Abstract
Lysyl-tRNA synthetase (LysRS) is packaged into human immunodeficiency virus type 1 (HIV-1) via its interaction with Gag, and this enzyme facilitates the selective packaging of tRNA3Lys, the primer for initiating reverse transcription, into HIV-1. The Gag/LysRS interaction is detected at detergent-resistant membrane but not in membrane-free cell compartments that contain Gag and LysRS. LysRS is found (i) in the nucleus, (ii) in a cytoplasmic high-molecular-weight aminoacyl-tRNA synthetase complex (HMW aaRS complex), (iii) in mitochondria, and (iv) associated with plasma membrane. The cytoplasmic form of LysRS lacking the mitochondrial import signal was previously shown to be efficiently packaged into virions, and in this report we also show that LysRS compartments in nuclei, in the HMW aaRS complex, and at the membrane are also not required as a primary source for viral LysRS. Exogenous mutant LysRS species unable to either enter the nucleus or bind to the cell membrane are still incorporated into virions. Many HMW aaRS components are not packaged into the virion along with LysRS, and the interaction of LysRS with p38, a protein that binds tightly to LysRS in the HMW aaRS complex, is not required for the incorporation of LysRS into virions. These data indicate that newly synthesized LysRS may interact rapidly with Gag before the enzyme has the opportunity to move to the above-mentioned cellular compartments. In confirmation of this idea, we found that newly synthesized LysRS is associated with Gag after a 10-min pulse with [35S]cysteine/methionine. This observation is also supported by previous work indicating that the incorporation of LysRS into HIV-1 is very sensitive to the inhibition of new synthesis of LysRS.
Human immunodeficiency virus type 1 (HIV-1) contains tRNA3Lys bound to the viral genomic RNA, and after infection of a cell by HIV-1, this tRNA3Lys is used to prime the reverse transcriptase-catalyzed synthesis of cDNA from the viral RNA genome template (26). The double-stranded viral DNA is translocated into the nucleus, where it integrates into the host cell's DNA, and codes for viral mRNA and proteins. Proteins comprising the viral structure include both the glycosylated envelope proteins (gp120 and gp41) and mature proteins resulting from the processing of the large precursor protein Gag (Pr55gag): matrix (MAp11), capsid (CAp24), and nucleocapsid (NCp7). Gag also contains C-terminal sequences for the p6 protein which, while not part of the viral structure, are believed to play an important role in viral processes related to the endosomal pathway, such as Gag trafficking and viral budding (1, 10, 40, 44). The precursor protein GagPol (Pr160GagPol) is also packaged into the virion and is processed into the three viral enzymes required in the HIV-1 life cycle, protease (PRp11), reverse transcriptase (RTp66/p51), and integrase (INp32). Gag alone is capable of forming extracellular Gag viruslike particles (VLPs) and facilitates the incorporation into virions of both viral genomic RNA and GagPol. Genomic RNA interacts with the nucleocapsid sequence in Gag (2, 9), and GagPol interacts with Gag through intermolecular interactions between homologous Gag sequences in both molecules (32, 37-39). Gag, GagPol, and genomic RNA assemble at the cell membrane containing viral envelope proteins, and, during budding from the cell, the viral protease PRp11 is activated and cleaves these two precursors into the proteins found in the mature virion (for a review of the HIV-1 life cycle, see reference 41).
The major cellular tRNALys isoacceptors, tRNA3Lys and tRNA1,2Lys, are selectively incorporated into HIV-1 during viral assembly (20). Their cognate aminoacyl-tRNA synthetase (aaRS), lysyl-tRNA synthetase (LysRS), is also incorporated into the virion (5). An HIV-1 population contains, on average, approximately 20 to 25 molecules of LysRS per virion (3), similar to the average number of tRNALys molecules per virion (17). Increasing the amount of tRNA3Lys incorporated into HIV-1 results in a viral population with increased levels of tRNA3Lys annealed to the viral RNA genome and increased infectivity (8). While LysRS can be packaged into VLPs composed only of Gag (5), GagPol is required for the additional packaging of tRNALys into these particles or into virions (21, 25). The reverse transcriptase domain in GagPol is important for its interaction with tRNALys (21). The sites of interaction between Gag and LysRS involve the C-terminal 54 amino acids in the capsid region in Gag with amino acids 207 to 259 in LysRS (19).
Several types of data favor a role for LysRS as the signal that targets tRNALys for incorporation into virions. LysRS is incorporated into viruses independently of tRNALys, i.e., it has been found to be packaged into VLPs composed only of Gag (5), and removal of tRNA binding sites on LysRS does not prevent its incorporation into Gag VLPs (19). Overexpression of LysRS in the cell results in a near doubling of the incorporation of both tRNALys and LysRS into HIV-1 (4, 8), and the ability of tRNALys to interact with LysRS is required for the incorporation of tRNALys into the virion (18). Further evidence that the cognate aaRS may serve to facilitate the selective incorporation of primer tRNA into retrovirus is provided by the observation that TrpRS, but not LysRS, is detected in Rous sarcoma virus, a virus that selectively incorporates and uses tRNATrp as a primer tRNA for reverse transcription (3). Based on this evidence, the selective packaging of the tRNALys isoacceptors into HIV-1 may result from a Gag/GagPol/genomic RNA complex interacting with a tRNALys/LysRS complex, with Gag interacting with both GagPol and LysRS, and GagPol interacting with the tRNALys, thereby stabilizing the presence of tRNALys in the complex.
The cellular source of viral LysRS is not known and was investigated in this work. In higher eukaryotes, LysRS is found both in the nucleus (27) and in a cytoplasmic high-molecular-weight (HMW) aaRS complex. Nuclear LysRS is not required as a source of viral LysRS, since, as shown herein, mutant LysRS unable to enter the nucleus is still packaged into the virus. The cytoplasmic HMW aaRS complex contains nine aaRSs: LysRS, arginyl-tRNA synthetase (ArgRS), prolyl-tRNA synthetase (ProRS), glutaminyl-tRNA synthetase (GlnRS), isoleucyl-tRNA synthetase (IleRS), methionyl-tRNA synthetase (MetRS), glutamyl-tRNA synthetase (GluRS), leucyl-tRNA synthetase (LeuRS), and aspartyl-tRNA synthetase (AspRS) (34). There are also three non-aaRS proteins, called p18, p38, and p43, found in this complex. p38, in particular, binds tightly to LysRS and may be a scaffold protein for the complex (34). In this report, we provide evidence that the HMW aaRS complex is not required as a source of viral LysRS based on the fact that, for those HMW aaRS components for which antibody is available, only LysRS reacts with Gag in the cytoplasm and is packaged into the virus. Furthermore, the incorporation of LysRS into virions does not depend upon its ability to interact with p38. In addition to nuclear and cytoplasmic LysRS, a small fraction of LysRS appears to be membrane bound in uninfected cells, but this membrane-bound LysRS does not seem to be required as a source of viral LysRS, since mutant LysRS unable to associate with the membrane in uninfected cells is still packaged into HIV-1.
(This work was performed by R.H. in partial fulfillment of the Ph.D. degree, McGill University, Montreal, Canada.)
MATERIALS AND METHODS
Plasmid construction.
BH10.P− is a simian virus 40-based vector that contains full-length wild-type HIV-1 proviral DNA with a single point mutation at position 25 of the protease region, converting Asp25 to Arg25. Transfection of BH10.P− produces noninfectious viral particles containing wild-type genomic RNA and the unprocessed precursor proteins Gag and GagPol (11). This construct was a gift from E. Cohen, University of Montreal. The plasmid hGag codes only for Gag and was constructed as previously described (16). The gene coding for this “humanized” Gag has had its codons optimized for mammalian cell codon usage, which results in more efficient translation and protein production and also makes nuclear export of the mRNA Rev-independent through modification of the inhibitory sequences (16, 33). Plasmid pM368 contains cDNA encoding full-length (1 to 597 amino acids) human LysRS, as previously described (35). In order to construct wild-type and mutant LysRS species, this cDNA was PCR amplified and digested with EcoRI, whose sites were placed in each of the PCR primers. These fragments were cloned into the EcoRI site of pcDNA1.0 Myc (Invitrogen, Carlsbad, Calif.). We used the following primers: for wild-type LysRS, 5′-CTCCGGGAATTCTAGCGGCCGTGCAGGCGGCCGAGGTG (forward primer) and 5′-AATTATGAATTCCTAGACAGAAGTGCCAACTGTTGTGCT (reverse primer; for Δ452-597, 5′-AATTATGAATTCCTACAGGAACTCCCCAACAAGCTTGTCAAGGAG; for Δ309-597, 5′-AATTATGAATTCCTAACCAACCACAAGCATCTTAGATAGAGTTC; for Δ260-597, 5′-AATTATGAATTCCTACTAATCTAAGAAACTTCTTATATA; and for Δ207-597, 5′-AATTATGAATTCCTAAGACAGCAGTGTGATTCATACGGAATGAT. The resulting constructs express Myc-tagged wild-type and mutant LysRS proteins. To construct plasmids coding for V5-tagged wild-type and Δ1-65 LysRS, plasmid pM368 was PCR amplified and digested with EcoRI and Xho1, whose sites were placed into each of the PCR primers. These fragments were cloned into pcDNA3.1 His/V5 (Invitrogen). We used the forward primers 5′-GATAGAGAATTCATGGCGGCCGTGCAGGCG (full length) and 5′-GACGGAGAATTCATGGGTCCTGAGGAAGAG (Δ1-65) and the reverse primer GAGCGACTCGAGAGAAGTGCCAACTGTTGT. The resulting constructs, once transfected into 293FT cells, express V5-tagged wild-type and mutant LysRS proteins.
