Abstract
The objective of this study was to evaluate the in vitro effects of flavonoid-rich plant extracts (PE) on ruminal fermentation characteristics and methane emission by studying their effectiveness for methanogenesis in the rumen. A fistulated Holstein cow was used as a donor of rumen fluid. The PE (Punica granatum, Betula schmidtii, Ginkgo biloba, Camellia japonica, and Cudrania tricuspidata) known to have high concentrations of flavonoid were added to an in vitro fermentation incubated with rumen fluid. Total gas production and microbial growth with all PE was higher than that of the control at 24 h incubation, while the methane emission was significantly lower (p<0.05) than that of the control. The decrease in methane accumulation relative to the control was 47.6%, 39.6%, 46.7%, 47.9%, and 48.8% for Punica, Betula, Ginkgo, Camellia, and Cudrania treatments, respectively. Ciliate populations were reduced by more than 60% in flavonoid-rich PE treatments. The Fibrobacter succinogenes diversity in all added flavonoid-rich PE was shown to increase, while the Ruminoccocus albus and R. flavefaciens populations in all PE decreased as compared with the control. In particular, the F. succinogenes community with the addition of Birch extract increased to a greater extent than that of others. In conclusion, the results of this study showed that flavonoid-rich PE decreased ruminal methane emission without adversely affecting ruminal fermentation characteristics in vitro in 24 h incubation time, suggesting that the flavonoid-rich PE have potential possibility as bio-active regulator for ruminants.
Keywords: Flavonoid-rich Plant, Methane Emission, Microbial Growth, Ruminal Fermentation, Rumen Anaerobic Microbes
INTRODUCTION
Agricultural greenhouse gas emission, mainly methane from ruminants, has currently been recognized as an important issue worldwide as it is a driver for global warming and climate change. Methane eructated from ruminants is considered to be one of the most important contributors to global warming, imposing an environmental burden that cannot be ignored. Meanwhile, it represents a loss of 2% to 15% of the gross energy intake (Johnson and Johnson, 1995; Ellis et al., 2007). Animal nutritionists have been studying manipulation of the rumen microbial ecosystem to reduce methane emission without the adverse effects on rumen function. There is a need to identify feed additives to modify ruminal fermentation characteristics and increase the efficiency of feed utilization, thereby inhibiting the ruminal methanogenesis. In recent years, essential oils (Benchaar, 2007), plant secondary metabolites such as condensed tannins and saponins (Pen et al., 2006; Bhatta et al., 2009) and dietary lipids (Dohme et al., 2001) have arisen as attractive rumen modifiers to improve rumen microbial metabolism as well as inhibit methane production in ruminants. The positive effects of flavonoid-rich plant extracts (PE) on methane emission and methanogens population in vitro as well as in vivo have been examined (Patra et al., 2006; Bodas et al., 2008; Patra and Saxena, 2010; Oskoueian et al., 2013; Becker et al., 2014). In addition, flavonoid supplementation could improve ruminal fermentation of dairy cows with increasing milk yield (Theodorou et al., 1994), protecting ruminal acidosis (Balcells et al., 2012), reducing methane emission and changing microbial populations such as protozoa and methanogen (Baker, 1999).
The objective of this study was to evaluate the effect of flavonoid-rich PE on the growth of rumen microorganisms using quantitative real-time polymerase chain reaction (PCR) assay and in vitro rumen fermentation with respect to methane emissions.
MATERIALS AND METHODS
Preparation of plant extracts
Plant extracts were obtained from Plant Extract Bank at the Korea Research Institute of Bioscience and Biotechnology (Daejeon, Korea). Plants were collected from fields in Korea (Table 1). Each plant was cut into small pieces and dried naturally under shade. The dried plant (100 g) were extracted with 99.9% methyl alcohol (1 L) using ultrasonic cleaner (Branson Ultrasonics corporation, Danbury, CT, USA) at room temperature for 3 days. After extraction, the solutions were filtered and the solvents were evaporated under vacuum. Stock solutions (20 mg/mL) of the extract were dissolved in dimethyl sulfoxide (Sigma-Aldrich Chemical Co., St. Louis, Mo, USA) and diluted using culture media immediately before experiments.
Table 1.
