Abstract
The VP40 matrix protein of Ebola virus buds from cells in the form of virus-like particles (VLPs) and plays a central role in virus assembly and budding. In this study, we utilized a functional budding assay and cotransfection experiments to examine the contributions of the glycoprotein (GP), nucleoprotein (NP), and VP24 of Ebola virus in facilitating release of VP40 VLPs. We demonstrate that VP24 alone does not affect VP40 VLP release, whereas NP and GP enhance release of VP40 VLPs, individually and to a greater degree in concert. We demonstrate further the following: (i) VP40 L domains are not required for GP-mediated enhancement of budding; (ii) the membrane-bound form of GP is necessary for enhancement of VP40 VLP release; (iii) NP appears to physically interact with VP40 as judged by detection of NP in VP40-containing VLPs; and (iv) the C-terminal 50 amino acids of NP may be important for interacting with and enhancing release of VP40 VLPs. These findings provide a more complete understanding of the role of VP40 and additional Ebola virus proteins during budding.
Ebola virus is a member of the Filoviridae family and is associated with recurrent outbreaks of deadly hemorrhagic fevers (8). Currently, there are no approved vaccines, nor are there antiviral therapeutics to prevent or treat individuals infected with Ebola virus (16). A better understanding of the molecular aspects of Ebola virus replication will be necessary for successful development of specific treatments for Ebola virus infection.
The VP40 matrix protein of Ebola virus is the most abundant virion protein and plays a key role in virus assembly and budding (12, 14, 28). For example, VP40 can bud as a filamentous virus-like particle (VLP) from mammalian cells in the absence of any other viral protein (12, 20). The ability of VP40 to bud as a VLP is due, in part, to the presence of L domains present at the N terminus of the protein (12, 14, 17). Viral L domains are thought to serve as docking sites for interactions with specific cellular proteins, and the resultant virus-host interactions are believed to facilitate efficient virus budding (for a review, see reference 9). Although VP40 and other viral matrix proteins (e.g., Gag and M proteins of retroviruses and rhabdoviruses, respectively) can bud independently from cells, additional viral proteins are undoubtedly important for the budding process. For example, the G protein of vesicular stomatitis virus (VSV) has been shown to be important for efficient budding. More specifically, the membrane-proximal stem, transmembrane domain, and cytoplasmic tail of VSV G appear to confer this efficiency (21, 25). In addition, alterations to the cytoplasmic tails of glycoproteins of other RNA viruses, such as influenza A, simian virus 5, and rabies virus, appear to result in poor budding despite an intact matrix protein (15, 19, 23).
In addition to VP40, the surface glycoprotein (GP), the nucleoprotein (NP), and the minor matrix protein (VP24) of Ebola virus have been implicated in virus assembly and budding (1, 11, 12). For example, GP and VP24 have been detected in a lipid-associated form in the media of transfected cells (11, 29). It has been suggested that localization of GP to lipid raft domains enhances budding of Ebola virus from these specialized microdomains (1). VP24 has been postulated to play a role in assembly and budding (11) and most recently in efficient assembly of viral nucleocapsids (13). While a role for Ebola virus NP in budding has not been specifically examined to date, data from studies of other negative-strand RNA viruses suggest a role for the nucleocapsid protein in the assembly process (5, 6). Together, GP, NP, and VP24 represent candidate viral proteins likely to contribute to virus budding.
In this report, we utilized our VP40 VLP budding assay to evaluate various combinations of GP/VP24/NP expression for their ability to enhance release of VP40 VLPs. Our results indicate that coexpression of GP and/or NP with VP40 leads to a reproducible enhancement of VLP release. In contrast, the combination of VP40 plus VP24 did not result in any detectable enhancement of VP40 budding; however, release of VP40 VLPs from cells expressing VP40 plus NP plus VP24 was greater than that from cells expressing only VP40 plus NP. Our results further suggest that GP- and NP-mediated enhancement of release of VLPs may be due to interactions with VP40.
MATERIALS AND METHODS
Cells.
Human 293T or COS-1 cells were maintained in Dulbecco minimal essential medium (DMEM) (Invitrogen/Life Technologies) supplemented with 10% fetal calf serum (Invitrogen/Life Technologies) and 1× penicillin-streptomycin (Invitrogen/Life Technologies).
Plasmids and antibodies.