Cell culture, transfection, and subcellular fractionation.
293FT cells (Invitrogen) were maintained in Dulbecco modified Eagle medium with 10% fetal bovine serum and antibiotics. Cells were transfected with protease-negative HIV-1 proviral DNA, wild-type Gag construct, or wild-type or mutant LysRS constructs by using Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer. Forty-eight hours posttransfection, cells, either transfected or untransfected, were lysed at 4°C in two ways: (i) with hypotonic medium, for which lysis was done by Dounce homogenization in 1.0 ml of hypotonic Tris-EDTA (TE) buffer (20 mM Tris-HCl [pH 7.4], 1 mM EDTA, 0.01% β-mercaptoethanol) supplemented with a protease inhibitor cocktail (Complete; Boehringer Mannheim), and (ii) with nonionic detergent, for which cells were lysed in 1.0 ml of TNT buffer (20 mM Tris-HCl [pH 7.5], 200 mM NaCl, 1% Triton X-100) supplemented with a protease inhibitor cocktail (Complete; Boehringer Mannheim). For either method, the cell homogenate was then centrifuged at 1,500 × g for 30 min to remove nuclei and unbroken cells. The supernatant (S1) was then centrifuged for 1 h at 100,000 × g in an SW 55Ti rotor (Beckman, Columbia, Md.) at 4°C, resulting in the S100 supernatant and the pellet (P100).
Further fractionation of the P100 fraction into membrane-free and membrane-bound protein was done by using a membrane flotation assay (36). The P100 was resuspended in 1 ml of 73% sucrose. Two milliliters of 65% sucrose in TNE (20 mM Tris [pH 7.8], 100 mM NaCl, 1 mM EDTA) was layered on top of the 73% sucrose, and 2 ml of 10% sucrose was layered on top of the 65% sucrose. The gradients were then centrifuged at 100,000 × g in a Beckman SW55 Ti rotor overnight at 4°C. Fractions (0.8 ml) were collected and diluted with an equal volume of 2× TNT, and each fraction was immunoprecipitated at 4°C, first with anti-integrase (anti-IN) and then with anti-capsid (anti-CA). Immunoprecipitates from each fraction were dissolved in sodium dodecyl sulfate (SDS) sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting.
Resolution of the P100 into free and membrane-associated protein complexes was also accomplished by using iodixanol (OptiPrep) gradient centrifugation as described by Lindwasser and Resh (24). The P100 fraction was adjusted to 0.75 ml of 50% iodixanol, and this volume was overlaid with 1.25 ml each of 40, 30, and 20% iodixanol and, finally, 0.5 ml of 10% iodixanol in TNT. After centrifugation at 170,000 × g for 4 h at 4°C, eight fractions were collected and diluted with an equal volume of 2× TNT, and each fraction was immunoprecipitated at 4°C, first with anti-IN and then with anti-CA. The immunoprecipitate from each fraction was dissolved in SDS sample buffer and analyzed by SDS-PAGE and Western blotting.
Viral isolation and subtilisin digestion.
Virions from cells transfected with protease-negative HIV-1 proviral DNA were harvested 64 h posttransfection and pelleted from culture medium by centrifugation in a Beckman 45 Ti rotor at 35,000 rpm for 1 h. The pellet was then purified by centrifugation in a Beckman SW41 rotor at 26,500 rpm for 1 h through 15% sucrose onto a 65% sucrose cushion. The band of purified virions was removed and pelleted in 1× TNE (20 mM Tris [pH 7.8], 100 mM NaCl, 1 mM EDTA) in a Beckman 45 Ti rotor at 40,000 rpm for 1 h. Sucrose gradient-purified virions were lysed by resuspension in 1× radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris [pH 7.4],100 mM NaCl, 1% deoxycholate, 0.1% SDS, 1% Nonidet P-40, protease inhibitor cocktail tablets [Boehringer Mannheim]). Part of the virus preparation was treated with the protease subtilisin before viral lysis. Subtilisin digestion assays were performed essentially according to Ott et al. (31). The purified virions were treated with 1 mg of subtilisin (Boehringer Mannheim)/ml in digestion buffer (10 mM Tris-HCl [pH 8], 1 mM CaCl2) for 16 h at 37°C. Subtilisin was inactivated by phenylmethylsufonyl fluoride. Treated and untreated viruses were then repelleted, resuspended in 2× loading buffer (120 mM Tris-HCl [pH 6.8], 20% glycerol, 4% SDS, 200 mM dithiothreitol, 0.002% [wt/vol] bromphenol blue), and subjected to 10% SDS-PAGE, followed by Western blot analysis.
Immunoprecipitation.
Protein complexes were immunoprecipitated from S1, S100, P100, or the different gradient fractions by using protein A-Sepharose beads cross-linked to rabbit anti-IN polyclonal antibodies (National Institutes of Health [NIH] AIDS Research and Reference Reagent Program), mouse anti-CA monoclonal antibodies (Cellular Products, Inc., Buffalo, N.Y.), rabbit anti-human lysyl-tRNA synthetase polyclonal antibodies (Pocono Rabbit Farm and Laboratory, Inc.), mouse anti-Myc monoclonal antibodies, or mouse anti-V5 monoclonal antibodies (Invitrogen). Antibodies were first cross-linked to the beads. Forty microliters of antibody and 400 μl of 50% (wt/vol) protein A-Sepharose (Pharmacia) were incubated together in 10 ml of 0.2 M-triethanolamine (pH 9). Fifty-two milligrams of dimethyl pimelimidate cross-linker (Pierce) was then added, and the mixture was incubated for 1 h at room temperature. The beads were then washed with 5 ml of 0.2 M triethanolamine (pH 9) and further incubated in 10 ml of 0.2 M triethanolamine for another 2 h at room temperature. Equal amounts of protein (approximately 200 to 500 μg, as determined by the Bio-Rad assay) were incubated with 30 μl of antibody cross-linked to protein A-Sepharose for 1 h at 4°C. The immunoprecipitate was then washed three times with TNT buffer and twice with phosphate-buffered saline (PBS). After the final supernatant was removed, 30 μl of 2× sample buffer (120 mM Tris-HCl [pH 6.8], 20% glycerol, 4% SDS, and 0.02% bromphenol blue) was added and the precipitate was then boiled for 5 min to release the precipitated proteins. After microcentrifugation, the resulting supernatant was analyzed by using Western blots.
Metabolic labeling of Gag/LysRS complexes.
Metabolic labeling was performed by using 60 μCi of Tran [35S]Cys/Met per ml of culture medium (obtained from ICN or NEN). Equal numbers of 293FT cells were either transfected with hGag alone or cotransfected with plasmids coding for hGag and for LysRS. Sixty-four hours posttransfection, culture medium was replaced for 1 h with serum-free, methionine/cysteine-free medium, followed by the addition of [35S]methionine/cysteine for 10 min. Cells were then resuspended in Dulbecco modified Eagle medium containing 10% fetal bovine serum and 100 μM cysteine and methionine and chased for various lengths of time. At specific time points, aliquots of cells were washed and lysed in 1.0 ml of TNT buffer at 4°C, and cell lysates were centrifuged at 1,500 × g for 30 min to remove nuclei and unbroken cells. The resulting S1 supernatants (1 ml) for the different time point chases were immunoprecipitated with either anti-CA, anti-LysRS, or anti-V5 for 1 h at 4°C. The immunoprecipitated proteins were then subjected to SDS-10% PAGE and autoradiography.
