0.05 | <0.05 | >0.05 | >0.05 | <0.05 | >0.05 | <0.01 | >0.05 |
| SNP8-15 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.05 | >0.05 | <0.05 | >0.05 | >0.05 | <0.01 | >0.05 |
| SNP9-15 | >0.05 | >0.05 | >0.05 | <0.05 | >0.05 | <0.05 | >0.05 | >0.05 | >0.05 | <0.05 | <0.05 | >0.05 | <0.01 | >0.05 |
| SNP10-15 | >0.05 | >0.05 | >0.05 | <0.05 | >0.05 | <0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.01 | >0.05 |
| SNP11-15 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.05 | >0.05 | >0.05 | <0.01 | >0.05 |
| SNP12-15 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.01 | >0.05 |
| SNP13-15 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.01 | >0.05 |
| SNP16-15 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | <0.05 | >0.05 | <0.05 | <0.05 | >0.05 | >0.05 | <0.01 | <0.01 |
CW, carcass weight; CCW, chilled carcass weight; DP, dressing percentage; RA, rib-eye area; RW, rib-eye width; RL, rib-eye length; FT, flank thickness; CT, chuck short rib thickness; CS, chuck short rib score; BFT, back-fat thickness; CF, chuck flap weight; FCS, fat color score; LCS, lean meat color score; MS, marbling score.
DISCUSSION
In the global scenario of increasing demand for protein sources and food, improving beef quality traits through marker assisted selection has received increasing attention (Pariacote et al., 1998; Lindholm-Perry et al., 2012; McClure et al., 2012). The use of effective molecular markers in selection for beef quality traits is a worthy approach that can bring improvement in beef quality (Hoey et al., 1995; Zan et al., 2007).
In the present study, fifteen SNPs in seven epigenetic-related genes and two meat quality candidate genes were detected in a Snow Dragon cattle population as well as thirteen other cattle breeds. Results of the study indicated that 8 out of 15 SNPs were significantly associated with carcass and beef quality traits. In these 8 SNPs, SNP1 (13154420A>G) and SNP15 (45219258C>T) were silent mutations found in the exon region of the DNMT1 and CAPN1 genes and were significantly associated with meat quality traits. DNMT3a and DNMT3b were found important genes in which SNPs were significantly associated with a couple of beef quality traits. Moreover, seven SNPs in the 5 genes (DNMT3L, AGO1, AGO2, HDAC5, and PRKAG3) were non-significantly associated with all of the beef carcass and quality traits in the Snow Dragon beef population.
The DNMT1, DNMT3a, and DNMT3b are DNA methylation-related genes and were found significantly associated with carcass traits related to beef quality. Recently, studies had indicated that gene expression of DNMT3a and DNMT3b were related to fat deposition depicting their potential role in adipose tissue (Kamei et al., 2010). Our recent studies on bovine DNMT3a and DNMT3b showed that DNMT3a was more highly expressed in liver and muscle tissues in beef cattle than DNMT3b, and both of the genes were found significantly associated with beef quality traits (Guo et al., 2012; Liu et al., 2012).
In the present study, the SNPs in DNMT1 and DNMT3a gene were significantly associated with many of the beef quality traits. Mutant allele “G” of SNP1 (13154420A>G) and wild type allele “A” of SNP4 (76198537A>G), associated with improvement in beef quality traits, can be used as potential markers for beef quality improvement. DNMT3b gene was revealed to be a noteworthy gene having 5 SNPs significantly associated with many beef quality traits. The significant influence of SNPs in DNMT3b gene on CF revealed that this gene could be a potential candidate gene in future studies for beef quality improvement.
The CAPN1 gene has been extensively studied and it has been reported that SNPs in this gene were significantly associated with carcass and meat quality traits, especially, with meat tenderness and MS in beef cattle (Page et al., 2004; Corva et al., 2007; van Eenennaam et al., 2007; Pinto et al., 2010). In the present study, SNP15 was chosen for combination genotype association with the rest of 14 SNPs to analyze the effects on beef quality traits. Interestingly, the combination genotype effect of SNP15 (45219258C>T) with all 14 other genotypes showed significant association with LCS. Therefore, it is proposed to consider this SNP as a genetic marker for selection of higher meat quality in beef cattle. Moreover, the results indicated that the effects of combination genotypes were higher than the single SNP effect on the corresponding phenotype.