Plasmids encoding VP40-WT, VP40-dPTA, VP40-Y13A, VP40-dPT/PY, and hemagglutinin (HA)-tagged VP24 have been described previously (11, 17). The NP open reading frame from Ebola virus (Zaire) was amplified by PCR and inserted into the EcoRI and XhoI sites of the pCAGGS/MCS vector by standard cloning techniques. Plasmid NPC-50, in which the C-terminal 50 amino acids of NP have been deleted, was generated by PCR and standard cloning techniques. The GP gene of EBOZ was inserted into the EcoRI and XhoI sites of the pCAGGS/MCS vector by standard PCR and cloning techniques. The sec-GP plasmid was constructed by PCR and encodes only the ectodomain of EBOZ GP. The VSV G gene was inserted into the EcoRI and XhoI sites of the pCAGGS/MCS vector by standard PCR and cloning techniques. The herpes simplex virus-1 (HSV-1) gD glycoprotein (kindly provided by R. Eisenberg, University of Pennsylvania) was amplified by PCR and inserted into the EcoRI and XhoI sites of the pCAGGS/MCS vector by standard cloning techniques. All plasmids and introduced mutations were confirmed by automated DNA sequencing.
Monoclonal antibodies against the VP40 and NP proteins of Ebola virus were kindly provided by J. Paragas (U.S. Army Medical Research Institute for Infectious Diseases, Fort Detrick, Md.). Monoclonal antibody against full-length GP of Ebola virus was kindly provided by P. Bates (University of Pennsylvania). Monoclonal antibody against HSV-1 gD (monoclonal antibody DL6) was kindly provided by R. Eisenberg (University of Pennsylvania). Monoclonal antibodies against the HA epitope tag (Roche Biochemicals, Indianapolis, Ind.), VSV G (Sigma-Aldrich, St. Louis, Mo.), horseradish peroxidase-linked anti-mouse and -rabbit (Molecular Probes), and horseradish peroxidase-linked anti-rat (Santa Cruz Biotechnology, Santa Cruz, Calif.) were used according to protocols of the suppliers. Fluorescein isothiocyanate-conjugated anti-rat and phycoerythrin-conjugated anti-mouse antibodies (Roche Biochemicals, Indianapolis, Ind.) were used according to the manufacturer's instructions.
Budding assay.
A functional budding assay for VP40 has been previously described (17). Briefly, 3 × 106 293T cells were transfected with 1.0 μg of the indicated plasmids using Lipofectamine (Invitrogen). At 24 h posttransfection (hpt), cells were metabolically labeled with [35S]Met-Cys (Perkin-Elmer) (100 μCi/ml) for 6 h. Culture medium was harvested and clarified at 2,500 rpm for 10 min. The supernatant was then centrifuged through a 20% sucrose cushion in STE buffer (0.01 M Tris-HCl [pH 8.0], 0.01 M NaCl, 0.001 M EDTA [pH 8.0]) for 2 h at 36,000 rpm (Beckman SW41 rotor) to isolate VLPs. VLPs were resuspended in STE buffer. Radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1.0% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) was added to transfected cells and resuspended VLPs. Twenty-five percent of the cell lysates and 100% of the VLPs were immunoprecipitated with appropriate antibodies and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). Protein bands were visualized by autoradiography and quantitated by phosphorimager analysis.
VLP flotation gradients.
293T cells were transfected with the indicated plasmids as described above. At 48 hpt, culture medium was harvested and VLPs were isolated as described above. Resuspended VLPs were then floated on a discontinuous sucrose gradient (80, 50, and 10% in STE buffer) by centrifugation at 36,000 rpm (Beckman SW41 rotor) for 18 h. Twelve 1.0-ml fractions were harvested from the top to the bottom of the gradient, and proteins were isolated by trichloroacetic acid precipitation and resuspended in 1× Laemmli sample buffer. Transfected cells were lysed in radioimmunoprecipitation assay buffer. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes for Western blotting. Membranes were blocked for 1 h and then incubated with the appropriate antibody for 1 h at room temperature. After several washes in 1× phosphate-buffered saline (PBS) containing 0.1% Tween 20, membranes were incubated with the appropriate horseradish peroxidase-linked secondary antibody for 1 h at room temperature. The membranes were washed as before, and proteins were visualized by chemiluminescence and autoradiography.
Immunofluorescence and confocal microscopy.
COS-1 cells were transfected with the indicated plasmids as described above. At 24 hpt, cells were washed with 1× PBS and fixed with cold methanol and stained with appropriate primary and secondary antibodies. Coverslips were mounted in ProLong Antifade (Molecular Probes, Ltd.). Microscopy was performed with a Bio-Rad Radiance 2000 confocal microscope using Lasersharp 2000 software (Bio-Rad, Ltd.).