Protein analysis.
Viral, cellular, or immunoprecipitated proteins were analyzed by SDS-PAGE (10% acrylamide) followed by blotting onto nitrocellulose membranes (Gelmann Science). Western blots were probed with (i) mouse monoclonal antibodies for CA; (ii) rabbit polyclonal antibodies for human lysyl-tRNA synthetase; (iii) rabbit polyclonal antibodies for MetRS, GlnRS, TrpRS, ArgRS, TyrRS, ProRS, IleRS, anti-p43, anti-p38, and anti-p18 (prepared as previously described [5, 23]); (iv) goat polyclonal antibodies for gp120 (NIH AIDS Research and Reference Reagent Program); (v) mouse monoclonal antibodies for Myc; (vi) mouse monoclonal antibodies for EF1α (Upstate, Lake Placid, N.Y.); (vii) mouse monoclonal antibodies for CD45 (Santa Cruz Biotechnology, Santa Cruz, Calif.); and (viii) mouse monoclonal antibodies for Na+/K+ ATPase (Biomol Research Lab, Plymouth Meeting, Pa.). These antibodies were used as the primary antibodies, and horseradish peroxidase-linked goat anti-mouse, donkey anti-rabbit, and rabbit anti-goat (Sigma, St. Louis, Missouri) were used as secondary antibodies. Antibody binding was detected by enhanced chemiluminescence (ECL kit; Pharmacia Amersham Biotech, Quebec, Canada). The sizes of the detected protein bands were estimated by using prestained HMW protein markers (New England Biolabs).
Nuclear extraction.
The cytoplasmic and nuclear extracts were prepared from the 293FT cells as described previously (28). Cells were suspended in L-buffer (PBS, 0.1% TritonX-100, 0.1% Nonidet P-40), and incubated on ice for 10 min or until they were determined to be >99% lysed by using trypan blue exclusion. Nuclei were pelleted by centrifugation at 1,000 × g for 10 min at 4°C. The supernatant fraction was collected and classified as cytoplasm. The nuclear pellet was purified from membrane contaminants by two rinses in L-buffer, passage through a 0.22-gauge needle three times, and passage through a 0.85 M sucrose cushion (15,000 rpm, microcentrifuge, 15 min). Nuclei in the pellet were lysed by sonication (30 s) in PBS prior to DNase treatment (100 units/200 μl, 45 min, 4°C). Nuclei were further sonicated twice at 4°C for 30 s each to make a nuclear lysate. Western blots of cytoplasmic and nuclear extracts were performed as described above by using rabbit polyclonal antibodies for YY1, mouse monoclonal antibodies for tubulin (Santa Cruz Biotechnology), and mouse monoclonal antibodies for V5 (Invitrogen).
RESULTS
Cellular localization of LysRS and the Gag/LysRS complex. (i) Detection of LysRS.
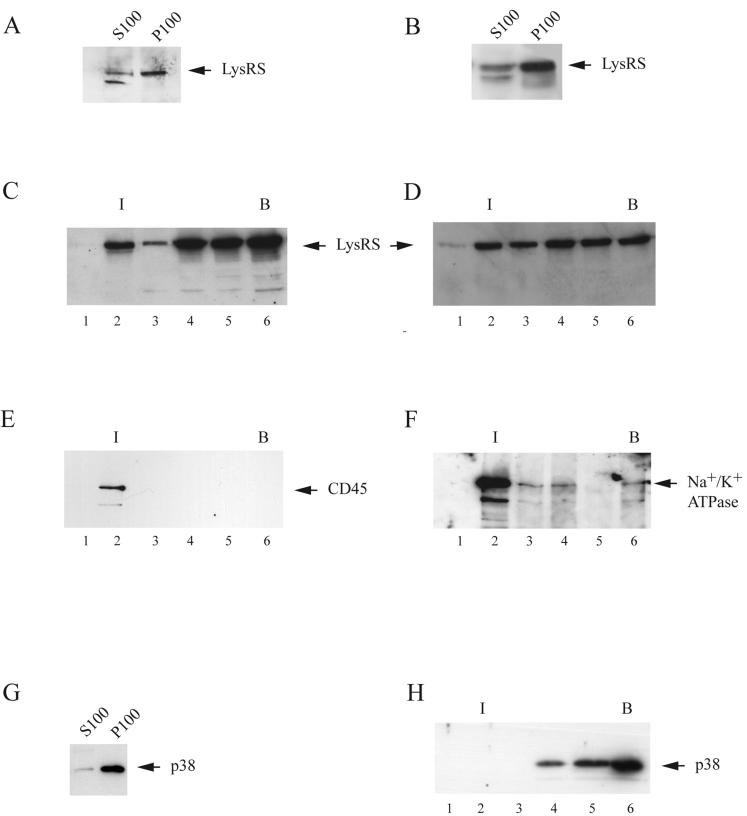
293FT cells were lysed in hypotonic buffer, and after low-speed centrifugation, the postnuclear supernatant was resolved by centrifugation at 100,000 × g into a membrane-free supernatant (S100) containing material soluble in the cytoplasm and a pelletable fraction containing insoluble material such as membranes and HMW complexes. Western blots of these fractions were probed with anti-LysRS. Panels A and B in Fig. 1 show that LysRS was found in both the S100 and the P100 fractions in uninfected cells (Fig. 1A) and in cells producing HIV-1 (Fig. 1B).
FIG. 1.
Cellular distribution of LysRS in 293FTcells lysed hypotonically. Uninfected or protease-negative HIV-1-transfected 293FT cells (cells transfected with BH10.P− DNA) were lysed at 4°C by Dounce homogenization in hypotonic TE buffer and, after clarification by centrifugation at 1,500 × g, were centrifuged at 100,000 × g for 1 h at 4°C. (A and B) For the detection of LysRS in these fractions, the pellet (P100) was suspended in 1.0 ml of TE buffer, equal to the S100 volume. Western blots of the S100 and P100 from uninfected cells (A) or HIV-1-transfected cells (B) were probed with anti-LysRS. (C and D) Sucrose flotation analysis. The P100 fractions from uninfected cells (C) and HIV-1-transfected cells (D) were resolved by discontinuous sucrose gradient centrifugation into membrane-bound and membrane-free proteins (flotation analysis). Aliquots of each gradient fraction were analyzed by Western blots probed with anti-LysRS. I, interface between 10 and 65% sucrose layers, where membrane-bound protein localizes; B, bottom fractions of gradient, where membrane-free protein remains during centrifugation. Fraction numbers increase from top to bottom of gradient. (E and F) Association of LysRS with membrane. Each fraction from sucrose gradients of uninfected cells was immunoprecipitated with anti-LysRS, and Western blots of the immunoprecipitates were probed with antibodies to the membrane markers CD45 (E) or Na+/K+ ATPase (F). (G and H). Cellular distribution of p38 in uninfected 293FT cells. (G) Western blots of the S100 and P100 probed with anti-p38. (H) The P100 fraction from uninfected cells was resolved by discontinuous sucrose gradient centrifugation into membrane-bound and membrane-free proteins. Aliquots of each gradient fraction were analyzed by Western blots probed with anti-p38.
The P100 fraction from either cell type was further resolved on sucrose flotation gradients (Fig. 1C and D), which resolve less dense complexes, such as those bound to membrane, from denser complexes free of membrane (29, 30). Gradient fractions were analyzed by Western blots probed with anti-LysRS, and it can be seen for both uninfected cells (Fig. 1C) and HIV-1-producing cells (Fig. 1D) that while most of the P100 LysRS is found in the more dense fractions, some LysRS floats to the 10%/65% sucrose interface and may be membrane-bound. Each fraction from gradients of uninfected cells was immunoprecipitated with anti-LysRS, and panels E and F in Fig. 1 show Western blots of the immunoprecipitates from uninfected cells probed with antibodies for the plasma membrane markers CD45 (Fig. 1E) and Na+/K+ ATPase (Fig. 1F). These markers were present in fraction 2, and their coprecipitation with LysRS is another indicator that LysRS in fraction 2 was associated with plasma membrane. A similar pattern was found by using HIV-1-producing cells (data not shown). The LysRS associated with fractions 4, 5, and 6 of the sucrose flotation gradient is most likely associated with the HMW aaRS complex, since, as shown in panels G and H of Fig. 1, this LysRS follows the distribution of p38, an important non-aaRS protein associated with the HMW aaRS complex. Thus, almost all p38 is found in the P100 fraction (Fig. 1G), and resolution of the P100 by sucrose flotation shows all detectable p38 banding at fractions 4, 5, and 6. Thus, pelletable LysRS can be detected in both the HMW aaRS and at the plasma membrane.