Initially the fixed effects of the genotype, sex, month-old and slaughtering date were used for data analysis in the model. The results revealed that only SNP had significant effect on the studied phenotypes whereas, all other fixed effects were non-significant effect. Considering the reduced model excluding all other fixed effects except the effect of the genotype changed the results but these results are in line with the previous results (the individual SNP effect remained significant on most of the traits under study). We have provided the fresh results as additional files (Supplementary Tables 1 and 2). These results show that the SNPs alone have significant effects on the phenotypes.
In PRKAG3 gene, SNPs are known to be correlated with gene expression and meat quality phenotypes and this gene has been proposed as a positional and functional candidate gene for quantitative trait loci exerting significant effects on the beef quality traits (Rohrer et al., 2012). More than 100 SNPs have been identified in the CAPN1 gene of cattle and out of these, 4 SNPs have revealed significant effects on meat tenderness (Smith et al., 2000; Juszczuket et al., 2004; Pag et al., 2004).
For the fourteen cattle population, we found that most wild type alleles had higher allelic frequencies in the population than the mutation alleles. These findings suggested that wild type alleles were not frequently mutated and maintained differential tendency for the native and imported breeds. The allelic and genotypic frequencies of Wagyu and Snow Dragon beef were in accordance with each other due to their common genetic background (As, Snow Dragon beef are the offspring of Wagyu and F1 crossbred cows of Limousin by Fuzhou yellow cattle). These results were in accordance with our previous study (Liu et al., 2012).
CONCLUSIONS
Amongst 9 screened and analyzed genes in the present study, SNPs in only 4 genes i.e. DNMT1, DNMT3a, DNMT3b, and CAPN1 were found significantly associated with carcass and beef quality traits. In the DNA methylation-related genes, 7 SNPs significantly associated with many of the carcass traits related to beef quality suggesting that these genes could be potential candidate genes for genetic improvement in beef quality traits. Moreover, the significant association of SNP15 in CPAN1 gene alone or in combination with all 14 other SNPs with lean meat score strongly qualify this SNP to be used as a genetic marker in selection for improvement in this particular beef quality trait. The SNPs found in the present study can be used in further studies to investigate the functional effects of these SNPs on meat quality traits.
Supplementary Data
ACKNOWLEDGMENTS
Authors are thankful to National-Level Conservation Farms of Yellow Cattle for providing cattle semen samples. The research was financially supported by the Earmarked Fund for Modern Agro-industry Technology Research System (CARS-37, 38), the National Natural Science Foundation of China (31272420), the Fund for Basic Research from the Ministry of Education of the People’s Republic of China (2011JS006), the Program for Changjiang Scholar and Innovation Research Team in University (IRT1191), the National Key Technologies R & D Program (2011BAD28B02), and Xuelong Biological Breeding Technology Industrialization Project funded by the National Development and Reform Commission (NDRC).
