Abstract
To investigate changes in cellular gene expression associated with malignant progression, we identified differentially expressed genes in a cottontail rabbit papillomavirus (CRPV) squamous carcinoma model employing New Zealand White rabbits. The technique of suppression subtractive cDNA hybridization was applied to pairs of mRNA isolates from CRPV-induced benign papillomas and carcinomas, with each pair derived from the same individual rabbit. The differential expression of 23 subtracted cDNAs was further confirmed by quantitative reverse transcription-PCR (RT-PCR) with additional biopsies. Eight papilloma-carcinoma pairs examined showed a constant upregulation of the transcripts for the multifunctional adaptor protein 14-3-3 ζ and the Y-box binding transcription factor YB-1, whereas transcripts for m-type calpain 2 and NB thymosin β, which are involved in cell motility and tissue invasion, as well as casein kinase 1 α, chaperonin, and annexin I, were found to be upregulated in the majority of the cases. RNA-RNA in situ hybridization and laser capture microdissection in combination with quantitative RT-PCR analysis verified the deregulated expression of the transcripts in the tumor cells. In contrast, CRPV E7 transcript levels remained rather constant indicating no requirement for a further upregulation of E7 expression following tumor induction. Small interfering RNA-mediated interference with expression of genes encoding YB-1, m-type calpain 2, or NB thymosin β in a CRPV-positive cell line established from New Zealand White rabbit keratinocytes resulted in decreased cell invasion in matrigel chamber assays.
While high-risk human papillomaviruses (HPVs) have been identified as necessary risk factors for the development of cervical cancer (7), the role of HPV in skin cancer, which is the most common cancer in Caucasians, is still undefined (3, 23). A first link between papillomaviruses and skin cancer was obtained in 1935 from an animal model system, when Rous and Beard (40) described the development of squamous cell carcinomas (SCCs) in rabbits after experimental infection with cottontail rabbit papillomavirus (CRPV). Initial benign warts develop into invasively growing SCCs without any further cocarcinogens within 6 to12 months in the vast majority of the animals (50). A similar situation has been described for humans, where in the rare genetic disorder epidermodysplasia verruciformis, patients develop flat HPV-induced warts which progress in up to 60% of the cases into mostly primary SCCs (36, 37). Skin cancer has also been recognized as a common side effect in long-term-immunosuppressed patients, who have a 65-fold-increased risk for this disease. Shortly after the onset of immunosuppressive therapy, HPV-induced warts develop, and after 20 years of immunosuppression, nonmelanocytic skin cancer can be found in up to 70% of the patients (8). As in the case of epidermodysplasia verruciformis patients, the lesions seem to progress from warts through dysplastic lesions into SCC (4, 16, 43). Although HPV-associated SCCs have been found in 65% (43) or 81% (4) of the cases, the mechanisms leading to the development of skin cancer and especially the role of papillomaviruses are not yet fully understood.
Thus far, the only model system for studying the progression of papillomavirus-induced skin tumors is the domestic rabbit infected with CRPV. Infection of domestic rabbits with CRPV particles or viral DNA leads to the development of local tumors within 3 to 6 weeks postinfection. These papillomas progress within 6 to 12 months in more than 80% of the cases into invasively growing and finally metastasizing carcinomas (45; S. Jeckel, unpublished data). Only in very few individual rabbits do papillomas persist in the benign state, and even fewer regress. This progressive behavior does not depend on any other known cofactor besides the viral infection, which makes CRPV an important model for papillomavirus-associated skin disorders in order to study the role of viral and cellular gene expression during cancer development.
Earlier studies using more descriptive methods, such as RNA-RNA in situ hybridization, showed no pronounced qualitative or quantitative changes in viral gene expression during malignant progression (50). The primary target cells localized in the bulge region of hair follicles already revealed rather high expression of E6 and E7 mRNAs (42), which remains a constant feature from papillomas through carcinomas to lung metastases (50, 52). Therefore it seems most likely that alterations in cellular gene expression further influence the malignant progression of papillomavirus-induced tumors.
In order to detect either progression-inhibiting or progression-promoting functions regarding carcinogenesis, we searched for genes which are differentially expressed in CRPV-induced papillomas versus carcinomas of domestic rabbits. The method of suppression subtractive cDNA hybridization (18) was used for a first screen, genes identified were subsequently quantified by real-time reverse transcription-PCR (RT-PCR), and their expression in tumor cells was confirmed by RNA-RNA in situ hybridization and laser capture microdissection-based quantitative RT-PCR analysis. Here we show that the malignant progression of CRPV-induced tumors is accompanied by an upregulation of genes that are directly involved in cell motility and tissue invasion. Some of these genes have already been described to be potential prognostic markers for other cancers and are regulated by or contribute to the extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) pathway. By transfection of small interfering RNAs (siRNAs) specific for the most prominent upregulated genes, such as those for the Y-box binding protein YB-1, calpain2, or thymosin β, in a CRPV-positive New Zealand White rabbit keratinocyte cell line, we observed decreased transmigration of the cells through matrigel.
MATERIALS AND METHODS
Specimens.
New Zealand White rabbits were inoculated with CRPV (6 × 106 CRPV particles per infection site) with a pedo jet injector (Ventron Medical Products) as described previously (9). For rabbits 1, 2, and 3, biopsies were taken at 13 months postinfection. Rabbits 4, 5, 6, and 7 were inoculated with 6 × 106 CRPV particles per infection site by use of a tattoo machine, and biopsies were taken at 3 months (for papillomas) and 13 months (for carcinomas) postinfection. Papillomas as well as carcinomas were taken from seven individual rabbits. All tumors developed without any further treatment. The biopsies were snap frozen after surgical excision and stored at −70°C until required. All biopsy samples used for the study were classified and reviewed for histopathological diagnosis by two pathology experts.
RNA extraction and mRNA isolation.