RESULTS
Budding of Ebola virus proteins as VLPs. To determine whether Ebola virus GP, NP, and VP24 contribute to VP40 VLP release, we first sought to determine whether GP, NP, and VP24 could be released from cells individually as lipid-containing particles. As expected, VP40 alone was released from cells as a VLP which was detected predominantly in the upper fractions of a flotation gradient (Fig. 1A; lanes 2 to 4). A minor amount of VP40 was detected in fractions 7 and 8, while no VP40 was detected at the bottom of the gradient (Fig. 1A). The presence of VP40 in the cell extract is shown as an expression control (Fig. 1A, lane 13). Consistent with previous findings of others (1, 29), Ebola virus GP alone was also released from cells, albeit at a level considerably less than that of VP40 (compare Fig. 1A, lanes 2 and 13, and Fig. 1B, lanes 2 and 13). Importantly, GP was detected in the same upper fractions as VP40 (Fig. 1B, lane 2). Conversely, neither VP24 nor NP was released as a VLP in this assay (Fig. 1C and D), although they were abundantly expressed in the cells (lane 13). VP40 was released from cells in greater abundance than either GP, NP, or VP24 and is therefore likely to function as the primary driving force for budding.
FIG. 1.
Flotation gradient analysis of Ebola virus protein-induced VLPs. Human 293T cells were transfected with VP40 (A), GP (B), VP24 (C), or NP (D), and VLPs were isolated from the culture medium at 48 hpt. VLPs were isolated by centrifugation through a 20% sucrose cushion and subsequently floated on a discontinuous (80-50-10%) sucrose gradient. One-milliliter fractions were taken from top (lane 1) to bottom (lane 12). Proteins in each fraction as well as in the cell lysate (lane 13) were isolated by trichloroacetic acid precipitation, separated by SDS-PAGE, and analyzed by Western blotting.
Coexpression of VP24 does not influence release of VP40 VLPs.
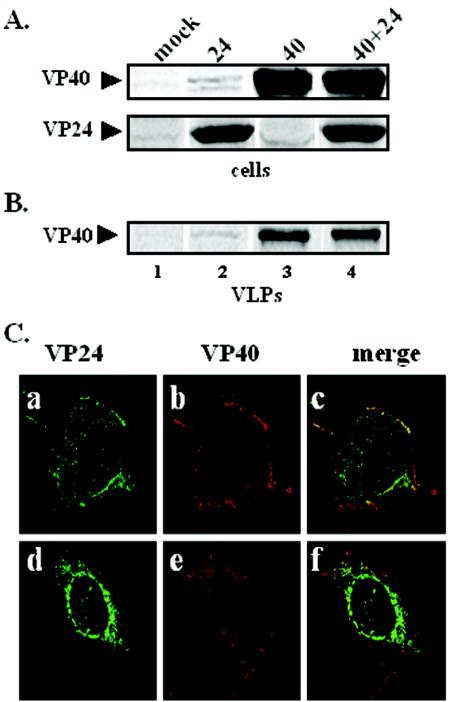
To determine whether expression of VP24 enhances release of VP40 VLPs, 293T cells were cotransfected with equivalent amounts of VP40 and VP24 expression plasmids, and VP40 VLPs were prepared from culture supernatants at 30 hpt. Cell extracts were immunoprecipitated with either anti-VP40 or anti-HA antisera to ensure that VP40 and VP24 were expressed appropriately in transfected cells (Fig. 2A). Mock-transfected cells served as a negative control (Fig. 2A, lane 1). Immunoprecipitation and phosphorimager analyses of VP40 VLPs from several independent experiments revealed that coexpression of VP24 had no significant effect on budding of VP40 VLPs, since equivalent amounts of VP40 were detected in the presence or absence of VP24 (Fig. 2B, lanes 3 and 4).
FIG. 2.
Functional budding assay and intracellular localization of VP40 and VP24. 293T cells were transfected with pCAGGS vector alone (lane 1), VP24 (lane 2), VP40 (lane 3), or VP24 plus VP40 (lane 4). VP40 and VP24 proteins were detected in cells (A), and VP40 was detected in budding VLPs (B) by immunoprecipitation and SDS-PAGE analysis at 30 hpt. (C) Cos-1 cells were cotransfected with VP40 and VP24, and fixed cells were visualized by confocal microscopy 24 hpt. VP24 is shown in green (panels a and c), VP40 is shown in red (panels b and e), and the merged images are shown in panels c and f. Yellow indicates colocalization.
To substantiate these results, confocal microscopy was used to determine whether VP40 and VP24 colocalized in cotransfected cells (Fig. 2C). As expected, VP40 was detected primarily on the plasma membrane and was often present in thin filamentous projections originating from the plasma membrane (Fig. 2C, panels b and e). In contrast, VP24 was present predominantly in a perinuclear pattern in the majority of cells, whereas localization of VP24 to the plasma membrane occurred to a much lesser extent (Fig. 2C, panels a and d). These results indicated that VP40 and VP24 did not colocalize in the majority of cotransfected cells (Fig. 2C, panel f); however, a limited number of cotransfected cells showed a small amount of colocalization at the plasma membrane (Fig. 2C, panel c). Taken together, these data suggest that VP24 likely does not interact directly with VP40, and coexpression of VP24 with VP40 does not influence release of VP40 VLPs.