Technical information regarding flavonoid-rich plant extracts used in the experimenta
| Stock No. | Botanical name | Scientific name | Family name | Partb |
|---|---|---|---|---|
| 034–052 | Pomegranate | Punica granatum | Punicaceae | L+S+F |
| 015–042 | Brich | Betula schmidtii | Betulaceae | S |
| 019–076 | Ginkgo | Ginkgo biloba | Ginkgoaceae | L |
| 001–053 | Camellia | Camellia japonica | Theaceae | L |
| 026–020 | Tricuspid cudrania | Cudrania tricuspidata | Moraceae | L |
Plant extracts were obtained from Plant Extract Bank (PEB) at Korea Research Institute of Bioscience and Biotechnology (Daejeon, Korea).
L: Leaf, S: Stem, F: Flower.
Ruminal inoculum and in vitro incubation
A fistulated Holstein cow was used as a donor of rumen fluid. Timothy and commercial concentrate in the ratio of 60:40 were fed at 2% of body weight twice a day (09:00 and 17:00). Water and mineral-vitamin block were allowed ad libitum. The rumen fluid was collected from the fistulated Holstein cow before morning feeding. Rumen liquor was filtered through four layers of cheesecloth before mixing with buffer and was maintained at 39°C. Fifteen mililiters of rumen fluid-buffer mixture, comprising McDougall buffer (McDougall, 1948) and rumen liquor in the ratio of 2 to 1, was dispensed anaerobically into 50 mL serum bottles containing 0.3 g of timothy substrate and PE (5% of substrate). The serum bottles were filled with O2-free N2 gas capped with a rubber stopper and held in a shaking incubator (Jeio Tech, SI-900R, Daejeon, Korea; 120× rpm) at 39°C for 72 h. The in vitro experiment was evaluated in triplicate run for data analysis using 90 serum bottles (6 treatments×5 incubation times×3 replication) with a completely randomized design.
Gas production measurement and analysis of gas profiles and ruminal fermentation
At the end of incubation, total gas production was measured according to the assay outlined by Theodorou et al. (1994). A detachable pressure transducer and a digital readout voltmeter (Laurel Electronics, Inc., Costa Mesa, CA, USA) were used to measure the headspace gas pressure of fermenting cultures. For the total gas production measurement, the transducer was modified in a way that it could be linked to the inlet of a disposable Luer-lock three-way stopcock (Theodorou et al., 1994). Gas pressure in the headspace was read from the display unit after insertion of the hypodermic syringe needle through the butyl rubber stopper above the culture medium. The headspace gas in the serum bottle was collected for analyzing methane and hydrogen by gas chromatography (Agilent Technologies HP 5890, Santa Clara, CA, USA) conducted using a TCD detector with a Column Carboxen 1006PLOT capillary column 30 m×0.53 mm (Supelco). The culture was subsampled for the analysis of pH (Mettler-Toledo, MP230, Greifensee, Switzerland), volatile fatty acid (VFA) concentration and genomic DNA extraction. The VFA analysis was performed with a HPLC (High Performance Liquid Chromatography, Agilent-1200, Waldbronn, Germany) equipped with column (300 mm×7.8 mm I.d. MetaCarb 87H, Varian, Palo Alto, CA, USA). In vitro DM disappearance rate was estimated by the modified method of nylon bag digestion process. Briefly, after incubation, the nylon bag with substrate was washed twice in a water-bath equipped with Heidolphs Rotamax 120 (Heidolph Instrument, Nuremberg, Germany) at 100×rpm for 30 min. Washed nylon bags were then dried to a constant weight at 60°C. Dry matter disappearance was determined by weight difference before and after incubation in the serum bottle.
Microbial growth rate
Incubated samples taken from each fermentation period were centrifuged at 3,000×rpm for 3 min to remove feed particles, and the supernatants were re-centrifuged at 14,000×rpm for 3 min to settle the pellets down. After that, sodium phosphate buffer (pH 6.5) was added to these precipitates and vortexed. Growth rates of total microbes were estimated as optical density (OD) values using spectrophotometer (Model 680, Bio-Rad Laboratories, Hercules, CA, USA) at 550 nm.