(ii) Detection of a Gag/LysRS complex in HIV-1-producing 293FT cells.
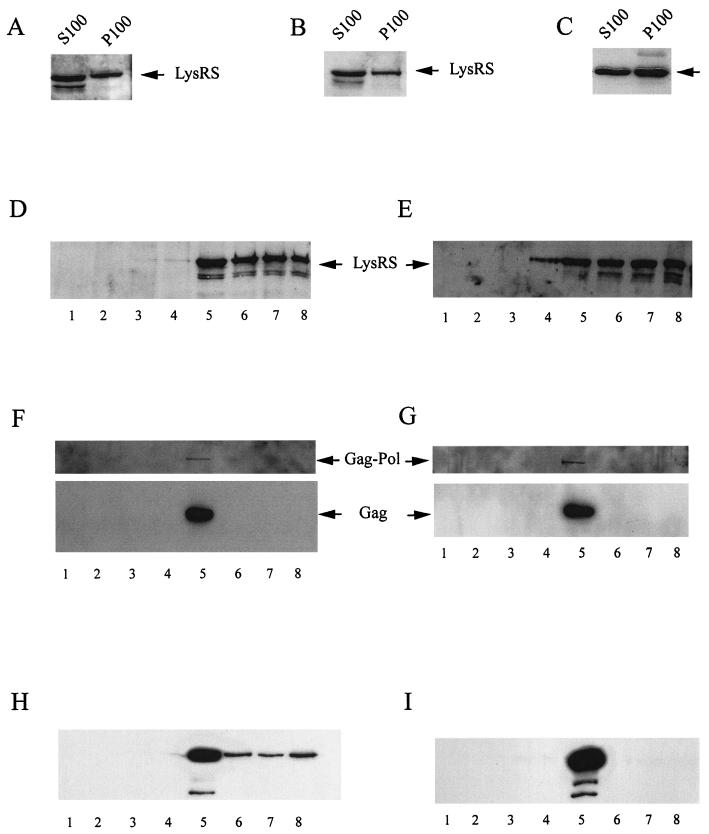
Gag is believed to assemble into polymeric Gag complexes at the cell membrane, and in HIV-1-transfected COS and HeLa cells, almost all steady-state Gag (14, 29, 43) and Gag/GagPol (13) are membrane-bound. A similar pattern is also found in 293FT cells (data not shown). We have searched for the presence of a Gag/LysRS complex in different fractions of lysates of 293FT cells producing HIV-1 by using the coimmunoprecipitation of Gag and LysRS with anti-LysRS as a sign of the Gag/LysRS interaction. To detect this interaction without the immunoprecipitation of large membrane complexes, most membrane was solubilized by lysing cells with 1% Triton X-100 at 4°C. Exposure of cells to Triton X-100 removes much of the envelope lipid associated with the Gag VLPs (7, 15). Uninfected and protease-negative HIV-1-transfected 293FT cells were lysed in the presence of 1% Triton X-100, and the lysates were resolved into S100 and P100 fractions. Exposure of uninfected cells to 1% Triton X-100 appeared to cause a moderate increase in the release of LysRS from the P100 to the S100 (compare Fig. 1A with Fig. 2A), but the release of LysRS into the S100 of HIV-1-producing cells was more dramatic (compare Fig. 1B with Fig. 2B). Much of the LysRS released into this S100 may have come from the HMW aaRS complex, for while the HMW aaRS marker protein p38 was entirely present in the P100 in cells lysed hypotonically (Fig. 1G), a Western blot of S100 and P100 from detergent-lysed cells probed with anti-p38 indicates that a significant fraction of p38 had now been released into the S100 (Fig. 2C).
FIG. 2.
Cellular distribution of LysRS in 293FT cells lysed with Triton X-100. Uninfected and protease-negative HIV-1-transfected 293FT cells were lysed at 4°C in Triton X-100 buffer, and the postnuclear supernatants were resolved into S100 and P100. (A, and B) Western blots of the S100 and P100 from uninfected cells (A) or HIV-1-transfected cells (B) were probed with anti-LysRS. (C) Western blots of the S100 and P100 from uninfected cells were probed with anti-p38. (D and E) The P100 fractions from uninfected cells (D) and HIV-1-transfected cells (E) were resolved on iodixanol gradients, and aliquots of each gradient fraction were analyzed by Western blots probed with anti-LysRS. Denser fractions found at the bottom of the gradient are represented by larger fraction numbers. (F and G) Each iodixanol gradient fraction from HIV-1-transfected cells was immunoprecipitated with either anti-IN (F) or anti-LysRS (G), and the immunoprecipitates were analyzed by Western blots probed with anti-reverse transcriptase and anti-CA. (H and I) 293FT cells were transfected with the plasmid hGag, which expresses only Gag. Cells were lysed at 4°C in Triton X-100 buffer, and the postnuclear supernatants were resolved into S100 and P100. The P100 was resolved on an iodixanol gradient, and gradient fractions were immunoprecipitated with either anti-CA (H) or anti-LysRS (I). Western blots of the immunoprecipitates were probed with anti-CA.
The P100 fractions from uninfected cells (Fig. 2D) and HIV-1-transfected cells (Fig. 2E) were resolved on iodixinol gradients. In these gradients, much of the pelletable LysRS was found near the bottom of the gradient (fractions 6, 7, and 8) and probably represents the pelletable, soluble HMW aaRS complex (the density of fraction 8 was 1.250) (13). However, some material was found in fractions 4 and 5 at densities at which Gag complexes (24) and Gag/GagPol complexes (13) also float (the density of fraction 5 was 1.185). Each gradient fraction from HIV-1-transfected cells was immunoprecipitated with either anti-integrase (Fig. 2F) or anti-LysRS (Fig. 2G), and Western blots of the immunoprecipitates were probed with anti-reverse transcriptase and anti-CA. As previously reported, anti-IN immunoprecipitates the Gag/GagPol complex, which has a buoyant density associated with fraction 5 and sometimes with fraction 4 (13). This Gag/GagPol complex was also immunoprecipitated with anti-LysRS (Fig. 2G), indicating that LysRS is part of this complex.
The fact that the Gag/GagPol/LysRS complex floats with a buoyant density lower than that of membrane-free protein suggests that this complex still contains some membrane. Further evidence for this idea, i.e., anti-IN immunoprecipitates which bring down the Gag/GagPol complex in fraction 5 also immunoprecipitate the cell membrane markers Fyn and CD59 and the viral envelope protein gp160, was previously presented (13). Fyn and CD59 are lipid raft membrane proteins, and while other work has indicated that many lipid raft-associated components have lower buoyant densities than polymeric Gag (7), some lipid raft and viral envelope proteins do remain associated with the Gag/GagPol/LysRS complex after exposure to Triton X-100.
The interaction of Gag with LysRS also occurs when only Gag is expressed in the cell. Transfected 293FT cells expressing only Gag were lysed in 1% Triton X-100, and the analysis of the P100 fraction on iodixanol gradients is shown in panels H and I of Fig. 2. Gradient fractions were immunoprecipitated with either anti-CA (Fig. 2H) or anti-LysRS (Fig. 2I), and Western blots of the immunoprecipitates were probed with anti-CA. Again, the LysRS interacts primarily with Gag floating in fraction 5. It can be noted in panel H that small amounts of Gag are also present in the cytoplasmic part of the gradient (fractions 6, 7, and 8), but panel I indicates that there is no detectable interaction of LysRS with this Gag. Gag and LysRS are also both present in the S100 fraction of the cell lysate, but failure to coimmunoprecipitate both molecules with either anti-CA or anti-LysRS indicates a lack of interaction between these molecules in this fraction (data not shown). These results indicate, therefore, that the Gag/LysRS interaction is first seen at the detergent-resistant membrane (i.e., that in the Gag/GagPol complex found in fraction 5 in the iodixanol gradients) and not for Gag found to be membrane-free in the cytoplasm, i.e., either in the more dense fractions of the iodixanol gradient or in the S100 supernatant. Previous work has shown that most Gag in the S100 is Gag released from the detergent-sensitive membrane by Triton X-100 (13).
Specificity of the LysRS/Gag interaction.