Coexpression of either GP or NP with VP40 enhances release of VP40 VLPs.
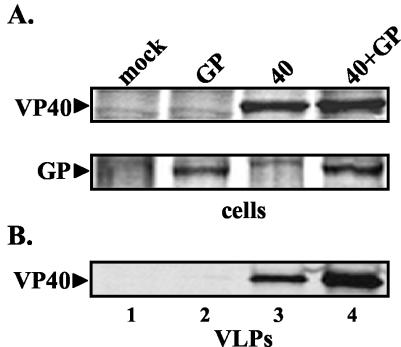
We next sought to determine whether coexpression of GP enhances release of VP40 VLPs. Human 293T cells were cotransfected with equivalent amounts of VP40 and GP expression constructs, and VP40 VLPs were detected by immunoprecipitation (Fig. 3). Immunoprecipitation and phosphorimager analyses of VP40 and GP from cell extracts demonstrated equivalent expression in appropriate samples (Fig. 3A). Immunoprecipitation of VP40 in VLP samples revealed that coexpression of GP and VP40 resulted in more efficient release (an average of fivefold) of VP40-containing VLPs (Fig. 3B, compare lanes 3 and 4). It should be noted that a similar level of enhancement of VP40 release was observed using VLPs purified through a flotation gradient and that VP40 and GP were routinely detected in identical fractions taken from these gradients (data not shown). Taken together, these findings suggest that enhancement of release of VP40 VLPs by GP is likely due, at least in part, to a specific mechanism and not simply to reported cytopathic effects of GP (30). Indeed, under these cotransfection conditions in both COS-1 and 293T cells, GP was found to be noncytopathic (26) as judged by cell viability assays (data not shown). Lastly, confocal microscopy was utilized to confirm that VP40 and GP colocalize heavily at the plasma membrane of transfected cells (data not shown).
FIG. 3.
Functional budding assay for coexpressed VP40 and GP. 293T cells were transfected with VP40 (40) or VP40 plus GP (40+GP), and the resulting VLPs were isolated from the culture medium at 30 hpt. VP40 and GP proteins were detected in cell extracts (A) and VP40 was detected in budding VLPs (B) by immunoprecipitation and SDS-PAGE analysis. A similar enhancement of VP40 budding was observed in the presence of GP when VLPs were purified further on a discontinuous flotation gradient (data not shown).
Similar cotransfection experiments were performed to determine the contribution of NP to budding of VP40 VLPs. Immunoprecipitation and phosphorimager analyses of cotransfected cells revealed that NP and VP40 were expressed at equivalent levels in the appropriate samples (Fig. 4A, lanes 1 to 4). We observed an average 3.5-fold increase in the level of VP40 present in VLPs budding from cells coexpressing the NP protein (Fig. 4B, compare lanes 3 and 4). Intriguingly, when the GP protein was expressed along with both NP and VP40, an additive increase (averaging greater than 40-fold) in VP40 VLPs was observed in repeated experiments (Fig. 5). Similarly, whereas VP24 alone had no effect on VP40 VLP release, coexpression of VP40 plus NP plus VP24 enhanced the release of VP40 VLPs to a greater extent than that observed from cells expressing only VP40 plus NP (Fig. 5). Taken together, these data indicate that in addition to VP40, other viral proteins contribute to the efficiency of VLP budding when coexpressed in specific combinations with VP40 (Fig. 5).
FIG. 4.
Functional budding assay for coexpressed VP40 and NP. 293T cells were transfected with pCAGGS vector alone (lane 1), NP (lane 2), VP40 (lane 3), or VP40 plus NP (lane 4). Radiolabeled cell extracts (A) and VLPs (B) were immunoprecipitated with appropriate antiserum, and immunoprecipitated proteins were analyzed by SDS-PAGE and autoradiography.
FIG. 5.
Relative release of VP40 VLPs from cells expressing various combinations of Ebola virus proteins. The amount of VP40 VLPs released from cells in the absence of other viral proteins was set at 1.0. The n-fold enhancement of VP40 VLP release (determined by phosphorimager analysis) is shown for cells cotransfected with VP40 plus VP24, VP40 plus NP, VP40 plus GP, VP40 plus NP plus VP24, and VP40 plus NP plus GP. The bars represent an average of at least three independent experiments. Empty vector was used when necessary to maintain an equivalent amount of total transfected DNA in each sample.
GP-enhanced release of VP40 VLPs is independent of L-domain function.