Quantitative polymerase chain reaction assays
DNA extraction
A high-speed reciprocal shaker which retains samples in screw-capped tubes containing silica beads was used for DNA extraction. Total nucleic acid was extracted from the incubated rumen samples by using the modified bead-beating protocol with the Soil kit (Macherey-nagel, Düren, Germany). Briefly 1.0 mL aliquot was taken from the incubated culture solution and was centrifuged at 3,000×rpm. Nucleic acid concentrations were measured by using a NanoDrop Spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
PCR primers
The PCR primer sets (Table 2) used in this study for amplification of total bacteria, Fibrobacter succinogenes, Ruminococcus albus, Ruminococcus flavefaciens, methanogenic archaea, and ciliate protozoa were from the published reports (Koike and Kobayashi, 2001; Denman and McSweeney, 2005; Skillman et al., 2006; and Denman et al., 2007). All microbial data were analyzed for calculating relative expressions to total bacteria (Denman and McSweeney, 2006).
Table 2.
PCR primer sets for real-time PCR assay
| Target species | Primer sequence (5′→3′) | References |
|---|---|---|
| Total bacteria | F: CGG CAA CGA GCG CAA CCC R: CCA TTG TAG CAC GTG TGT AGC C |
Denman and McSweeney (2006) |
| Fibrobacter succinogenes | F: GTT CGG AAT TAC TGG GCG TAA A R: CGC CTG CCC CTG AAC TAT C |
Denman and McSweeney (2006) |
| Ruminococcus albus | F: CCC TAA AAG CAG TCT TAG TTC G R: CCT CCT TGC GGT TAG AAC A |
Koike and Kobayashi (2001) |
| Ruminocuccus flavefaciens | F: CGA ACG GAG ATA ATT TGA GTT TAC TTA GG R: CGG TCT CTG TAT GTT ATG AGG TAT TAC C |
Denman and McSweeney (2006) |
| Methanogenic archaea | F: TTC GGT GGA TCD CAR AGR GC R: GBA RGT CGW AWC CGT AGA ATC C |
Denman et al. (2007) |
| Ciliate protozoa | F: GAG CTA ATA CAT GCT AAG GC R: CCC TCA CTA CAA TCG AGA TTT AAG G |
Skillman et al. (2006) |
PCR, polymerase chain reaction.
Quantitative Real-time PCR
Quantitative PCR assays for enumeration of microbes were performed according to the methods described by Denman and McSweeney (2006) and Denman et al. (2007) on a real-time PCR Machine (CFX96 Real-Time system, BIO RAD, Hercules, CA, USA) using the SYBR Green Supermix (QPK-201, Toyobo Co., LTD., Tokyo, Japan). The values of cycle threshold (Ct) after real-time PCR were used to determine fold change (number of fold difference) of different microbial population relative to control without additives. Abundance of these microbes was expressed by the equation: relative quantification = 2−ΔCt(Target)−ΔCt(Control), where Ct represents threshold cycle. All quantitative (q) PCR reaction mixtures (final volume of 20 μL) contained forward and reverse primers, the SYBR Green Supermix and DNA template. A negative control without the template DNA was used in every qPCR assay for each primer. The PCR amplification of the target DNA, included the annealing and the extension temperature, was conducted following the references in Table 2.
Total polyphenol and total flavonoid concentration
Total polyphenol concentration
Total polyphenol content in the PE was determined with Folin-Ciocalteu reagent using modified method by Velioglu et al. (1998) in a 96 well plate. Reaction mixture consisted of 10 μL of PE, 180 μL of Na2CO3 (2%), 10 μL of Folin-Ciocalteu reagent (1:1 with water) and 10 μL distilled water. Total polyphenolic content in PE was estimated as OD values using enzyme-linked immunosorbent assay (ELISA) reader (Spectra Max reader M5, Molecular Devices, Sunnyvale, CA, USA) at 750 nm after 30 min incubation (37°C). The standard calibration plot was generated at 750 nm using known concentration of gallic acid.
Total flavonoid concentration
The aluminum chloride method modified by Jia et al. (1999) was used for the determination of total flavonoid content of PE. Aliquots of PE solutions were taken and made up the volume 100 μL with methanol. Then, 7.5 μL of NaNO (5%), 15 μL of AlCl3 (10%), 100 μL of NaOH (1 M) and 25 μL of distilled water were added sequentially. Total flavonoid content in PE was estimated as OD values using ELISA reader (Spectra Max reader M5, Molecular Devices, Sunnyvale, CA, USA) at 510 nm after 30 min incubation. The standard calibration plot was generated at 510 nm using known concentration of catechin.