Total RNA was extracted from biopsies by using Trizol reagent (Invitrogen, Karlsruhe, Germany). Prior to reverse transcription, RNA was treated with RQ1 RNase-free DNase (Promega, Mannheim, Germany) or DNase I (amplification grade; Invitrogen). An mRNA separator kit (Clontech, Heidelberg, Germany) was used to isolate mRNA from total RNA according to the manufacturer's protocols.
Suppression subtractive cDNA hybridization.
Isolation of differentially expressed genes was performed by the suppression subtractive cDNA hybridization technique (18) with a PCR-Select cDNA subtraction kit (Clontech) and 1 μg of mRNA per biopsy. The resulting cDNA fragments were cloned into the pT-Adv vector (AdvanTAge PCR cloning kit; Clontech).
DNA sequencing and database searching.
DNA sequencing of the cloned cDNA fragments was carried out by using the M13 reverse primer GAAACAGCTATGACCATG, the T7 promoter primer GTAATACGACTCACTATAGGG, and the BigDye terminator cycle sequencing kit (Applied Biosystems, Warrington, United Kingdom) with an ABI PRISM 310 Genetic Analyzer (Applied Biosystems). Sequences were compared to sequences in GenBank of the National Center for Biology Information (NCBI) by using the programs BLASTN and TBLASTX.
Recombinant plasmids.
For generating the standard curve to quantify transcripts in biopsy samples by quantitative RT-PCR, plasmid standards were constructed. To obtain cDNAs specific for each transcript, ready-to-use RT-PCR beads (Amersham Pharmacia Biotech Inc.) were used with 500 ng of total rabbit RNA and 0.3 μM gene-specific primers (Table 1) in a conventional RT-PCR under the following conditions: reverse transcription at 42°C for 30 min; denaturation at 95°C for 10 min; and 45 cycles of denaturation at 95°C for 20 s, annealing at 55°C for 20 s, and extension at 72°C for 40 s. PCR products were cloned into pCR2.1-TOPO (Invitrogen). The identities of the cDNAs were verified by sequencing.
TABLE 1.
Oligonucleotide sequences
| Use | Name | Sequence
|
GenBank accession no. | Position
|
||
|---|---|---|---|---|---|---|
| Forward | Reverse | Forward | Reverse | |||
| Quantitative RT-PCR | rh 14-3-3 | CTGCAACGATGTACTGTCTC | CCTCCTTCTCCTGCTTCAGCT | D87660.1 | 414 | 864 |
| Annexin I | ACGAAGACTTGGCTGATACGG | CCTTTGGTTTCATCCAGGATG | U24656.1 | 617 | 1031 | |
| β-Actin | CATTGCCGACAGGATCCAGA | GACTCGTCGTACTCCTGCTTG | BC014861.1 | 998 | 1148 | |
| Calgranulin C | GAAGCTGATCACCACGGAAC | GCGCTTGCTACTCTTTGTGG | AF091848.1 | 69 | 253 | |
| rh calmodulin | GATGACACGCAAAGTGAAGACC | AGGCATAACCCAGATGTTCC | U94728.1 | 2109 | 2405 | |
| rh calpain 2 | CCTCGGCAAAAACTTCTTCCT | CCGGAATCCATCGTCAATGT | M13797 | 507 | 772 | |
| Casein kinase 1 α | TGGCCATCAACATCACCAA | TGACGACCAATACCCATTAGGA | U59166.1 | 478 | 826 | |
| rh chaperonin | GCAAAAGTAGCAACAGCCCA | TGGCCAAAATGGAGTCCAC | AF068483.1 | 232 | 536 | |
| rh connexin 30 | CGCGTCCCACCCGAAGAGTATC | CGTGGACCAGCTGACCTAC | from cDNA clone from SSHa | |||
| rh destrin | GGCCAAATGCCTGTTTTGATG | CAATTCCAAGATGCAAGGTG | NM_006870.2 | 776 | 1231 | |
| rh hnRNP for JKTBP | ATAGTGCCTATGGTGGTGATC | CCCTCGAGATGCCTTGCCAT | from cDNA clone from SSH | |||
| rh ileal sodium-dependent bile acid ransporter | GCCAGTGCGCCGTGCTGATCT | CAATGATGGCAGGCGCCATGGC | from cDNA clone from SSH | |||
| mRNP particle major protein p50/YB-1 | CGTCGACCACAGTATTCCAAC | GTCAACGGGCAAAAAGCAAGC | U16821.1 | 732 | 1232 | |
| rh NB thymosin β | CGAAGGAAACTATCCAGCAGG | CAGGCCTAAAGCCAAGACTC | D82345.1 | 189 | 297 | |
| rh PA-FABP | GCACCTTGGGAGAGAAGTTTG | CCTTCTCATAGACCCGAGTAC | M94856.1 | 243 | 444 | |
| rh ribosomal protein L27 | CAGATTCTTGGACGGGTGGT | GGAATCATGACACGGGAGGT | from cDNA clone from SSH | |||
| Quantitative real-time PCR | E7 | AGAAAGGCCCTATGCAGTGTC | AGGGAAAGCGATGCGGATA | NC_001541.1 | 1113 | 1334 |
| Hybridization probe CRPV-FL | CTATCAGCTTCGTCTGCGTCTGTGC-FL | NC_001541.1 | 1259 | |||
| Hybridization probe CRPV-LC | LC-Red640-CCAGAAGCCATAAGAACCTTGAATCG | NC_001541.1 | 1285 | |||
| α-Tubulin | CTGGAGCACTCTGATTGTGC | CTGTGGTGTTGCTCAGCATG | BC056169 | 642 | 1206 | |
| Target sequence for siRNA generation | mRNP particle major protein p50/YB-1 | GAAGGTCATCGCAACGAAG | U16821.1 | |||
| rh calpain 2 | GAACTAAGGTTCCACGGAC | M13797 | ||||
| rh NB thymosin β | GAGTCTTGGCTTTAGGCCT | D82345.1 | ||||
SSH, suppression-subtractive DNA hybridization.