We hypothesized that the L domains of VP40 likely were not necessary for GP-mediated enhancement, since the L domains mediate interactions with specific host proteins. Mutations that disrupt the L-domain motifs of VP40 have been shown to impair release of VP40 VLPs to various degrees (17). Cells were cotransfected with GP and a series of previously described VP40 L-domain mutants (Fig. 6). As predicted, release of L-domain dPT/PY, dPTA, and Y13A mutants was enhanced by GP an average of eightfold as determined by phosphorimager analysis (Fig. 6A, compare lanes 1 and 2, 3 and 4, and 5 and 6). Consistent with our previous report, the dPT/PY mutant buds much less efficiently than either the dPTA or Y13A mutant, even in the presence of GP (Fig. 6A, compare lanes 2, 4, and 6). Therefore, while expression of GP leads to more efficient release of all VP40 L-domain mutants (Fig. 6B), GP expression does not rescue the L-domain deficiency and thus does not permit all of the L-domain mutants to bud to equivalent levels (Fig. 6A).
FIG. 6.
Budding of VP40 L-domain mutants in the absence or presence of GP. (A) 293T cells were transfected with VP40 L-domain dPT/PY (lane 1), dPTA (lane 3), or Y13A (lane 5) mutants alone or along with GP (lanes 2, 4, and 6, respectively). VP40 was detected in budding VLPs by immunoprecipitation and SDS-PAGE analysis. (B) Levels of VP40 detected in VLPs were quantitated by phosphorimager analysis. The amount of each L-domain mutant alone was normalized to 1.0.
Specific enhancement of VP40 VLP release by Ebola virus GP.
Enhancement of virus budding by glycoproteins often results from direct interactions between the viral matrix protein and the transmembrane and/or cytoplasmic domains of the glycoprotein (7, 15, 25). To determine whether the enhanced release of VP40 VLPs by Ebola virus GP was specific to GP, cells were cotransfected with VP40 and heterologous glycoproteins VSV G and HSV-1 gD (Fig. 7A). Expression controls for VP40 and glycoproteins GP, G, and gD were performed by immunoprecipitation (Fig. 7A, upper panel). Analysis of flotation gradient purified VP40 VLPs released from the cotransfected cells revealed that similar amounts of VP40 budded from cells receiving no glycoprotein, VSV G, and HSV-1 gD (Fig. 7A, lower panels). In contrast, release of VP40 was clearly enhanced as expected in the presence of Ebola virus GP (Fig. 7A, lower panel). These results indicate that the enhancement of VP40 VLP release is not observed in the presence of heterologous glycoproteins and thus suggests some level of specificity for GP-enhanced VP40 VLP release. It should be noted that under these assay conditions, expression of Ebola virus GP did not result in cell death. However, a role for GP-mediated cell rounding in the enhancement of VP40 VLP release cannot be ruled out.
FIG. 7.
Effect of heterologous glycoproteins on VP40 VLP release. (A) 293T cells were transfected with VP40 alone (40), VP40 plus GP (40 + GP), VP40 plus VSV G (40 + G), or VP40 plus HSV-1 gD (40 + gD). Expression controls for VP40 and the various glycoproteins in cells are shown (upper panel). VLPs from the above-transfected cells were isolated and purified by a discontinuous flotation gradient. Immunoprecipitation of VP40 from upper fractions 2 and 3 are shown for each transfection condition (lower panel), and the amount of VP40 present was determined by phosphorimager analysis. (B) Effect of the secreted GP on VP40 VLP release. 293T cells were transfected with pCAGGS (lane 1), VP40 (lane 2), VP40 plus WT GP (lane 3), or VP40 and sec-GP (lane 4). Expression controls for VP40 and the various forms of GP in cells are shown (upper panel). The amount of VP40 in isolated VLPs was determined by immunoprecipitation and phosphorimager analysis (bottom panel).
Furthermore, we sought to determine whether a secreted form of Ebola virus GP (sec-GP), which contains the ectodomain and mucin-rich region of GP, would enhance release of VP40 VLPs. Expression controls for each protein are shown (Fig. 7B, upper panel). Analysis of VP40 VLPs released from these cells revealed that sec-GP was unable to enhance VP40 VLP release (Fig. 7B, lower panel, compare lanes 2, 3, and 4). Indeed, phosphorimager analysis confirmed these findings in repeated experiments. These results indicate that the membrane-bound form of GP is required for enhancement of VP40 VLP release.
Potential interaction of VP40 and NP in VLPs.