Statistical analysis
Data were analyzed using the general linear model procedure of the Statistical Analysis System Institute, Inc. (SAS Institute, 2002). The effects of PE on total gas production, gas profiles, pH, VFA, and microbial growth were compared to the controls and significant differences between treatment means were examined using Duncan’s multiple comparison tests. A p<0.05 was considered to indicate statistical significance.
RESULTS AND DISCUSSION
In vitro ruminal fermentation characteristics
The concentration of total polyphenol and total flavonoid in PE used in this experiment is shown in Figure 1. In particular, the concentration of total flavonoid was higher in Pomegranate and Birch extracts compared with others. The in vitro ruminal fermentation profiles are shown in Table 3. pH did not significantly show a difference (p< 0.05) with the addition of all PE except at 72 h incubation time. Our study supports the previous studies that pH was not significant different in in vitro ruminal incubation with flavonoids such as flavone, myricetin, naringin, catechin, rutin, quercetin and kaempferol (Oskoueian et al., 2013) as well as flavonoid, tannin and essential oil (Bodas et al., 2008). The tVFA concentrations were decreased or increased by flavonoid-rich PE according to incubation times (6 h, 12 h, 24 h, 48 h, and 72 h) but were not affected significantly as compared to control. Although dry matter (DM) disappearance was not significantly different except with the addition of Cudrania tricuspidata at 24 h incubation and pomegranate at 72 h incubation (p<0.05), DM disappearance in added flavonoid-rich PE was lower than that of the control at 6 h, 12 h, and 24 h incubation. The effect of flavonoid-rich PE on gas production and gas profiles is shown in Table 4. In particular, total gas production in added all PE was higher than that of the control at 24 h incubation, while the methane emission was significantly lower (p<0.05) than that of the control. This finding supports the finding that flavonoid-rich PE reduced methane emission by 4.7% to 14% after 24 h incubation (Bodas et al., 2008). The decreased methane emission may be due to the changes in ciliate protozoan community (Figure 3B).
Figure 1.
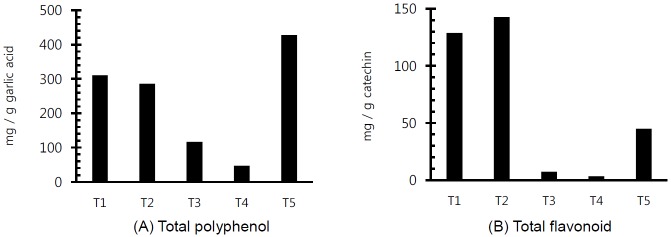
Concentration of total polyphenol (A) and total flavonoid (B) in flavonoid-rich plant extracts used in the experiment (T1, Pomegranate; T2, Birch; T3, Ginkgo; T4, Camellia; T5, Cudrania tricuspidata).
Table 3.
Effects of flavonoid-rich plant extracts on ruminal fermentation characteristics during 72 h incubation
| Treatments | SEM | p-value | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Control | Pomegranate | Birch | Ginkgo | Camellia | Cudrania tricuspidata | |||
| ----------------------------------------------------------- pH ------------------------------------------------------ | ||||||||
| 6 h | 7.43a | 7.31a | 7.39a | 7.45a | 7.47a | 7.52a | 0.07 | 0.061 |
| 12 h | 7.07a | 7.06a | 7.04a | 7.08a | 7.15a | 7.15a | 0.04 | 0.143 |
| 24 h | 6.67 a | 6.71 a | 6.71 a | 6.69 a | 6.69 a | 6.71 a | 0.04 | 0.637 |
| 48 h | 6.45 a | 6.46 a | 6.47 a | 6.47 a | 6.52 a | 6.49 a | 0.05 | 0.523 |