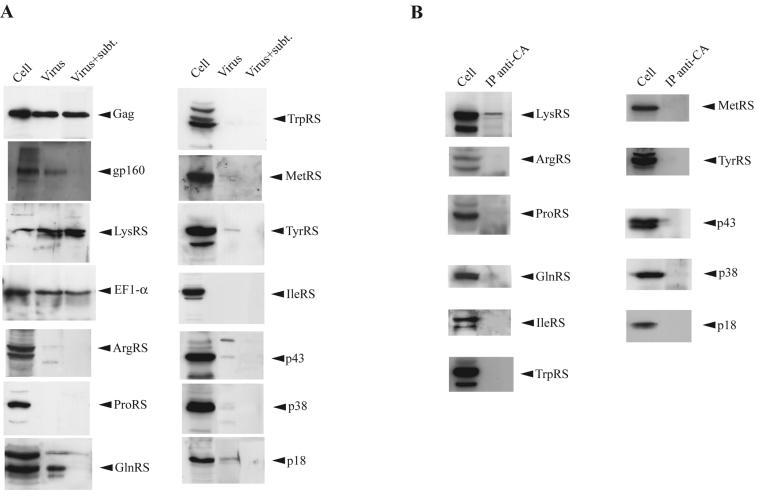
We tested for the presence in HIV-1 of aaRSs and aaRS-associated proteins found in the HMW aaRS complex and detected only LysRS. 293FT cells were transfected with protease-negative HIV-1 proviral DNA, and 64 h later cells and viruses were harvested. Cells were lysed in 1% Triton X-100 buffer (TNT). Viruses were either lysed directly in RIPA buffer or, prior to lysing, were treated with the protease subtilisin to remove cytoplasmic proteins bound nonspecifically to the outside of the viral membrane (31). Figure 3A shows Western blots of cellular and viral lysates probed with antibodies to different proteins. EF1α has been reported to be incorporated into HIV-1 (6), and EF1α, Gag, and LysRS are shown to be present in virions even after treatment of the viruses with subtilisin. On the other hand, the external viral envelope protein whose electrophoretic mobility corresponds to gp160 (5, 13) is digested with this enzyme. In addition to LysRS, blots for detecting five other aaRSs and the three aaRS-associated proteins p18, p38, and p43, all of which are found in the HMW aaRS complex, failed to detect any of these proteins within the virion. Additionally, TrpRS and TyrRS, which are not found in the HMW aaRS complex, are also not incorporated into the virion.
FIG. 3.
The selective interaction of LysRS with Gag and its incorporation into HIV-1. (A) Detection of aaRSs and non-aaRSs in HIV-1. Viruses produced from 293FT cells transfected with protease-negative HIV-1 proviral DNA were pelleted from cell culture medium and purified by centrifugation through 15% sucrose onto a 65% sucrose cushion. Purified viruses were either left untreated or treated with the protease subtilisin (subt.) before viral lysis. Cell and viral lysates were analyzed by Western blots. Blots were probed with antibodies to the proteins shown and were analyzed by enhanced chemiluminescence by using anti-mouse or anti-rabbit as a secondary antibody. (B) Detection of aaRSs and non-aaRSs in a cytoplasmic Gag immunoprecipitate (IP). 293FT cells transfected with protease-negative HIV-1 proviral DNA were lysed in TNT buffer 64 h posttransfection. After removal of nuclei and broken cells at 1,500 × g, Gag was immunoprecipitated from the clarified lysate with anti-CA. After heating the immunoprecipitate to release the precipitated proteins, the precipitate was analyzed by Western blots, which were probed with antibodies to the proteins shown and were analyzed by enhanced chemiluminescence by using anti-rabbit as a secondary antibody.
The cytoplasmic interaction between Gag and LysRS reflects the specific incorporation of LysRS into viral particles. The lysate from detergent-treated cells was treated with anti-CA to immunoprecipitate Gag from the cytoplasm. Western blots of the Gag immunoprecipitate were probed with antibodies to various proteins, as shown in Fig. 3B. These results indicate a clear correlation between the ability of the proteins to be packaged into virions and their ability to form a cytoplasmic complex with Gag, i.e., only LysRS is detected in the Gag immunoprecipitate.
LysRS in the cytoplasmic HMW aaRS complex is not required as a source of viral LysRS.
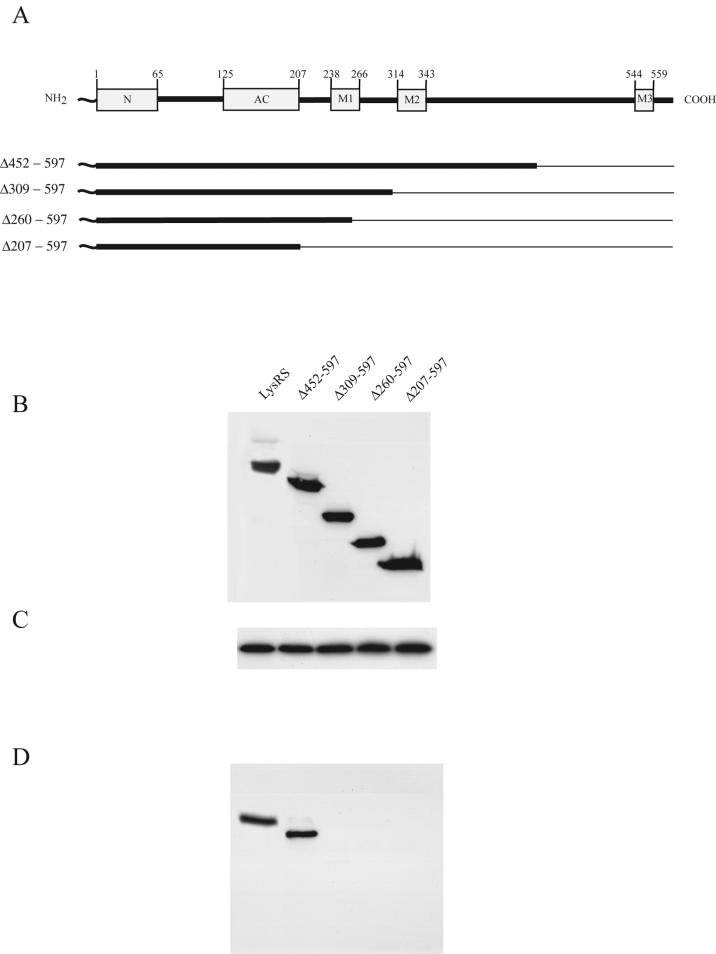
The function of the cytoplasmic HMW aaRS complex is not known, but the absence of many of its components from the virion suggests that it may not be the cellular source of viral LysRS. In this respect, the absence of p38 in the virus is significant. During the formation of the HMW aaRS complex, p38 is believed to act as a scaffold protein for binding to the aaRSs (22, 34). In vitro assembly studies indicate that LysRS binds with the highest affinity to p38 and that the p38/LysRS binding interaction facilitates binding of the other aaRSs both to p38 and to each other (34). To establish whether association with p38 is a prerequisite for LysRS packaging, we have determined whether LysRS mutants that are unable to interact with p38 will still be packaged into HIV-1. In a previous work (19), 293FT cells were cotransfected with protease-negative HIV-1 proviral DNA and plasmids coding for wild-type and mutant LysRS variants, and the ability of the mutant LysRS species to be incorporated into virions was monitored. Wild-type LysRS contains 597 amino acids, and we observed that N-terminal Myc-tagged LysRS mutants require amino acids 208 to 259 for binding to Gag and incorporation into virions (19). Thus, LysRS containing C-terminal deletions that do not extend into these sequences (Δ452-597, Δ309-597, and Δ260-597) are packaged into viruses, while C-terminal deletion mutant Δ207-597 LysRS is not incorporated. Wild-type and C-terminal deletion mutant LysRS species are shown in Fig. 4A. The abilities of these mutant LysRS variants to interact with p38 have been examined herein. 293FT cells were transfected with the plasmids coding for wild-type or mutant LysRS species, which were tagged at their N termini with Myc. Cells were harvested and lysed with TNT buffer, and the ability of the LysRS mutant variants to be immunoprecipitated from the cell lysate with anti-p38 was examined. Panels B and C of Fig. 4 show Western blots of cell lysate probed with either anti-Myc (Fig. 4B) or anti-p38 (Fig. 4C), demonstrating the cytoplasmic expression of the mutant LysRS species and p38, respectively. Figure 4D shows Western blots of the anti-p38 immunoprecipitates from these cell lysates probed with anti-Myc. Figure 4D indicates that an interaction between p38 and either wild-type or Δ452-597 LysRS occurs but that no interaction with p38 was detected with LysRS species containing larger C-terminal deletions. Since two of these species, Δ309-597 and Δ260-597, have previously been shown to be packaged into HIV-1 (19), the ability of LysRS to interact with p38 is not required for its incorporation into virions, indicating that the HMW aaRS complex is an unlikely source of viral LysRS.