REFERENCES
- Allan MF, Thallman RM, Cushman RA, Echternkamp SE, White SN, Kuehn LA, Casas E, Smith TPL. Association of a single nucleotide polymorphism in SPP1 with growth traits and twinning in a cattle population selected for twinning rate. J Anim Sci. 2007;85:341–347. doi: 10.2527/jas.2006-460. [DOI] [PubMed] [Google Scholar]
- Casas E, White SN, Wheeler TL, Shackelford SD, Koohmaraie M, Riley DG, Chase CC, Johnson DD, Smith TPL. Effects of calpastatin and micro-calpain markers in beef cattle on tenderness traits. J Anim Sci. 2006;84:520–525. doi: 10.2527/2006.843520x. [DOI] [PubMed] [Google Scholar]
- Corva P, Soria L, Schor A, Villarreal E, Cenci MP, Motter M, Mezzadra C, Melucci L, Miquel C, Pavan E, Depetris G, Santini F, Naon JG. Association of CAPN1 and CAST gene polymorphisms with meat tenderness in Bos taurus beef cattle from Argentina. Genet Mol Biol. 2007;30:1064–1069. [Google Scholar]
- Dekkers JCM, Hospital F. The use of molecular genetics in the improvement of agricultural populations. Nat Rev Genet. 2002;3:22–32. doi: 10.1038/nrg701. [DOI] [PubMed] [Google Scholar]
- Ge W, Davis ME, Hines HC, Irvin KM, Simmen RCM. Association of a genetic marker with blood serum insulin-like growth factor-I concentration and growth traits in Angus cattle. J Anim Sci. 2001;79:1757–1762. doi: 10.2527/2001.7971757x. [DOI] [PubMed] [Google Scholar]
- Gill JL, Bishop SC, McCorquodale C, Williams JL, Wiener P. Association of selected SNP with carcass and taste panel assessed meat quality traits in a commercial population of Aberdeen Angus-sired beef cattle. Genet Sel Evol. 2009;41:36. doi: 10.1186/1297-9686-41-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Liu X, Xu X, Wu M, Zhang X, Li Q, Liu W, Zhang Y, Wang Y, Yu Y. The expression levels of DNMT3a/3b and their relationship with meat quality in beef cattle. Mol Biol Rep. 2012;39:5473–5479. doi: 10.1007/s11033-011-1349-2. [DOI] [PubMed] [Google Scholar]
- Hoey AJ, Reich MM, Davis G, Shorthose R. Beta 2-adrenoceptor densities do not correlate with growth, carcass quality, or meat quality in cattle. J Anim Sci. 1995;73:3281–3286. doi: 10.2527/1995.73113281x. [DOI] [PubMed] [Google Scholar]
- Juszczuket KE, Sakowski T, Flisikowski K, Wicinska K, Oprzadek J, Rosochacki SJ. Bovine mu-calpain (CAPN1) gene: New SNP within intron 14. J Appl Genet. 2004;45:457–460. [PubMed] [Google Scholar]
- Kamei Y, Suganami T, Ehara T, Kanai S, Hayashi K, Yamamoto Y, Miura S, Ezaki O, Okano M, Oqawa Y. Increased expression of DNA methyltransferase 3a in obese adipose tissue: Studies with transgenic mice. Obesity. 2010;18:314–321. doi: 10.1038/oby.2009.246. [DOI] [PubMed] [Google Scholar]
- Li WF, Li JY, Gao X, Xu SZ, Yue WB. Association analysis of PRKAG3 gene variants with carcass and meat quality traits in beef cattle. Afr J Biotechnol. 2012;11:1855–1861. [Google Scholar]
- Lindholm-Perry AK, Kuehn LA, Snelling WM, Smith TPL, Ferrell CL, Jenkins TG, King DA, Shackelford SD, Wheeler TL, Freet HC. Genetic markers on BTA14 predictive for residual feed intake in beef steers and their effects on carcass and meat quality traits. Anim Genet. 2012;43:599–603. doi: 10.1111/j.1365-2052.2011.02307.x. [DOI] [PubMed] [Google Scholar]
- Liu X, Guo XY, Xu XZ, Wu M, Zhang X, Li Q, Ma PP, Zhang Y, Wang CY, Geng FJ, Qin CH, Liu L, Shi WH, Wang YC, Yu Y. Novel single nucleotide polymorphisms of the bovine methyltransferase 3b gene and their association with meat quality traits in beef cattle. Genet Mol Res. 2012;11:2569–2577. doi: 10.4238/2012.June.29.1. [DOI] [PubMed] [Google Scholar]