| 72 h | 6.48b | 6.47b | 6.68a | 6.65a | 6.74a | 6.66a | 0.07 | 0.002 |
| ---------------------------------------------------- tVFA (mM) --------------------------------------------------- | ||||||||
| 6 h | 30.96 a | 37.02 a | 30.66 a | 32.46 a | 30.90 a | 28.74 a | 4.73 | 0.424 |
| 12 h | 37.99b | 39.66ab | 39.09b | 40.05ab | 38.27b | 42.28a | 1.45 | 0.037 |
| 24 h | 64.08a | 60.46a | 59.93a | 61.72a | 62.49a | 62.16a | 1.87 | 0.168 |
| 48 h | 78.21a | 73.37abc | 74.79ab | 69.03bc | 67.43c | 70.87bc | 3.77 | 0.041 |
| 72 h | 75.80a | 76.05a | 64.92ab | 66.36ab | 57.51b | 70.21a | 6.49 | 0.033 |
| --------------------------------------------------- DM degradability (%) ---------------------------------------- | ||||||||
| 6 h | 22.61a | 22.17a | 21.91a | 22.10a | 21.90a | 20.58b | 0.57 | 0.018 |
| 12 h | 25.94 a | 25.59 a | 24.18 a | 24.65 a | 23.90 a | 27.67 a | 2.83 | 0.617 |
| 24 h | 31.19a | 30.67a | 30.90a | 29.85a | 29.72a | 28.50a | 1.10 | 0.096 |
| 48 h | 40.69 a | 41.88 a | 41.78 a | 39.65 a | 41.03 a | 40.80 a | 1.29 | 0.362 |
| 72 h | 41.59b | 46.17a | 32.82c | 41.78b | 34.72c | 43.04b | 1.57 | 0.000 |
SEM: standard error of the mean; tVFA, total volatile fatty acids; DM, dry matter.
Means in the same row with different superscripts differ significantly (p<0.05).
Table 4.
Effects of flavonoid-rich included plant extracts on gas production and gas profiles during 72 h incubation
| Treatments | SEM | p-value | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Control | Pomegranate | Birch | Ginkgo | Camellia | Cudrania tricuspidata | |||
| ---------------------------------------------- Total gas (mL/g DM) --------------------------------------- | ||||||||
| 6 h | 142.9ab | 143.0ab | 143.7a | 144.0a | 144.0a | 142.0b | 0.46 | 0.061 |
| 12 h | 170.3b | 179.1a | 181.1a | 179.2a | 177.1a | 175.5ab | 2.02 | 0.033 |
| 24 h | 218.9 a | 221.0 a | 220.3 a | 220.4 a | 220.4 a | 219.5 a | 2.33 | 0.989 |
| 48 h | 258.5 b | 272.2 a | 237.8 c | 277.9 a | 245.1 c | 275.5 a | 3.97 | <.0001 |
| 72 h | 274.3b | 294.9a | 237.8c | 278.1b | 245.1c | 276.8b | 3.18 | 0.006 |
| ---------------------------------------------------- CH4 (mL/g DM) ------------------------------------------ | ||||||||
| 6 h | 1.70 a | 1.63 a | 2.09 a | 1.57 a | 1.60 a | 1.47 a | 0.26 | 0.610 |
| 12 h | 6.24a | 6.25a | 5.46a | 5.59a | 5.63a | 5.47a | 0.45 | 0.632 |
| 24 h | 15.87a | 8.31b | 9.59b | 8.46b | 8.27b | 8.12b | 1.35 | 0.010 |
| 48 h | 29.90ab | 22.81ab | 16.99b | 32.91a | 22.61ab | 35.70a | 5.20 | 0.170 |
| 72 h | 39.64ab | 44.96a | 30.16c | 40.27ab | 35.82bc | 37.69abc | 2.36 | 0.017 |
| ---------------------------------------------------- CO2 (mL/g DM) ----------------------------------------- | ||||||||
| 6 h | 19.73a | 16.55ab | 18.90ab | 15.42ab | 16.04ab | 14.81b | 1.72 | 0.384 |
| 12 h | 44.08a | 52.74a | 48.39a | 49.05a | 49.75a | 48.43a | 3.37 | 0.643 |
| 24 h | 77.47a | 59.12a | 74.65a | 64.28a | 67.89a | 62.48a | 9.23 | 0.705 |
| 48 h | 102.1ab | 109.0ab | 74.99d | 93.65bc | 78.14cd | 116.6a | 5.41 | 0.001 |
| 72 h | 104.4a | 115.6a | 103.9a | 110.5a | 107.81a | 100.5a | 6.30 | 0.613 |
SEM, standard error of the mean; DM, dry matter.
Means in the same row with different superscripts differ significantly (p<0.05).