FIG. 4.
Interaction of wild-type or mutant LysRS with p38. Plasmids coding for wild-type or C-terminal deletion mutant LysRS tagged at the N terminus with Myc were transfected into 293FT cells. (A) Wild-type and mutant LysRS variants tested. The cartoon at the top shows the various LysRS domains and the amino acid positions (numbers) at which they occur. The unnumbered N-terminal squiggle represents Myc. Deleted amino acid sequences are shown graphically as thin lines and are listed to the left of each mutant. The N-terminal domain (N), the anticodon binding domain (AC) and motifs 1, 2, and 3 (M1, M2, and M3) are sequence elements characteristic of class II tRNA synthetases and are associated with tRNA binding (N and AC), LysRS dimerization (M1), and aminoacylation (M2 and M3). (B and C) Western blots of lysates of cells transfected with plasmids coding for the different LysRS species. Blots were probed with either anti-Myc (B) or anti-p38 (C). (D) Cell lysates were treated with anti-p38, and the p38 immunoprecipitate was analyzed by Western blots probed with anti-Myc.
LysRS contains an N-terminal nuclear localization signal whose deletion does not prevent the incorporation of LysRS into virions.
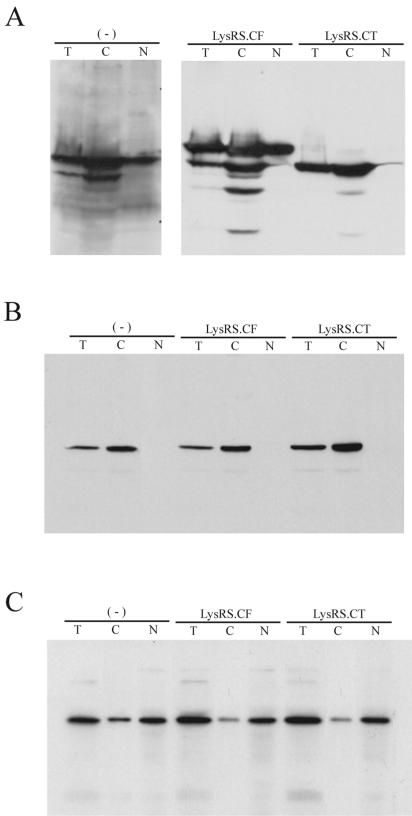
LysRS has also been found in the nucleus (27) and could be a source of viral LysRS. However, we have found that a mutant LysRS species unable to enter the nucleus is still packaged into the virion. We have previously demonstrated that a truncated LysRS missing the N-terminal 65 amino acids is efficiently packaged into HIV-1 (4). Herein, we show that the removal of the N-terminal sequence prevents this truncated LysRS from entering the nucleus. 293FT cells were transfected with plasmids coding for either full-length LysRS (LysRS.CF) or the truncated LysRS (LysRS.CT); the “C” in the designations indicates that the LysRS has been C-terminally tagged with the 14-amino-acid V5 epitope. Cells were lysed in 0.1% NP-40, and Western blots were used to examine either the total lysate or cell lysate fractionated by low speed centrifugation into nuclear and cytoplasm compartments. Figure 5A shows the distribution of wild-type and mutant LysRS species in the cell. The first three lanes show the detection of endogenous LysRS in nontransfected cells by using anti-LysRS and show that while both full-length and smaller LysRS species appear in the total lysate and in the cytoplasm, as previously described (5), only the full-length LysRS can be seen in the nuclear fraction. In the next six lanes, anti-V5 was used to detect exogenous LysRS.CF and LysRS.CT in the different cell fractions. The expression of LysRS.CF in the cell does result in the generation of some smaller peptides in the cytoplasm, but clearly only the full-length LysRS.CF is seen in the nucleus. Since the smaller fragments must contain the C-terminal tag V5 to be detected by anti-V5, the smaller fragments may have resulted from N-terminal deletions. In fact, as shown in the last three lanes, experimental deletion of the N-terminal 65 amino acids (LysRS.CT) results in the inability of this truncated LysRS to migrate to the nucleus. Fig. 5B and C represent controls for the purity of the nuclear and cytoplasmic preparations, i.e., the known cytoplasmic protein, alpha tubulin, was not detected in the nuclear fraction (Fig. 5B), while the nuclear transcription factor, YYI, was primarily found in the nucleus (Fig. 5C).
FIG. 5.
Distribution of wild-type and mutant LysRS between nuclei and cytoplasm. 293FT cells were either nontransfected (−) or transfected with a plasmid coding for full-length LysRS (pLysRS.CF) or a truncated LysRS missing the N-terminal 65 amino acids (pLysRS.CT). The exogenous LysRS species contain C-terminal V5. Cells were lysed in PBS buffer containing 0.1% Nonidet P-40 and 0.1% Triton X-100 as described in Materials and Methods. Nuclei were pelleted from the total cell lysate by centrifugation at 1,000 × g for 10 min and purified as described in Materials and Methods, and the nuclear extract was prepared by sonication. Total cell lysate (T), nuclear extract (N), and the postnuclear supernatant (C) were analyzed by Western blotting. (A) The distribution of endogenous LysRS in nontransfected cells and of LysRS.CF and LysRS.CT in transfected cells. Endogenous LysRS was detected with anti-LysRS, while LysRS.CF and LysRS.CT were detected with anti-V5. (B) A Western blot similar to that shown in panel A but probed with anti-tubulin. (C) A Western blot similar to that shown in panel A but probed with anti-YYI, a nuclear transcription factor.
Membrane-bound LysRS is not required as the source of viral LysRS.
Figure 1 provides evidence that some LysRS in the cell is found at the plasma membrane. However, as shown in Fig. 6, migration of LysRS to the plasma membrane is not required for its incorporation into virions. 293FT cells were transfected with plasmids coding for either wild-type LysRS, LysRS with an N-terminal deletion (Δ1-207), or LysRS with a C-terminal deletion (Δ260-597 or Δ207-597). All LysRS species contain an N-terminal Myc tag, and Fig. 6A shows the cytoplasmic expression of these species in Western blots of uninfected cell lysate probed with anti-Myc. The P100 from each postnuclear supernatant was isolated and resolved by sucrose flotation. The membrane-associated fraction (fraction 2) from each gradient was immunoprecipitated with anti-Myc and resolved by Western blots probed with anti-Myc (Fig. 6B). Both wild type LysRS and Δ260-597 LysRS were found in fraction 2 (remaining associated with membrane), but neither the Δ1-207 nor the Δ207-597 LysRS was retained at the membrane. Since it has previously been shown that the Δ1-207 LysRS is incorporated into virions (19), the association of LysRS with membrane does not appear to be required for its packaging into HIV-1. Nevertheless, the major Gag/LysRS interaction was detected at the membrane.
FIG. 6.

Ability of wild-type and mutant LysRS to bind to cell membrane. Uninfected 293FT cells were transfected with plasmids coding for either wild type LysRS, N-terminal deletion mutant LysRS (Δ1-207), or C-terminal deletion mutant LysRS (Δ260-597 or Δ207-597). All LysRS species contain an N-terminal Myc tag. Cells were lysed hypotonically, and the cytoplasmic expression of these species, as analyzed in Western blots of uninfected cell lysate probed with anti-Myc, is shown (A). The P100 from each postnuclear supernatant was isolated and resolved by sucrose flotation. The membrane-associated fraction (fraction 2) from each gradient was immunoprecipitated with anti-Myc and resolved by Western blots probed with anti-Myc (B).
Newly synthesized LysRS interacts rapidly with newly synthesized Gag.