To begin to probe the mechanism of NP-enhanced VP40 VLP release, we rationalized that an NP-VP40 interaction might mediate the observed increase in VP40 VLP release. We examined the ability of wild-type (WT) NP and a deletion mutant (NPC-50) of NP lacking the C-terminal 50 amino acids to enhance release of VP40 VLPs (Fig. 8). Cells were either mock transfected, transfected with VP40 and NP, or transfected with VP40 and NPC-50 (Fig. 8A). As expression controls, cell extracts were immunoprecipitated with anti-VP40 antiserum (Fig. 8A, lanes 1, 3, and 5) or anti-NP antiserum (lanes 2, 4, and 6). Although VP40-specific antiserum pulled down both VP40 and NP (lane 3) and VP40 and NPC-50 (lane 5), this may be due to cross-reactivity of the antiserum. Anti-NP antibody pulled down NP (lane 4) and NPC-50 (lane 6) but not VP40 (lanes 4 and 6).
FIG. 8.
The C-terminal region of NP is important for recruitment into VP40 VLPs. (A) 293T cells were mock transfected (lanes 1 and 2) of transfected with VP40 plus NP (lanes 3 and 4) or VP40 plus NPC-50 (lanes 5 and 6). Cell extracts were immunoprecipitated with either anti-VP40 antiserum (lanes 1, 3, and 5) or anti-NP antiserum (lanes 2, 4, and 6). (B) VLPs were isolated from the culture medium and immunoprecipitated with anti-VP40 antiserum. Immunoprecipitated proteins were analyzed by SDS-PAGE and autoradiography. (C) VLPS were isolated from 293T cells transfected with NP alone (lane 1), NPC-50 (lane 2), VP40 plus NP (lane 3), or VP40 plus NPC-50 (lane 4). VLPs were immunoprecipitated with anti-NP antiserum, and proteins were analyzed by SDS-PAGE and autoradiography.
Interestingly, coexpression of NP with VP40 resulted in the expected increase in VP40 VLP release (Fig. 8B, compare lanes 1 and 2); however, coexpression of NPC-50 did not result in enhanced release of VP40 VLPs (Fig. 8B, compare lanes 1 and 3). Phosphorimager analysis was used to confirm these findings in repeated experiments (data not shown). These data suggest that the extreme C-terminal region of NP may be important for enhancing VP40 VLP budding. If NP does in fact interact with VP40, we might expect to observe NP in VP40-containing VLPs. Indeed, WT NP was detected in VLPs released from cells expressing NP and VP40 (Fig. 8C, lane 3) but not from cells expressing NP alone (Fig. 8C, lane 1). Moreover, NPC-50 was not detected in VLPs released from cells expressing NPC-50 alone (Fig. 8C, lane 2) or both NPC-50 and VP40 (Fig. 8C, lane 4). These data suggest that VP40 can recruit NP, but not NPC-50, into budding VLPs.
DISCUSSION
The prominent role of the Ebola virus VP40 protein in budding is well established (14, 17, 20, 28); however, the mechanism(s) by which VP40 achieves this final step of the viral life cycle remains unclear. In addition to the clear role of the VP40 L domains in mediating interactions with host proteins to promote the formation of VP40 VLPs (12, 27), we present evidence that efficient budding of VP40 VLPs also requires the concerted effort of additional viral proteins, including, GP, NP, and VP24.
Flotation gradient analysis of budding VLPs indicates that VP40 is the major virus budding protein. Coexpression of VP40 and VP24 did not result in any significant change in the amount of VP40 VLPs released from transfected cells. Furthermore, VP40 and VP24 exhibited little to no colocalization as determined by confocal microscopy (Fig. 2). These data strongly suggest that VP24 alone does not influence VP40 budding and that VP24 does not directly interact with VP40. However, the possibility that a physical and/or functional interaction between VP40 and VP24 occurs in Ebola virus-infected cells cannot be ruled out.
In contrast to VP24, we were able to demonstrate that coexpression of NP and VP40 enhanced VP40 VLP release in a functional budding assay. Indeed, cotransfection of NP and VP40 consistently resulted in an average 3.5-fold increase in the amount of VP40 released as VLPs (Fig. 4B). Although this increase is fairly low, it is highly reproducible and thus is likely to be significant. NP was detected in VP40 VLPs, and initial results suggest that NP may be recruited to budding VLPs by interacting with VP40 via its C-terminal region (Fig. 8C). This result is not surprising, since there is precedent for NP incorporation into matrix protein induced particles in other virus systems, such as human parainfluenza virus type 1 (5, 6). In addition, the matrix protein of simian virus 5 was released at levels comparable to those of infectious virus only when coexpressed with at least one of two glycoproteins and nucleoprotein (24). Putative interactions between NP and VP40 may function to stabilize VP40 at the plasma membrane, perhaps accounting for the observed NP-mediated enhancement of VP40 VLP release. Alternatively, NP-mediated enhancement of budding may result from the ability of NP to promote interactions between VP40 and cytoskeletal elements, undoubtedly important in the overall assembly and budding processes. Experiments are currently under way to identify further the regions of NP and VP40 that may be involved in protein-protein interactions that promote budding.