Figure 3.
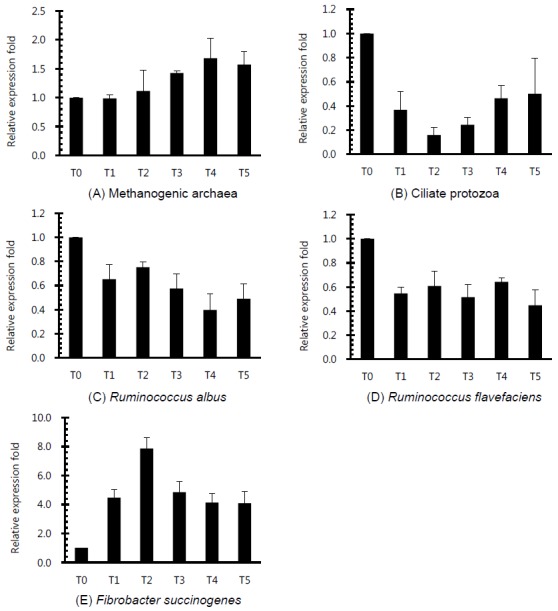
Relative quantification analysis of rumen microorganism populations in vitro ruminal fermentation by the addition of different plant extracts after 24 h incubation (T0, Control; T1, Pomegranate; T2, Birch; T3, Ginkgo; T4, Camellia; T5, Cudrania tricuspidata).
The in vitro change in microbial diversity in the rumen
Rumen bacterial growth with the addition of flavonoid-rich PE is shown in Figure 3. Although flavonoids are widely known to possess antifungal, antiviral and antibacterial activities (Cushnie and Lamb, 2005), the result of microbial growth under supplement of all PE was higher than that in control for 72 h incubation (Figure 2), which may be the cause of total gas production increase. The ciliate protozoa community with the addition flavonoid-rich PE was decreased more than that of the control (Figure 3A). Patra et al. (2006) reported that extracts containing phenolics decreased the ruminal methane emission and protozoa count although they appeared not effective against ruminal methanogenesis. This study also showed that ciliate protozoan populations were reduced by more than 60% in flavonoid-rich PE treatments. Ciliate protozoa are an important key in methanogenesis in the rumen as methanogens attach to their surface. Flavonoid-rich PE reduced the ciliated-associated methanogens population and hence decreased the methane emission. Patra and Saxena (2010) reported that flavonoids gave direct effects against methanogens, and reduced protozoa related with ruminal methanogenesis. The Fibrobacter succinogenes diversity under all flavonoid-rich PE was shown to increase, while the Ruminococcus albus and Ruminococcus flavefaciens populations under all PE decreased as compared with control. In particular, the F. succinogenes community with the addition of Birch extract increased to a greater extent than that of others (Figure 3C, 3D, and 3E). R. albus, one of ruminal fibrolytic bacteria, is a very promising bacteria to produce hydrogen (H2) from energy forage, with the potential of utilizing the cellulosic and hemicellulosic biomass (Ntaikou et al., 2008). Latham and Wollin (1977) reported that succinic acid is produced by R. flavefaciens culture as a major fermentation product with acetic and formic acids, H2, and CO2. H2 is the critical concern to the microbial ecosystem in ruminants. H2 produced during enteric fermentation is the precursor of methane emission from ruminants and the regulation of H2 rather than methane is the key to control ruminant methane emission. The formation of propionate from succinate would result in a lower availability of H2 for the methanogenesis. The ruminal microbe population may show that PE with flavonoid influences ruminal methanogenesis in this study.
Figure 2.
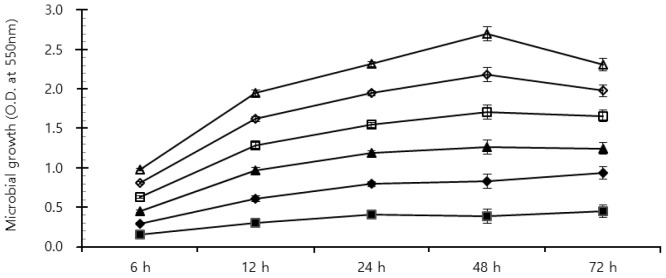
Effects of flavonoid-rich included plant extracts on growth rate of ruminal microbes in fermentation after time-scheduled batch incubation (■: control ◆: Pomegranate, ▲: Birch, □: Ginkgo, ⋄: Camellia, Δ: Cudrania tricuspidata).