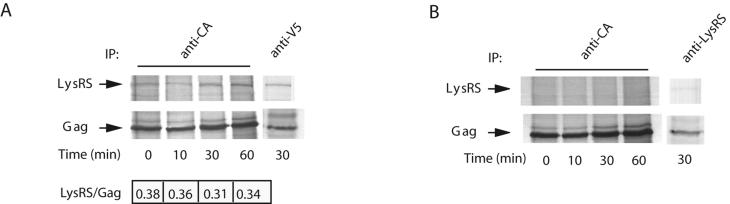
Our results indicate the possibility that newly synthesized LysRS may interact with Gag before LysRS moves to the other cell compartments being discussed. To examine the kinetics of the LysRS/Gag interaction, 293FT cells were transfected with the plasmid coding for hGag or cotransfected with this plasmid plus the plasmid coding for wild-type LysRS tagged with the V5 epitope. Sixty-four hours posttransfection, cells were placed in serum-free, cysteine/methionine-free culture medium and, after 1 h, labeled for 10 min by the addition of [35S]cysteine/methionine. Cells were then washed and resuspended in normal culture medium plus serum, and aliquots of cells were removed for a chase of up to 1 h. Cells were lysed in TNT buffer, and cell lysate immunoprecipitates were produced by using either anti-CA, anti-V5, or anti-LysRS. The labeled immunoprecipitates were then analyzed by one-dimensional SDS-PAGE and autoradiography, and the results are shown in Fig. 7. Figure 7A represents cells transfected with both hGag and LysRS, while Fig. 7B represents cells transfected with hGag alone. The labeled bands in Fig. 7A are identified as LysRS and Gag by their comigration with similar bands detected by probing the same blot with anti-V5 and anti-CA (data not shown). The top band is further identified as LysRS by its immunoprecipitation with either anti-CA or anti-V5 (lanes 1 to 4 and lane 5, respectively) and by the fact that this band is barely detectable in Fig. 7B, in which only endogenous LysRS is present. The interaction of LysRS seen in panel A is rapid, occurring at the earliest observed time point (0-min chase time, i.e., immediately after a 10-min pulse). Although the labeling of LysRS increased somewhat over the next 60-min chase, this trend was accompanied by a similar increase in Gag labeling, and the LysRS/Gag ratio, given at the bottom of Fig. 7A, remained constant. This increase in labeled Gag and LysRS during the chase most likely indicates an incomplete chase of the radioactive pool of [35S]Cys/Met.
FIG. 7.
Kinetics of interaction between newly synthesized LysRS and Gag. 293FT cells were either cotransfected with hGag and LysRS cDNA (A) or transfected with hGag alone (B). Cells were metabolically labeled 64 h posttransfection with [35S]methionine/cysteine for 10 min and then chased for various lengths of time in medium containing 100 μM cysteine and methionine as described in Materials and Methods. Aliquots of cells obtained at various times were lysed in TNT buffer and centrifuged at 1,500 × g to produce the S1 supernatant. Gag/LysRS complexes were then immunoprecipitated from this supernatant and analyzed by one-dimensional SDS-PAGE and autoradiography. (A) One-dimensional SDS-PAGE and autoradiography of immunoprecipitates from the S1 supernatants of cells cotransfected with hGag and LysRS. Lanes 1 to 4 show anti-CA immunoprecipitates from cells lysed at 0 to 60 min postlabeling. Lane 5 shows an anti-V5 immunoprecipitate from cells lysed 30 min postlabeling. The ratios of the signals for LysRS/Gag are shown below the lanes. (B) One-dimensional SDS-PAGE and autoradiography of immunoprecipitates from the S1 supernatants of cells transfected with hGag alone. Lanes 1 to 4 show anti-CA immunoprecipitates from cells lysed at 0 to 60 min postlabeling. Lane 5 shows an anti-LysRS immunoprecipitate from cells lysed 30 min postlabeling.
DISCUSSION
LysRS can be detected at several locations in the cell, including the nucleus, a cytoplasmic HMW aaRS complex, and the cell membrane. An interaction between HIV-1 Gag and LysRS was first detected at the detergent-resistant membrane containing both lipid raft proteins and viral envelope protein. Exogenous LysRS can be expressed in the cell and distinguished from endogenous LysRS by specific tags or size. N-terminal deletions in LysRS which prevent LysRS from either entering the nucleus (Fig. 5) or associating with the membrane (Fig. 6) do not inhibit the incorporation of the mutant LysRS into virions.
Several pieces of evidence also indicate that the HMW aaRS complex in the cytoplasm is not required as the source of viral LysRS. We investigated six aaRSs and three non-aaRSs associated with this complex and found that only LysRS is packaged into the virus (Fig. 3A) and interacts with Gag in the cytoplasm (Fig. 3B). This finding implies that viral LysRS either was never part of the HMW aaRS complex or was removed from it prior to viral packaging. Since formation of this complex appears to depend upon an initial tight binding of LysRS to p38 (the scaffold protein of the HMW aaRS to which other aaRSs will eventually bind as well [34]), we tested the abilities of mutant LysRS species known to be packaged into Gag VLPs (19) to also bind to p38 and found that LysRS binding to p38 was not required for the incorporation of LysRS into Gag VLPs or viruses (Fig. 4).
Mitochondrial LysRS is also not required as a source of viral LysRS. The human LysRS gene contains 15 exons (42). Both human and mitochondrial forms of LysRS are made by alternate splicing from a single primary RNA transcript. The mRNA for the cytoplasmic form excludes exon 2, while the mRNA for the mitochondrial form contains exon 2. Upon translation and processing, exon 1 codes for the amino-terminal sequences in the cytoplasmic LysRS, while exon 2 codes for the amino-terminal sequences in the mitochondrial form. The carboxy 576 amino acids encoded by exons 3 to 15 are identical in both LysRS forms, but the 49 amino acids at the N terminus of the mitochondrial form, representing the putative mitochondrial targeting signal, contain no sequence overlap with the 21 N-terminal amino acids of the cytoplasmic form. It has been shown that mitochondrial LysRS expressed from an exogenous plasmid migrates to the mitochondria but that the cytoplasmic form stays in the cytoplasm (42). Since we have previously shown that the cytoplasmic form of LysRS is efficiently packaged into HIV-1 and Gag VLPs (4, 19), the mitochondrial source of LysRS is clearly not required for the incorporation of LysRS into viruses. On the other hand, it is not known whether the mitochondrial form of LysRS can also be packaged into HIV-1, for while the mitochondrial form of LysRS is 29 amino acids larger than the cytoplasmic form, attempts to resolve these forms electrophoretically have failed (L. Kleiman, unpublished data), and antibodies specific for each type have not been reported (I. A. Tarassov, personal communication).
In this report, we have studied the ability of mutant LysRS species unable to enter certain cellular pools to retain the ability to be incorporated into HIV-1. None of the known cellular LysRS pools investigated seem to be required for LysRS packaging into virions. However, if multiple cellular pools of LysRS contribute to viral LysRS, then the elimination of one of these pools might not necessarily be reflected in a reduction in LysRS incorporated into virions. Evidence against this possibility is the fact that the removal of the N-terminal 207 amino acids in LysRS does not prevent this truncated form from being incorporated into virions (19), even though this mutant LysRS now lacks the nuclear localization signal (Fig. 5), the membrane-binding signal (Fig. 6), and the putative mitochondrial localization signal (42).
It is therefore likely that the separate pools of LysRS studied here do not contribute to viral LysRS because a part of the pool of newly synthesized LysRS might rapidly interact with Gag before moving to the other pools. Several data reported here and elsewhere directly support this view. First, the data in Fig. 7 indicate that the interaction of newly synthesized LysRS with Gag occurs rapidly, i.e., after a 10-min pulse of radioactive label. Second, when small interfering RNA specific for LysRS was used to inhibit the new synthesis of LysRS, the reduction of LysRS incorporation into virions mirrored the rapid reduction in newly synthesized LysRS as opposed to the much slower reduction in the total LysRS present in the cell (12). Third, studies on the kinetics of assembly of HIV-1 in COS7 cells have indicated that after a similar pulse with [35S]Cys/Met for 10 min, all newly synthesized GagPol and approximately one third of newly synthesized, membrane-bound Gag are found at detergent-resistant membrane, the type of membrane believed to be the site of HIV-1 budding (13). Since both GagPol (21) and LysRS (4) are required for the incorporation of tRNALys incorporation into virions, the movement of LysRS into virions would be predicted to follow that of GagPol, a prediction that the data in Fig. 7 support. Finally, cellular LysRS and tRNALys are likely to be present in great excess over the amount packaged into virions, since the infection of cells with HIV-1 does not noticeably reduce cell replication. And yet, LysRS seems to represent a limiting factor for tRNALys incorporation into virions, i.e., overexpression of LysRS results in up to a twofold increase in the incorporation of both LysRS and the major tRNALys isoacceptors into virions (8). It seems likely, therefore, that the LysRS that interacts with Gag may come from a small pool separate from the bulk cytoplasmic pool, i.e., a part of the newly synthesized pool of LysRS.
Acknowledgments
This work was supported by grants from the Canadian Institutes for Health Research (CIHR), the National Institutes of Health (Bethesda, Md.), and a CIHR doctoral research award to R.H.