The siRNA expression vector (pSUPER) for generating gene-specific siRNAs has been described previously (10). The constructs pSUPER-p50, pSUPER-calpain 2, pSUPER-thymosin β, and pSUPER-luc were derived by single-strand annealing of the oligonucleotides (12.5 μg each) of the target sequences (for sequences, see Table 1) and subsequent cloning of the double-stranded oligonucleotides into the pSUPER-vector between the BglII and SalI restriction sites or the BglII and HindIII restriction sites (for pSUPER-luc). Unique target sequences for mRNP particle major protein p50/YB-1, the rabbit homologue of calpain 2 (rh calpain 2), and rh NB thymosin β were identified by siRNA finder (Ambion). The target sequence for the luciferase gene has been described previously (20). Sequences were verified by DNA sequencing.
Quantitative RT-PCR.
Prior to quantitative PCR, DNase I-treated total RNA was reverse transcribed by using the TaqMan reverse transcription reagent kit (Applied Biosystems) and oligo(dT) primers for 1 h at 42°C. Each quantitative PCR mixture included 50 ng of cDNA and 0.4 μM sequence-specific primers (for sequences, see Table 1), with a final MgCl2 concentration of 4 mM. PCR was carried out either with the LightCycler-Fast-Start Master SYBR Green I kit (Roche, Mannheim, Germany) in the LightCycler instrument (Roche) or with the SYBR Green PCR core reagents kit (Applied Biosystems) in the 5700 sequence detection system (Applied Biosystems); the two systems showed identical results. In the LightCycler the reaction mix was denatured at 95°C for 10 min and amplified with 45 cycles of denaturation at 95°C for 10 s, annealing at 52°C for 10 s, and extension at 72°C for 20 s. When the 5700 sequence detection system was used, the reaction mixture was denatured at 95°C for 5 min and amplified with 45 cycles of denaturation at 95°C for 20 s, annealing at 52°C for 20 s, and extension at 72°C for 40 s. In order to quantify the absolute amount of transcripts in the biopsy samples, standard curves were generated with 10-fold dilutions of plasmids containing cloned cDNA fragments. Following quantitative PCR amplification, melting curves were determined for the amplicons of the plasmid standards and the biopsy samples. By comparison of the melting points of the amplicons of the biopsy samples and the plasmid standards, the specificity of the amplicons could be verified. Absolute transcript numbers of the genes were normalized to β-actin transcripts determined for each cDNA.
DNA extraction and real-time PCR.
DNA from biopsies was isolated by proteinase K digestion in 0.1 M EDTA (pH 8.0)-0.05 M Tris (pH 8.0)-0.5% sodium dodecyl sulfate followed by phenol-chloroform extraction and ethanol precipitation. CRPV copy numbers were determined by quantitative real-time PCR from 30 ng of total DNA with 0.25 μM sequence-specific fluorescently labeled probes and 0.5 μM sequence-specific primers (Table 1) derived from the CRPV E7 gene with a final MgCl2 concentration of 3 mM. PCR was carried out with the LightCycler system under following conditions: denaturation at 95°C for 10 min and then amplification with 40 cycles of denaturation at 95°C for 10 s, annealing at 55°C for 10 s, and extension at 72°C for 20 sec. CRPV copy numbers were normalized to the cell numbers obtained by measuring α-tubulin copies in 30 ng of DNA with the LightCycler system.
Laser capture microdissection.
Serial 5.0-μm cryostat sections were mounted on uncoated glass slides and stored at −70°C. Immediately before dissection, sections were fixed with 95 and 70% ethanol (15 s each), stained with hematoxylin for 30 s, rinsed in deionized water, dehydrated for 15 s in 70% ethanol, stained with eosin for 5 s, and dehydrated for 15 s in 95% ethanol followed by 1 min in xylene. The sections were air dried and microdissected with a PixCell II laser capture microdissection system (Arcturus Engineering, Mountain View, Calif.). Approximately 100,000 of epidermal papilloma and carcinoma cells were microdissected and stored on microdissection caps at −70°C until required. Total RNA was isolated by using Trizol reagent (Invitrogen). Four caps with papilloma cells and four caps with carcinoma cells were pooled for isolation of total RNA.
Construction of probes for in situ hybridization analysis.
To construct in vitro transcription vectors for the synthesis of mRNA-specific riboprobes, the RT-PCR products obtained as described above were cloned into in vitro transcription vector pSP72 (Promega). The correct orientation of the inserts was confirmed by DNA sequence analysis. All clones were linearized by restriction digestion prior to in vitro transcription. In vitro RNA synthesis (sense and antisense orientations) (Riboprobe Combination System SP6/T7; Promega) in the presence of 35S-UTP (50 μCi of a 400-mCi/mmol preparation) and the appropriate RNA polymerase was followed by alkaline hydrolysis to reduce the probe length to approximately 300 nucleotides (15). The quality of the RNAs was controlled by polyacrylamide gel electrophoresis and autoradiography. The probes were applied at a concentration of 5 × 105 cpm per section.
RNA-RNA in situ hybridization.