In conclusion, the results of this study indicate that flavonoid-rich PE appears to have a potential possibility as bio-active regulator for ruminants with decreasing ruminal methane emission. Future studies need to be aimed at finding a suitable effective dose of PE for inhibiting ruminal methanogenesis.
ACKNOWLEDGMENTS
This research was supported by Bio-industry Technology Development Program, Ministry of Agriculture, Food and Rural Affairs, and Cooperative Research Program for Agriculture Science & Technology Development, Rural Development Administration, Republic of Korea. E. T. Kim was supported by Postdoctoral Fellowship Program of National Institute of Animal Science, Rural Development Administration, Republic of Korea.
REFERENCES
- Baker SK. Rumen methanogens, and inhibition of methanogenesis. Aust J Agric Res. 1999;50:1293–1298. [Google Scholar]
- Balcells J, Aris A, Serrano A, Seradj AR, Crespo J, Devant M. Effects of an extract of plant flavonoids (Bioflavex) on rumen fermentation and performance in heifers fed high-concentrate diets. J Anim Sci. 2012;90:4975–4984. doi: 10.2527/jas.2011-4955. [DOI] [PubMed] [Google Scholar]
- Becker PM, Van Wikselaar PG, Franssen MCR, De Vos RCH, Hall RD, Beekwilder J. Evidence for a hydrogen-sink mechanism of (+) catechin-mediated emission reduction of the ruminant greenhouse gas methane. Metabolomics. 2014;10:179–189. [Google Scholar]
- Benchaar C, Petit HV, Berthiaume R, Ouellet DR, Chiquette J, Chouinard PY. Effects of essential oils on digestion, ruminal fermentation, rumen microbial populations, milk production, and milk composition in dairy cows fed alfalfa silage or corn silage. J Dairy Sci. 2007;90:886–897. doi: 10.3168/jds.S0022-0302(07)71572-2. [DOI] [PubMed] [Google Scholar]
- Bhatta R, Uyeno Y, Tajima K, Takenaka A, Yabumoto Y, Nonaka I, Enishi O, Kurihara M. Difference in the nature of tannins on in vitro ruminal methane and volatile fatty acid production and on methanogenic archaea and protozoal populations. J Dairy Sci. 2009;92:5512–5522. doi: 10.3168/jds.2008-1441. [DOI] [PubMed] [Google Scholar]
- Bodas R, Lopez S, Fernandez M, Garcia-Gonzalez R, Rodrıguez AB, Wallace RJ, Gonzalez JS. In vitro screening of the potential of numerous plant species as antimethanogenic feed additives for ruminants. Anim Feed Sci Technol. 2008;145:245–258. [Google Scholar]
- Cushnie TPT, Lamb AJ. Antimicrobial activity of flavonoids. Int J Antimicrob Agents. 2005;26:343–356. doi: 10.1016/j.ijantimicag.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denman SE, McSweeney CS. Quantitative (real-time) PCR. In: Makkar HPS, McSweeney CS, editors. Methods in Gut Microbial Ecology for Ruminants. Springer; Dordrecht, The Netherlands: 2005. pp. 105–115. [Google Scholar]
- Denman SE, McSweeney CS. Development of a Real-Time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol Ecol. 2006;58:572–582. doi: 10.1111/j.1574-6941.2006.00190.x. [DOI] [PubMed] [Google Scholar]
- Denman SE, Tomkins NW, McSweeney CS. Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol Ecol. 2007;62:313–322. doi: 10.1111/j.1574-6941.2007.00394.x. [DOI] [PubMed] [Google Scholar]
- Dohme F, Machmuller A, Wasserfallen A, Kreuzer M. Ruminal methanogenesis as influenced by individual fatty acids supplemented to complete ruminant diets. Lett Appl Microbiol. 2001;32:47–51. doi: 10.1046/j.1472-765x.2001.00863.x. [DOI] [PubMed] [Google Scholar]
- Ellis JL, Kebreab E, Odongo NE, McBride BW, Okine EK, France J. Prediction of methane production from dairy and beef cattle. J Dairy Sci. 2007;90:3456–3467. doi: 10.3168/jds.2006-675. [DOI] [PubMed] [Google Scholar]
- Jia Z, Tang M, Wu J. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999;64:555–559. [Google Scholar]