- Liu R. PhD Thesis. China Agricultural University; Beijing, China: 2007. Fine Maping Milk Production Traits QTL on BTA6 in Chinese Holstein with SNP Markers. [Google Scholar]
- Marshal DM. Genetics of meat quality. In: Fries R, Ruvinsky A, editors. Genetics of Cattle. CABI Publishing; Oxford, UK: 1999. pp. 605–636. [Google Scholar]
- McClure MC, Ramey HR, Rolf MM, McKay SD, Decker JE, Chapple RH, Kim JW, Taxis TM, Weaber RL, Schnabel RD, Taylor JF. Genome-wide association analysis for quantitative trait loci influencing Warner–Bratzler shear force in five taurine cattle breeds. Anim Genet. 2012;43:662–673. doi: 10.1111/j.1365-2052.2012.02323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page BT, Casas E, Quaas RL, Thallman RM, Wheeler TL, Shackelford SD, Koohmaraie M, White SN, Bennett GL, Keele JW, Dikeman ME, Smith TP. Association of markers in the bovine CAPN1 gene with meat tenderness in large crossbred populations that sample influential industry sires. J Anim Sci. 2004;82:3474–3481. doi: 10.2527/2004.82123474x. [DOI] [PubMed] [Google Scholar]
- Pariacote F, Van Vleck LD, Hunsley RE. Genetic and phenotypic parameters for carcass traits of American Shorthorn beef cattle. J Anim Sci. 1998;76:2584–2588. doi: 10.2527/1998.76102584x. [DOI] [PubMed] [Google Scholar]
- Pinto LFB, Ferraz JBS, Meirelles FV, Eler JP, Rezende FM, Carvalho ME, Almeida HB, Silva RCG. Association of SNPs on CAPN1 and CAST genes with tenderness in Nellore cattle. Genet Mol Res. 2010;9:1431–1442. doi: 10.4238/vol9-3gmr881. [DOI] [PubMed] [Google Scholar]
- Qin C. PhD Thesis. China Agricultural University; Beijing, China: 2008. QTL Mapping of Important Economic Traits on BTA3in Chinese Holstein. [Google Scholar]
- Rohrer GA, Nonneman DJ, Miller RK, Zerby H, Moeller SJ. Association of single nucleotide polymorphism (SNP) markers in candidate genes and QTL regions with pork quality traits in commercial pigs. Meat Sci. 2012;92:511–518. doi: 10.1016/j.meatsci.2012.05.020. [DOI] [PubMed] [Google Scholar]
- Ryan MT, Hamill RM, O’ Halloran OAM, Davey GC, McBryan J, Mullen AM, McGee C, Gispert M, Southwood OI, Sweeney T. SNP variation in the promoter of the PRKAG3 gene and association with meat quality traits in pig. BMC Genet. 2012;13:66. doi: 10.1186/1471-2156-13-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SC, Chung ER. SNP detection of carboxypeptidase E gene and its association with meat quality and carcass traits in Korean cattle. Asian Australas J Anim Sci. 2007;20:328–333. [Google Scholar]
- Smith TP, Casas E, Rexroad CE, Kappes SM, Keele JW. Bovine CAPN1 maps to a region of BTA29 containing a quantitative trait locus for meat tenderness. J Anim Sci. 2000;78:2589–2594. doi: 10.2527/2000.78102589x. [DOI] [PubMed] [Google Scholar]
- Turek-Plewa J, Jagodzinski PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10:631–647. [PubMed] [Google Scholar]
- White SN, Casas E, Wheeler TL, Shackelford SD, Koohmaraie M, Riley DG, Chase CC, Johnson DD, Keele JW, Smith TPL. A new single nucleotide polymorphism in CAPN1 extends the current tenderness marker test to include cattle of Bos indicus, Bos taurus, and crossbred descent. J Anim Sci. 2005;83:2001–2008. doi: 10.2527/2005.8392001x. [DOI] [PubMed] [Google Scholar]
- van Eenennaam AL, Li J, Thallman RM, Quaas RL, Dikeman ME, Gill CA, Franke DE, Thomas MG. Validation of commercial DNA tests for quantitative beef quality traits. J Anim Sci. 2007;85:891–900. doi: 10.2527/jas.2006-512. [DOI] [PubMed] [Google Scholar]
- Zan L, Zhang J, Liu X. Association study on AGPAT6 intron3 polymorphism and milk performance of dairy cattle. Sci Agric Sin. 2007;40:1498–1503. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