For RNA-RNA in situ hybridization, serial cryostat sections of biopsy material mounted on aminopropylsilane-coated slides were fixed in 4% paraformaldehyde in phosphate-buffered saline and then dehydrated through graded ethanol solutions. Dried sections were then acetylated in 0.1 M triethanolamine-0.25% acetic anhydride for 10 min, washed in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and preincubated for 2 h at 42°C covered by solution I, which contained 45% formamide, 0.6 M NaCl, 2.5× Denhardt's solution, 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 0.1% (vol/vol) sodium dodecyl sulfate, and 0.15 mg of tRNA per ml. After removal of the prehybridization solution, the tissue sections were incubated for 16 h at 42°C with 35S-labeled antisense riboprobes (5 × 105 cpm) in solution I containing 10% (wt/vol) dextran sulfate. The incubated slides were washed in 50% formamide-1× SSC at 55°C, treated with RNase A (10 μg/ml), washed five times for 20 min each in 0.1× SSC at 55°C, dehydrated, and finally coated with Kodak NTB-2 emulsion. All slides from one experiment were developed after the same exposure time (10 days) to allow the direct comparison of signal strengths obtained with different probes. Hematoxylin-and-eosin-counterstained sections were evaluated and photographed with a Zeiss Axiovert 200 microscope with a dark- or bright-field condenser. The level of background signals for RNA hybridization was determined by using the corresponding sense riboprobes.
Cell culture.
Primary keratinocytes from New Zealand White rabbits were isolated from rabbit skin epithelium as described previously for the isolation of human keratinocytes from human foreskin (41). Primary cell cultures were maintained in supplemented keratinocyte serum-free medium (KSFM) (Invitrogen). To obtain CRPV-containing keratinocytes, cells were grown to 70% confluency on a 60-mm-diameter dish and infected with CRPV by incubation with 4.8 × 106 CRPV particles diluted in 2.5 ml of KSFM for 16 h. Virus was removed the next day, and cells were further maintained in KSFM. At passage 18, the presence of integrated CRPV DNA was confirmed by Southern blot analysis (data not shown). The cell line was designated AVS.
Transient matrigel invasion assay.
Approximately 5 × 105 AVS cells were seeded into 60-mm-diameter dishes. The next day cells were transfected in the presence of 5 μl of Lipofectamine (Invitrogen) with 750 ng of siRNA expression vector. Matrigel invasion assays were carried out 72 h after transfection in triplicates with Biocoat matrigel invasion chambers (Becton Dickinson Labware, Bedford, Mass.). Prior to use, the upper and lower compartments were filled with 0.5 ml of prewarmed KSFM for rehydrating the matrigel layer and incubated for 2 h at 37°C. The medium in the lower compartment was replaced by 0.5 ml of conditioned KSFM harvested from AVS cells after 24 h of incubation and supplemented with 10% fetal bovine serum (25, 32, 46). The medium in the upper compartment was replaced by 1.5 × 105 transfected cells in 0.5 ml of KSFM. After 48 h, the medium in both chambers was replaced by KSFM containing 0.5 mg of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) per ml, which was left for 2 h. After removal of the MTT-containing medium, the upper surface of the invasion chamber insert membrane was scrubbed five times in each direction with a cotton swab, and the membrane was removed from the invasion chamber insert with a scalpel. The cotton swab and the membrane were each placed in a tube containing 1 ml of dimethyl sulfoxide (DMSO), and the optical density was measured at 595 nm. The extinction of the cotton swab containing DMSO is a direct indicator of the number of cells which remained in the upper chamber (a), and the extinction of the membrane containing DMSO is a direct indicator of the number of cells which transmigrated through the matrigel and the membrane (b), as previously described (44). Percent invasion is expressed as the extinction b divided by the total extinction a + b.
RESULTS
Identification of differentially expressed genes by suppression subtractive cDNA hybridization.
To exclude differences in gene expression resulting from comparing different rabbit individuals, we used histologically verified papilloma-carcinoma pairs taken simultaneously at 13 months postinfection from the same rabbits, using a total of two rabbits (rabbits 1 and 2) for a first screen. Total mRNA was isolated, and differentially expressed transcripts in papillomas versus carcinomas were identified by the technique of suppression subtractive cDNA hybridization (18). This technique is based on the selective amplification of cDNAs which are derived from transcripts abundant in one of two tissues compared (the tester) and the suppression of amplification of transcripts which are present in equal amounts in the two tumor types. To identify candidates for papilloma-associated genes, cDNA from papillomas was used as a tester. Vice versa, to identify candidates for carcinoma-associated genes we used cDNA from carcinomas as a tester. The resulting 92 cDNAs were sequenced, and nucleic acid homology searches using GenBank of NCBI revealed that 49 genes were unique. Thirteen of the 49 unique genes shared more than 85% homology to known rabbit genes; 23 other cDNAs showed homology only to human, mouse, rat, or dog genes and are therefore considered to be rabbit homologues (rh's) of these genes (Table 2); and the remaining 13 cDNAs showed no homology to any sequences in the NCBI GenBank. We performed the following analysis with 15 candidate genes. Negative selection criteria for known genes were involvement in cell metabolism or being a pseudogene.
TABLE 2.