- Johnson KA, Johnson DE. Methane emissions from cattle. J Anim Sci. 1995;73:2483–2492. doi: 10.2527/1995.7382483x. [DOI] [PubMed] [Google Scholar]
- Koike S, Kobayashi Y. Development and use of competitive PCR assays for the rumen cellulolytic bacteria: Fibrobacter succinogenes, Ruminococcus albus and Ruminococcus flavefaciens. FEMS Microbiol Ecol. 2001;204:361–366. doi: 10.1111/j.1574-6968.2001.tb10911.x. [DOI] [PubMed] [Google Scholar]
- Latham MJ, Wolin MJ. Fermentation of cellulose by Ruminococcus flavefaciens in the presence and absence of Methanobacterium ruminantium. Appl Environ Microbiol. 1977;34:297–301. doi: 10.1128/aem.34.3.297-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougall EI. Studies on ruminant saliva. 1. The composition and output of sheep’s saliva. Biochem J. 1948;43:99–109. [PMC free article] [PubMed] [Google Scholar]
- Ntaikou I, Gavala HN, Kornaros M, Lyberatos G. Hydrogen production from sugars and sweet sorghum biomass using Ruminococcus albus. Int J Hydrogen Energy. 2008;33:1153–1163. [Google Scholar]
- Oskoueian E, Abdullah N, Oskoueian A. Effects of flavonoids on rumen fermentation activity, methane production, and microbial population. Biomed Res Int. 2013:8. doi: 10.1155/2013/349129. Article ID 349129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra AK, Kamra DN, Agarwa N. Effect of plant extracts on in vitro methanogenesis, enzyme activities and fermentation of feed in rumen liquor of buffalo. Anim Feed Sci Technol. 2006;128:276–291. [Google Scholar]
- Patra AK, Saxena J. A new perspective on the use of plant secondary metabolites to inhibit methanogenesis in the rumen. Phytochemistry. 2010;71:1198–1222. doi: 10.1016/j.phytochem.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Pen B, Sar C, Mwenya B, Kuwaki K, Morikawa R, Takahashi J. Effects of Yucca schidigera and Quillaja saponaria extracts on in vitro ruminal fermentation and methane emission. Anim Feed Sci Technol. 2006;129:175–186. [Google Scholar]
- Institute SAS. SAS User’s Guide. SAS Institute Inc; Cary, NC, USA: 2002. [Google Scholar]
- Skillman LC, Evans PN, Strömpl C, Joblin KN. 16S rDNA directed PCR primers and detection of methanogens in the bovine rumen. Lett Appl Microbiol. 2006;42:222–228. doi: 10.1111/j.1472-765X.2005.01833.x. [DOI] [PubMed] [Google Scholar]
- Skillman LC, Toovey AF, Williams AJ, Wright AG. Development and validation of a real-time PCR method to quantify rumen protozoa and examination of variability between Entodinium populations in sheep offered a hay-based diet. Appl Environ Microbiol. 2006;72:200–206. doi: 10.1128/AEM.72.1.200-206.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedesco D, Tava A, Galletti S, Tameni M, Varisco G, Costa A, Steidler S. Effects of silymarin, a natural hepatoprotector, in periparturient dairy cows. J Dairy Sci. 2004;87:2239–2247. doi: 10.3168/jds.S0022-0302(04)70044-2. [DOI] [PubMed] [Google Scholar]
- Theodorou MK, Williams BA, Dhanoa MS, McAllan AB, France J. A simple gas production method using a pressure transducer to determine the fermentation kinetics of ruminant feeds. Anim Feed Sci Technol. 1994;48:185–197. [Google Scholar]
- Velioglu YS, Mazza G, Cao L, Oomah BD. Antioxidant activity and total phenolics in selected fruit, vegetables, and grain products. J Agric Food Chem. 1998;46:4113–4117. [Google Scholar]
- Zhou Z, Meng Q, Yu Z. Effects of methanogenic inhibitors on methane production and abundances of methanogens and cellulolytic bacteria in in vitro ruminal cultures. Appl Environ Microbiol. 2011;77:2634–2639. doi: 10.1128/AEM.02779-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