REFERENCES
- 1.Basyuk, E., T. Galli, M. Mougel, J. M. Blanchard, M. Sitbon, and E. Bertrand. 2003. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev. Cell 5:161-174. [DOI] [PubMed] [Google Scholar]
- 2.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging, p. 177-218. In H. G. Krausslich (ed.), Morphogenesis and maturation of retroviruses, vol. 214. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 3.Cen, S., H. Javanbakht, S. Kim, K. Shiba, R. Craven, A. Rein, K. Ewalt, P. Schimmel, K. Musier-Forsyth, and L. Kleiman. 2002. Retrovirus-specific packaging of aminoacyl-tRNA synthetases with cognate primer tRNAs. J. Virol. 76:13111-13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cen, S., H. Javanbakht, M. Niu, and L. Kleiman. 2004. Ability of wild-type and mutant lysyl-tRNA synthetase to facilitate tRNALys incorporation into human immunodeficiency virus type 1. J. Virol. 78:1595-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cen, S., A. Khorchid, H. Javanbakht, J. Gabor, T. Stello, K. Shiba, K. Musier-Forsyth, and L. Kleiman. 2001. Incorporation of lysyl-tRNA synthetase into HIV-1. J. Virol. 75:5043-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cimarelli, A., and J. Luban. 1999. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 73:5388-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding, L., A. Derdowski, J. J. Wang, and P. Spearman. 2003. Independent segregation of human immunodeficiency virus type 1 Gag protein complexes and lipid rafts. J. Virol. 77:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabor, J., S. Cen, H. Javanbakht, M. Niu, and L. Kleiman. 2002. Effect of altering the tRNA3Lys concentration in human immunodeficiency virus type 1 upon its annealing to viral RNA, GagPol incorporation, and viral infectivity. J. Virol. 76:9096-9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geigenmüller, U., and M. L. Linial. 1996. Specific binding of human immunodeficiency virus type 1 (HIV-1) Gag-derived proteins to a 5′ HIV-1 genomic RNA sequence. J. Virol. 70:667-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goff, A., L. S. Ehrlich, S. N. Cohen, and C. A. Carter. 2003. Tsg101 control of human immunodeficiency virus type 1 Gag trafficking and release. J. Virol. 77:9173-9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Göttlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo, F., S. Cen, M. Niu, H. Javanbakht, and L. Kleiman. 2003. Specific inhibition of the synthesis of human lysyl-tRNA synthetase results in decreases in tRNALys incorporation, tRNA3Lys annealing to viral RNA, and viral infectivity in HIV-1. J. Virol. 77:9817-9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halwani, R., A. Khorchid, S. Cen, and L. Kleiman. 2003. Rapid localization of Gag/GagPol complexes to detergent-resistant membrane during the assembly of HIV-1. J. Virol. 77:3973-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm, K., K. Weclewicz, R. Hewson, and M. Suomalainen. 2003. Human immunodeficiency virus type 1 assembly and lipid rafts: Pr55gag associates with membrane domains that are largely resistant to Brij98 but sensitive to Triton X-100. J. Virol. 77:4805-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, Y., W.-P. Kong, and G. J. Nabel. 2001. Human immunodeficiency virus type I-specific immunity after genetic immunization is enhanced by modification of Gag and Pol expression. J. Virol. 75:4947-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, Y., J. Mak, Q. Cao, Z. Li, M. A. Wainberg, and L. Kleiman. 1994. Incorporation of excess wild type and mutant tRNA3Lys into HIV-1. J. Virol. 68:7676-7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Javanbakht, H., S. Cen, K. Musier-Forsyth, and L. Kleiman. 2002. Correlation between tRNALys3 aminoacylation and incorporation into HIV-1. J. Biol. Chem. 277:17389-17396. [DOI] [PubMed] [Google Scholar]
- 19.Javanbakht, H., R. Halwani, S. Cen, J. Saadatmand, K. Musier-Forsyth, H. G. Gottlinger, and L. Kleiman. 2003. The interaction between HIV-1 Gag and human lysyl-tRNA synthetase during viral assembly. J. Biol. Chem. 278:27644-27651. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, M., J. Mak, A. Ladha, E. Cohen, M. Klein, B. Rovinski, and L. Kleiman. 1993. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J. Virol. 67:3246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khorchid, A., H. Javanbakht, M. A. Parniak, M. A. Wainberg, and L. Kleiman. 2000. Sequences within Pr160gag-pol affecting the selective packaging of tRNALys into HIV-1. J. Mol. Biol. 299:17-26. [DOI] [PubMed] [Google Scholar]
- 22.Kim, J. Y., Y. Kang, J. Lee, H. Kim, Y. Park, Y. Ko, and S. Kim. 2002. p38 is essential for the assembly and stability of macromolecular tRNA synthetase complex: implications for its physiological significance. Proc. Natl. Acad. Sci. USA 99:7912-7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, T., S. G. Park, J. E. Kim, W. Seol, Y.-G. Ko, and S. Kim. 2000. Catalytic peptide of human glutaminyl-tRNA synthetase is essential for its assembly to the aminoacyl-tRNA synthetase complex. J. Biol. Chem. 275:21768-21772. [DOI] [PubMed] [Google Scholar]
- 24.Lindwasser, O. W., and M. D. Resh. 2001. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 75:7913-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mak, J., M. Jiang, M. A. Wainberg, M.-L. Hammarskjold, D. Rekosh, and L. Kleiman. 1994. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J. Virol. 68:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mak, J., and L. Kleiman. 1997. Primer tRNAs for reverse transcription. J. Virol. 71:8087-8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nathanson, L., and M. P. Deutscher. 2000. Active aminoacyl-tRNA synthetases are present in nuclei as a high molecular weight multienzyme complex. J. Biol. Chem. 275:31559-31562. [DOI] [PubMed] [Google Scholar]
- 28.Neufeld, K. L., and R. L. White. 1997. Nuclear and cytoplasmic localizations of the adenomatous polyposis coli protein. Proc. Natl. Acad. Sci. USA 94:3034-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98:13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ono, A., J. M. Orenstein, and E. O. Freed. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 74:2855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ott, D. E., L. V. Coren, D. G. Johnson, R. C. I. Sowder, L. O. Arthur, and L. E. Henderson. 1995. Analysis and localization of cyclophilin A found in the virions of human immunodeficiency virus type 1 MN strain. AIDS Res. Hum. Retrovir. 11:1003-1006. [DOI] [PubMed] [Google Scholar]
- 32.Park, J., and C. D. Morrow. 1992. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into virus-like particles. J. Virol. 66:6304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu, J. T., R. Song, M. Dettenhofer, C. Tian, T. August, B. K. Felber, G. N. Pavlakis, and X. F. Yu. 1999. Evaluation of novel human immunodeficiency virus type 1 Gag DNA vaccines for protein expression in mammalian cells and induction of immune responses. J. Virol. 73:9145-9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson, J.-C., P. Kerjan, and M. Mirande. 2000. Macromolecular assemblage of aminoacyl-tRNA synthetases: quantitative analysis of protein-protein interactions and mechanism of complex assembly. J. Mol. Biol. 304:983-994. [DOI] [PubMed] [Google Scholar]
- 35.Shiba, K., T. Stello, H. Motegi, T. Noda, K. Musier-Forsyth, and P. Schimmel. 1997. Human lysyl-tRNA synthetase accepts nucleotide 73 variants and rescues E. coli double-defective mutant. J. Biol. Chem. 272:22809-22816. [DOI] [PubMed] [Google Scholar]
- 36.Simon, J. H. M., E. A. Carpenter, R. A. M. Fouchier, and M. H. Malim. 1999. Vif and the p55gag polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J. Virol. 73:2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, A. J., M. I. Cho, M. L. Hammarskjöld, and D. Rekosh. 1990. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into virus-like particles. J. Virol. 64:2743-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, A. J., N. Srivivasakumar, M.-L. Hammarskjöld, and D. Rekosh. 1993. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J. Virol. 67:2266-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srinivasakumar, N., M.-L. Hammarskjöld, and D. Rekosh. 1995. Characterization of deletion mutations in the capsid region of HIV-1 that affect particle formation and Gag-Pol precursor incorporation. J. Virol. 69:6106-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 41.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 42.Tolkunova, E., H. Park, J. Xia, M. P. King, and E. Davidson. 2000. The human lysyl-tRNA synthetase gene encodes both the cytoplasmic and mitochondrial enzymes by means of an unusual alternative splicing of the primary transcript. J. Biol. Chem. 275:35063-35069. [DOI] [PubMed] [Google Scholar]
- 43.Tritel, M., and M. D. Resh. 2000. Kinetic analysis of human immunodeficiency virus type 1 assembly reveals the presence of sequential intermediates. J. Virol. 74:5845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]