Database information of the cDNA clones examined
| Accession no. | Best matches in NCBI databasea | % Identity | Transcript increaseb |
|---|---|---|---|
| AF091848.1 | Oryctolagus cuniculuscalgranulin C mRNA, partial cds | 100 | 1 |
| NM_005621.1 | Homo sapiens S100 calcium binding protein A12 (calgranulin C) (S100A12) mRNA | 80 | |
| U16821.1 | O. cuniculusmRNP particle major protein p50 mRNA, complete cds | 98 | 3 |
| J03827.1 | YB-1 mRNA | 95 | 3 |
| L37516.1 | H. sapiens DNA binding protein B pseudogene (PSDBPB1) gene, complete cds | 95 | |
| U09494.1 | Papillomavirus sylvilagiWashington B (E7) gene, complete cds | 97 | 1 |
| M13797.1 | O. cuniculuscalcium-dependent protease large subunit m-type mRNA | 97 | 3 |
| M23254.1 | H. sapiens calpain 2 (m/II) large subunit (CAPN2) mRNA | 85 | |
| NM_001748.3 | Human Ca2-activated neutral protease large subunit (CANP) mRNA, complete cds | 85 | |
| U59166.1 | O. cuniculus casein kinase 1 alpha mRNA, complete cds | 95 | 3 |
| D89092.1 | H. sapienshnRNP JKTBP mRNA, complete cds | 95 | 2 |
| D89678.1 | H. sapiens mRNA for A + U-rich element RNA binding factor, complete cds | 95 | |
| U24656.1 | O. cuniculusannexin I gene, complete cds | 94 | 2 |
| X05908.1 | H. sapiens mRNA for lipocortin | 92 | |
| U94728.1 | H. sapienscalmodulin (CALM2) gene, exons 3 to 6, and complete cds | 94 | 2 |
| AJ388516.1 | C. familiarismRNA for ribosomal protein L27 | 94 | 2 |
| D87660.1 | M. musculusmRNA for 14-3-3 ζ, complete cds | 91 | 3 |
| D78647.1 | M. musculus mRNA for phospholipase A2, complete cds | 91 | |
| M94856.1 | H. sapiensfatty acid binding protein homologue (PA-FABP) mRNA, complete cds | 89 | 2 |
| X74801.1 | H. sapiensCctg mRNA for chaperonin | 89 | 2 |
| NM_006870 | H. sapiens destrin (actin-depolymerizing factor) (DSTN) mRNA | 87 | 2 |
| AJ005585.1 | H. sapiensCx30 gene | 86 | 1 |
| D82345.1 | H. sapiensmRNA for NB thymosin β, complete cds | 85 | 3 |
Boldface indicates the best match; other entries are the second or third best matches found for the same cDNA sequences in the NCBI database.
The numbers indicate the increase of the transcripts in the carcinomas of the first three papilloma-carcinoma pairs examined. 3, increase in three of three carcinomas; 2, increase in two of three carcinomas and no change or decrease in the other carcinoma; 1, increase in only one of three carcinomas and no change or decrease in the other carcinomas. cds, coding sequence.
Verification of the differential expression by quantitative RT-PCR.
In order to verify and quantify the differential expression of the transcripts identified by suppression subtractive cDNA hybridization, their expression levels in the papilloma-carcinoma pairs of rabbits 1 and 2 and in another histologically verified papilloma-carcinoma pair of a third rabbit was examined by quantitative RT-PCR. First, total RNA obtained from all biopsies was DNase I treated and reverse transcribed by using oligo(dT) priming. Each subsequent quantitative PCR was carried out with 50 ng of cDNA and SYBR Green and the primers listed in Table 1. To obtain the absolute transcript numbers of the genes examined, a standard curve with 10-fold dilutions of target DNA was generated in parallel to each PCR as described in Materials and Methods. The specificity of the amplicons obtained during each PCR with cDNAs of the biopsy samples was verified by comparison of their melting points with the melting points of amplicons obtained with target DNAs of the plasmid standards. The absolute transcript numbers of each gene examined were normalized to β-actin transcript numbers determined for the same cDNA. The quotient of the relative transcript numbers determined for each papilloma-carcinoma pair gives the x-fold transcript increase or decrease for all investigated genes in the carcinomas. Interestingly, we could not in any case verify a differential overexpression of certain genes in papillomas relative to carcinomas. In contrast, the transcripts of 5 of the 15 genes examined proved to be increased (as defined by a ≥2-fold difference) in all three carcinomas: the rh to 14-3-3 ζ, the Ca2+-dependent m-type protease (rh to human calpain 2), casein kinase 1 α, mRNP particle major protein p50 (rh to the human transcription factor YB-1 (21), and rh NB thymosin β. The transcripts of seven genes examined proved to be increased in only two of three of the carcinomas, and three genes showed an increase of their transcripts in only one carcinoma. Transcripts of two genes with no homology to sequences in the NCBI GenBank showed an increase in two carcinomas. The other genes with no homology to sequences in the NCBI GenBank remained unchanged or showed an increase in transcript number in only one carcinoma and are therefore not included in Table 2.
In order to substantiate our findings for the genes that were upregulated in all three papilloma-carcinoma pairs, their transcript numbers were determined by quantitative RT-PCR with five additional histologically verified papilloma-carcinoma pairs, where papillomas and carcinomas were taken at 3 and 13 months postinfection, respectively. In addition we analyzed the transcript levels of rh chaperonin, annexin I, rh ribosomal protein L27, and rh PA-FABP, which were found to be upregulated in two of three carcinomas in the first screen, as well as the transcript for CRPV E7, which was found to have increased numbers in only one carcinoma. This analysis confirmed, in general, an increase of the transcripts for rh 14-3-3 ζ and mRNP particle major protein p50/YB-1 in all carcinomas, while transcripts for rh calpain 2 and casein kinase 1 α were found to be increased in seven of eight carcinomas, transcripts for rh chaperonin were found to be increased in six of eight carcinomas, and transcripts for rh NB thymosin β, annexin I, and rh ribosomal protein L27 were found to be increased in only five of eight carcinomas (Table 3). In contrast, the transcripts for CRPV E7 were only moderately increased in two carcinomas but remained unchanged or decreased in six carcinomas. When calculating the mean relative expression changes of all papilloma-carcinoma pairs, the most prominent increases were found for the transcripts for mRNP particle major protein p50/YB-1 (4.4-fold), rh calpain 2 (5.5-fold), and rh 14-3-3 ζ (3.8-fold).
TABLE 3.