Interestingly, we found that the combination of VP40, NP, and VP24 resulted in a slightly, but reproducibly, larger enhancement of VP40 VLP budding than that observed with only VP40 and NP (Fig. 5). Based upon these findings and data of others (12), it is tempting to speculate that VP24 may serve as a bridging or linking protein between VP40 and NP. VP24 has been shown to be a component of Ebola virus nucleocapsids (15), and thus, our finding that VP24 may indirectly enhance budding of VP40 VLPs only in the presence of NP would be consistent with these findings.
Consistent with previous reports, Ebola virus GP was found to be present in budding VP40 VLPs. Findings that (i) GP colocalized with VP40 on the plasma membrane (data not shown), (ii) coexpression of GP resulted in a significant increase in the amount of VP40 present in the upper fractions of a flotation gradient, and (iii) GP was not found to be cytopathic in our transfection assays (data not shown) all strongly suggest that the enhancement effect of GP is likely due to a specific mechanism. Our finding that the heterologous glycoproteins VSV G and HSV-1 gD were unable to enhance VP40 VLP release are consistent with this hypothesis. We were able to demonstrate clearly that the GP-mediated enhancement of VP40 release was not dependent on the presence of L domains within VP40, since we observed a significant increase in budding of several L-domain mutants of VP40 in the presence of GP (Fig. 6). These results support the idea that VP40 is the major driving force for budding and, importantly, indicate that there are separable viral and host-mediated (10, 18) mechanisms to augment release of VP40 VLPs.
We have begun to probe the mechanism of GP-enhanced VP40 VLP release. GP has been demonstrated to localize to lipid raft domains on the plasma membrane (1). Indeed, accumulating evidence suggests that lipid rafts play a role in the assembly of many viruses (for a review, see reference 2). VP40 appears to be localized to lipid rafts in Ebola virus-infected cells (1); however, whether recruitment of VP40 to rafts is achieved solely by interactions with host factors such as TSG101 (17) or by interactions with viral proteins remains less clear. One possible mechanism for GP-mediated enhancement of budding is that GP may also recruit a population of VP40 to lipid raft domains that may represent active sites of budding. However, results from raft gradient flotation analysis indicated that the population of VP40 present in lipid raft fractions in VP40-transfected cells was not increased significantly when these cells were cotransfected with GP (J. M. Licata and R. N. Harty, unpublished data). In addition, GP-enhanced release of VP40 VLPs was not affected by disrupting lipid raft formation by treating cells with methyl-β-cyclodextrin (data not shown). Although lipid raft localization does not appear to play a major role in mediating GP-enhanced budding of VP40 VLPs, the concentration of GP to particular regions of the membrane is likely important for budding. For example, evidence suggests that VSV G does not localize to lipid raft domains (22, 31); however, VSV G does appear to be concentrated in specific lipid microdomains which may serve as assembly sites for progeny virions (4). Concentration of viral proteins in these regions has been postulated to allow for modification of the lipid environment to promote virus budding or to facilitate membrane curvature required for virus budding (3).
Current efforts are focused on identifying regions within GP and VP40 that may physically interact to promote release of VP40 VLPs. Our data indicate that the membrane-bound form of GP is required for the observed enhancement of VP40 VLP release (Fig. 7B). One possibility is that the transmembrane and/or cytoplasmic tail of GP is important for budding. Studies with VSV indicate that while a specific sequence of the cytoplasmic tail of VSV G protein is not required for glycoprotein-enhanced release of M-induced VLPs, there is a requirement for a specific cytoplasmic domain length (25). More recently, the membrane-proximal stem of VSV G was found to be required for efficient viral budding (21). For example, a G-stem protein containing this region in addition to the transmembrane and cytoplasmic tails was sufficient to enhance virus release (21). The membrane-proximal stem has been proposed to act by facilitating membrane curvature to induce the budding process (25). An in-depth mutational analysis of the cytoplasmic tail, transmembrane region, and proximal stem region of Ebola virus GP is currently under way in an attempt to characterize further those regions that function to enhance VP40 VLP budding. A better understanding of these Ebola virus protein-protein interactions will complement our existing knowledge of Ebola virus-host interactions and provide a more complete insight into the mechanism of Ebola virus budding.
Acknowledgments
We thank P. Bates, J. Paragas, and R. Eisenberg for generously providing reagents and for helpful discussions. We also thank Meghan Matukonis for construction of the Ebola virus NP expression plasmid.
J.M.L. and R.F.J. are supported by NIH Training Grant AI007324. R.N.H. is supported in part by a Health Research Faculty Development Block Grant.