Summary of differential expression of selected genes in CRPV-induced papillomas versus carcinomas determined by real-time RT-PCR
| Carcinoma | Fold change in regulation in carcinomaa
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rh 14-3-3 ζ | mRNP particle major protein p50/YB-1 | rh calpain 2 | Casein kinase 1 α | rh chaperonin | Annexin I | rh NB thymosin β | rh ribosomal protein L27 | rh PA-FABP | CRPV E7 | |
| 1 | +2 | +3 | +2 | +3 | +4 | 1 | +2 | +5 | +3 | +2 |
| 2 | +6 | +23 | +16 | +4 | +5 | +3 | +6 | +2 | +14 | 1 |
| 3 | +5 | +16 | +2 | +2 | −2 | +5 | +5 | 1 | 1 | −2 |
| 4 | +7 | +5 | +9 | +3 | 1 | −3 | 1 | −3 | −174 | −2 |
| 5 | +3 | +2 | +11 | 1 | +6 | +4 | −3 | +2 | 1 | +3 |
| 6 | +3 | +3 | 1 | +3 | +3 | +3 | +6 | +4 | +36 | −2 |
| 7 | +2 | +2 | +2 | +3 | +2 | 1 | 1 | 1 | −16 | 1 |
| 8 | +2 | +2 | +2 | +3 | +3 | +4 | +3 | +3 | +2 | 1 |
| Avg | +3.8 | +4.4 | +5.5 | +2.6 | +2.6 | +2 | +2.4 | +1.6 | NAb | NA |
Positive and negative values indicate up- and downregulation, respectively, in the carcinoma.
Na, not applicable.
Verification of the differential expression in tumor cells by RNA-RNA in situ hybridization and laser capture microdissection-based quantitative RT-PCR.
In order to confirm that transcripts found to be differentially expressed by quantitative RT-PCR are indeed localized within the tumor cells, RNA-RNA in situ hybridization analyses were performed for selected transcripts with frozen sections of the papilloma-carcinoma pairs of rabbits 1, 2, and 3. The results obtained with antisense RNA probes for CRPV E7, rh 14-3-3 ζ, and annexin I (data not shown) and for rh NB thymosin β, mRNP p50 particle major protein/YB-1, and casein kinase 1 α (Fig. 1, 2, and 3) indicated expression of the transcripts exclusively in epithelial tissue in all biopsies, with the exception of rh NB thymosin β, which resulted, in addition, in some spotted signals also in the connective tissue (Fig. 1B and 2B). Antisense RNA probes for rh calpain 2 did not produce any signal in the in situ hybridization analysis, most probably because of the low transcript numbers observed by quantitative RT-PCR analysis (ranging from 0.15 to 2.5 transcripts per β-actin transcript, compared to, e.g., 4.8 to 110 mRNP particle major protein p50/YB-1 transcripts per β-actin transcript in the papilloma-carcinoma pairs investigated). CRPV E7, rh NB thymosin β, the particle major protein p50/YB-1, and rh 14-3-3 ζ were expressed mainly in the basal layers of the papillomas, whereas annexin I and casein kinase 1 α were expressed throughout the epithelium. When comparing papillomas with carcinomas, an increase of transcripts for thymosin β and p50/YB-1 was observed, which was especially highly pronounced in the invading part of the carcinoma tissue of rabbit 2 (Fig. 2 and 3). In contrast, the transcript for casein kinase 1 α showed differences in expression levels in only one carcinoma (Fig. 3) compared to the papilloma shown in Fig. 1.
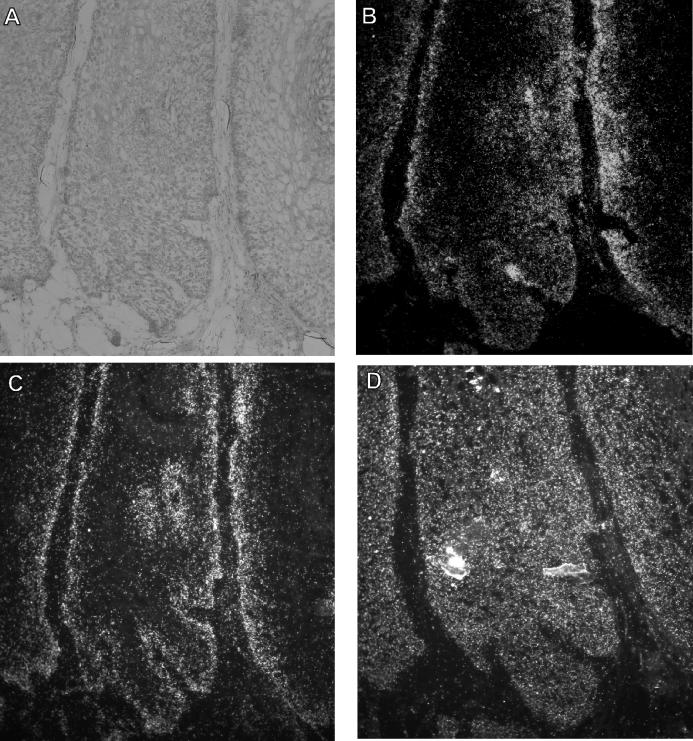
FIG. 1.
RNA-RNA in situ hybridization of a CRPV-induced papilloma. (B to D) Dark-field illumination of serial tissue sections, from a specimen taken 13 months after infection from rabbit 1, that were incubated with riboprobes specific for rh NB thymosin β (B), mRNP particle major protein p50/YB-1 (C), and casein kinase 1 α (D). (A) Corresponding histopathology of the papilloma shown by bright-field illumination after staining with hematoxylin and eosin. The silver grains generated in the film emulsion after exposure to the 35S-labeled probes are visible as white grains under the dark-field illumination.
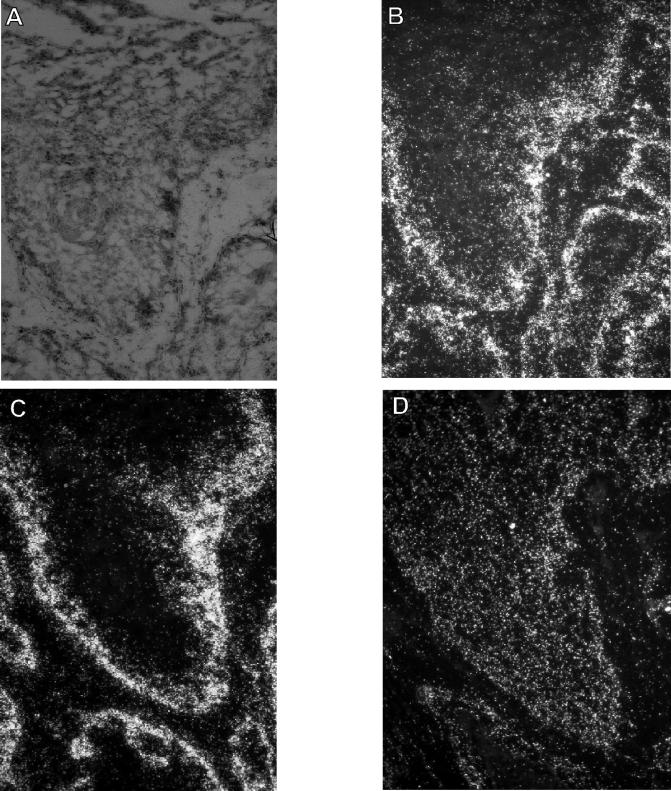
FIG. 2.
RNA-RNA in situ hybridization of a well-differentiated CRPV-induced carcinoma. (B to D) Dark-field illumination of serial tissue sections, from a specimen taken 13 months after infection from rabbit 1, that were incubated with riboprobes specific for rh NB thymosin β (B), mRNP particle major protein p50/YB-1 (C), and casein kinase 1 α (D). (A) Corresponding histopathology of the carcinoma shown by bright-field illumination after staining with hematoxylin and eosin. The silver grains generated in the film emulsion after exposure to the 35S-labeled probes are visible as white grains under the dark-field illumination.
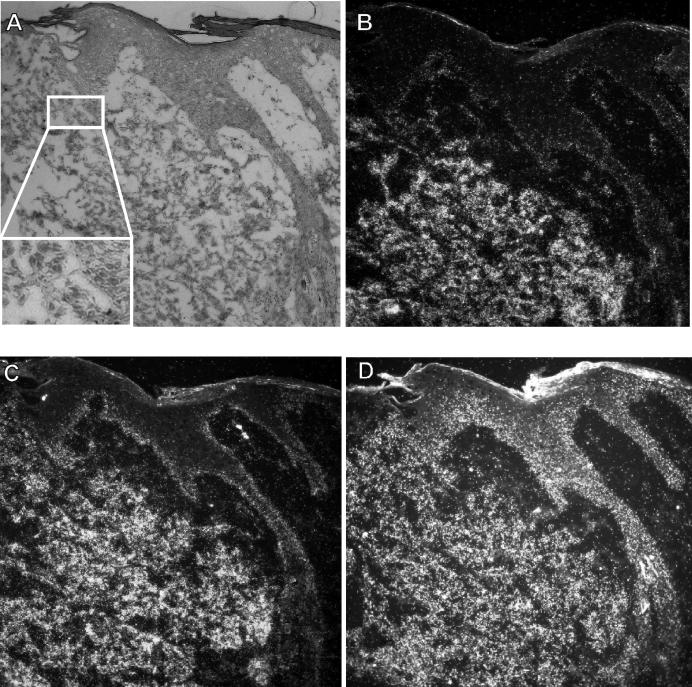
FIG. 3.
RNA-RNA in situ hybridization of the upper epithelium of a poorly differentiated CRPV-induced carcinoma. (B to D) Dark-field illumination of serial tissue sections, from a specimen taken 13 months after infection from rabbit 2, that were incubated with riboprobes specific for rh NB thymosin β (B), mRNP particle major protein p50/YB-1 (C), and casein kinase 1 α (D). (A) Corresponding histopathology of the carcinoma shown by bright-field illumination after staining with hematoxylin and eosin. The silver grains generated in the film emulsion after exposure to the 35S-labeled probes are visible as white grains under the dark-field illumination. The magnified insert in panel A shows that the loose tissue in the lower part of the panel consists of invasive tumor cells.
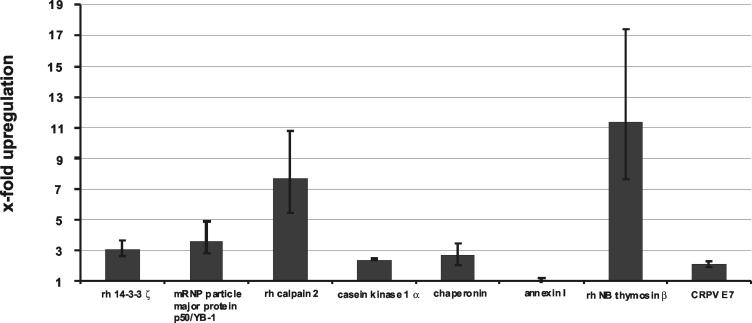
To obtain more quantitative evidence for the upregulation of the transcripts within the carcinoma cells, frozen sections of a papilloma-carcinoma pair were used for laser capture microdissection. For papilloma sections the basal layers to the spinous layers of the papilloma epithelium was dissected from the tissue, leaving connective tissue and highly differentiated papilloma epithelium, whereas for the carcinoma only the deeply invading part was taken in order to be able to compare gene expression in the less differentiated cells of the papilloma to that in the carcinoma cells. Total RNA was isolated from the dissected material, and determination of cDNA synthesis, quantitative PCR, and determination of transcript number were carried out as described above. Figure 4 summarizes the relative differences in gene expression in the papilloma-carcinoma pair, showing that the increase of all cellular transcripts of interest in the carcinoma could be verified, with the exception of annexin I. The transcripts for rh 14-3-3 ζ, rh calpain 2, casein kinase 1 α, mRNP particle major protein p50/YB-1, and rh chaperonin were increased in the carcinoma cells to roughly the same extent as observed when comparing undissected tissue of the different papilloma-carcinoma pairs. Transcripts of CRPV E7 and annexin I were not increased or were only moderately increased in the microdissected carcinoma cells. Interestingly, the transcripts of rh NB thymosin β showed a greater increase in the microdissected epidermis of the carcinoma than in total biopsy material, which could be due to the irregular spotted expression of thymosin observed also in the connective tissue as demonstrated by in situ hybridization analysis (Fig. 1B and 2B).