REFERENCES
- 1.Bavari, S., C. M. Bosio, E. Wiegand, G. Ruthel, A. B. Will, T. W. Geisbert, M. Hevey, C. Schmaljohn, A. Schmaljohn, and M. J. Aman. 2002. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 195:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briggs, J. A., T. Wilk, and S. D. Fuller. 2003. Do lipid rafts mediate virus assembly and pseudotyping? J. Gen. Virol. 84:757-768. [DOI] [PubMed] [Google Scholar]
- 3.Brown, E. L., and D. S. Lyles. 2003. A novel method for analysis of membrane microdomains: vesicular stomatitis virus glycoprotein microdomains change in size during infection, and those outside of budding sites resemble sites of virus budding. Virology 310:343-358. [DOI] [PubMed] [Google Scholar]
- 4.Brown, E. L., and D. S. Lyles. 2003. Organization of the vesicular stomatitis virus glycoprotein into membrane microdomains occurs independently of intracellular viral components. J. Virol. 77:3985-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coronel, E. C., K. Gopal Murti, T. Takimoto, and A. Portner. 1999. Human parainfluenza virus type 1 matrix and nucleoprotein genes transiently expressed in mammalian cells induce the release of virus-like particles containing nucleocapsid-like structures. J. Virol. 73:7035-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coronel, E. C., T. Takimoto, K. Gopal Murti, N. Varich, and A. Portner. 2001. Nucleocapsid incorporation into parainfluenza virus is regulated by specific interaction with matrix protein. J. Virol. 75:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosson, P. 1996. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 15:5783-5788. [PMC free article] [PubMed] [Google Scholar]
- 8.Feldmann, H., and H. D. Klenk. 1996. Marburg and Ebola viruses. Adv. Virus Res. 47:1-52. [DOI] [PubMed] [Google Scholar]
- 9.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 11.Han, Z., H. Boshra, J. O. Sunyer, S. H. Zwiers, J. Paragas, and R. N. Harty. 2003. Biochemical and functional characterization of the Ebola virus VP24 protein: implications for a role in virus assembly and budding. J. Virol. 77:1793-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, Y., L. Xu, Y. Sun, and G. J. Nabel. 2002. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol. Cell 10:307-316. [DOI] [PubMed] [Google Scholar]
- 14.Jasenosky, L. D., G. Neumann, I. Lukashevich, and Y. Kawaoka. 2001. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J. Virol. 75:5205-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin, H., G. P. Leser, J. Zhang, and R. A. Lamb. 1997. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 16:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klenk, H. D. 2000. Will we have and why do we need an Ebola vaccine? Nat. Med. 6:1322-1323. [DOI] [PubMed] [Google Scholar]
- 17.Licata, J. M., M. Simpson-Holley, N. T. Wright, Z. Han, J. Paragas, and R. N. Harty. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77:1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 19.Mebatsion, T., M. Konig, and K. K. Conzelmann. 1996. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell 84:941-951. [DOI] [PubMed] [Google Scholar]
- 20.Noda, T., H. Sagara, E. Suzuki, A. Takada, H. Kida, and Y. Kawaoka. 2002. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 76:4855-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robison, C. S., and M. A. Whitt. 2000. The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J. Virol. 74:2239-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheiffele, P., A. Rietveld, T. Wilk, and K. Simons. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038-2044. [DOI] [PubMed] [Google Scholar]
- 23.Schmitt, A. P., B. He, and R. A. Lamb. 1999. Involvement of the cytoplasmic domain of the hemagglutinin-neuraminidase protein in assembly of the paramyxovirus simian virus 5. J. Virol. 73:8703-8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitt, A. P., G. P. Leser, D. L. Waning, and R. A. Lamb. 2002. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J. Virol. 76:3952-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schnell, M. J., L. Buonocore, E. Boritz, H. P. Ghosh, R. Chernish, and J. K. Rose. 1998. Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 17:1289-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simmons, G., R. Wool-Lewis, F. Baribaud, R. Netter, and P. Bates. 2002. Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J. Virol. 76:2518-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Timmins, J., G. Schoehn, S. Ricard-Blum, S. Scianimanico, T. Vernet, R. W. Ruigrok, and W. Weissenhorn. 2003. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 326:493-502. [DOI] [PubMed] [Google Scholar]
- 28.Timmins, J., S. Scianimanico, G. Schoehn, and W. Weissenhorn. 2001. Vesicular release of ebola virus matrix protein VP40. Virology 283:1-6. [DOI] [PubMed] [Google Scholar]
- 29.Volchkov, V. E., V. A. Volchkova, W. Slenczka, H. D. Klenk, and H. Feldmann. 1998. Release of viral glycoproteins during Ebola virus infection. Virology 245:110-119. [DOI] [PubMed] [Google Scholar]
- 30.Yang, Z. Y., H. J. Duckers, N. J. Sullivan, A. Sanchez, E. G. Nabel, and G. J. Nabel. 2000. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 6:886-889. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]