FIG. 4.
Determination of the relative differences in transcript numbers in microdissected tissue of a papilloma versus a carcinoma for rh 14-3-3 ζ, mRNP particle major protein p50/YB-1, rh calpain 2, casein kinase 1 α, rh chaperonin, annexin I, rh NB thymosin β, and CRPV E7. Absolute transcript numbers were determined by quantitative RT-PCR and normalized to the absolute transcript numbers of the housekeeping β-actin gene, which were determined for each cDNA. The quotient of the relative transcript numbers determined for the papilloma-carcinoma pair gives the x-fold increase of transcript numbers for all investigated genes in the carcinoma. At least two experiments were performed with each cDNA. Error bars indicate standard deviations.
Determination of viral load.
To investigate a possible correlation between viral genome copy number and the extent of cellular gene deregulation, the viral loads in the biopsies of rabbits 1, 2, and 3 were determined by real-time PCR. CRPV genome copy numbers were standardized for α-tubulin gene copy numbers. The results, given as copies per cell, do not support a correlation between increased transcription of specific cellular genes and viral copy number (Table 4).
TABLE 4.
Summary of viral load in biopsy material
| Rabbit | Viral loada (mean ± SD) in:
|
Relative difference in viral load | |
|---|---|---|---|
| Papilloma | Carcinoma | ||
| 1 | 5.7 ± 1.6 | 6.5 ± 1.3 | +1.1 |
| 2 | 4.8 ± 1.1 | 23.4 ± 4.9 | +4.8 |
| 3 | 12.8 ± 3.1 | 4.3 ± 1.2 | −2.9 |
Viral loads in papillomas and carcinomas are expressed as the number of CRPV genomes per cell as determined by real-time PCR.
Inhibition of cell invasion by gene silencing of mRNP particle major protein p50/YB-1, rh calpain 2, or rh NB thymosin β.
The human counterparts of mRNP particle major protein p50/YB-1, rh calpain 2, and rh NB thymosin β have already been shown to be involved in cell invasion in several other experimental systems (26, 31, 33). We therefore analyzed whether these rabbit genes also contribute to the invasion of CRPV-positive rabbit keratinocytes in tissue culture. Because primary epithelial rabbit cells cannot be used in the matrigel invasion assay, we generated a CRPV-positive rabbit cell line by infection of primary keratinocytes of a New Zealand White domestic rabbit with CRPV particles designated AVS. These cells showed an invasion rate of approximately 50% in matrigel invasion assays. To interfere with the expression of mRNP particle major protein p50/YB-1, rh calpain 2, and rh NB thymosin β in AVS cells, gene-specific siRNA expression vectors (pSUPER-p50, pSUPER-calpain 2, pSUPER-thymosin β) were constructed. In addition an siRNA expression vector directed against the luciferase gene (pSUPER-luc) was constructed, which specifically reduced luciferase expression in reporter assays (data not shown). AVS cells were transfected with 750 ng of either pSUPER-p50, pSUPER-calpain 2, or pSUPER-thymosin β or with the parental expression vector pSUPER. To control for nonspecific effects of biologically active siRNA, AVS cells were also transfected with pSUPER-luc. Transfected cells were subjected to a matrigel invasion assay at 72 h posttransfection. In addition, we extracted total RNA and determined the ratios of transcript levels of p50/YB-1, thymosin β, and calpain 2 to β-actin transcripts by quantitative real-time RT-PCR, which clearly demonstrated a reduction of transcript levels by the specific siRNA constructs, from 3-fold for thymosin β and calpain 2 to 20-fold in the case of p50/YB-1, in comparison to pSUPER or pSUPER-luc.
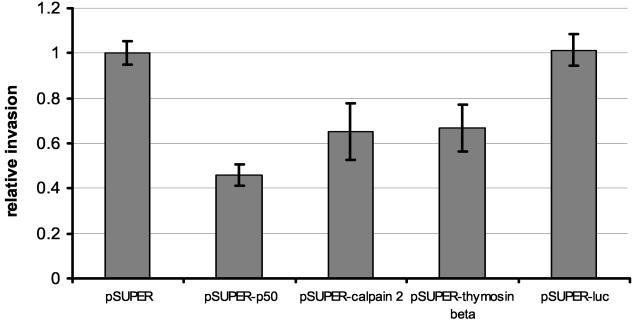
Cells transfected with pSUPER or pSUPER-luc showed a similar invasion rate of approximately 50%, indicating that expression of a biologically active siRNA per se does not influence the invasive behavior of AVS cells (Fig. 5). In contrast, expression of siRNA specific for mRNP particle major protein p50/YB-1 led to a reduction of cell invasion to 23% (Fig. 5), whereas expression of siRNA specific to rh calpain 2 or rh NB thymosin β reduced invasion to 33% (Fig. 5). Taken together, these results suggest that mRNP particle major protein p50/YB-1, rh calpain 2, and rh NB thymosin β contribute to the invasion of CRPV-positive cells in vitro and therefore are likely to play a role in progression in vivo.
FIG. 5.
Transfection of siRNA expression vectors specific for either mRNP particle major protein p50/YB-1, rh calpain 2, or rh NB thymosin β reduces cell invasion in matrigel invasion assays. A 750-ng amount of each siRNA expression vector containing target sequences for either mRNP particle major protein p50/YB-1, rh calpain 2, rh NB thymosin β, or the luciferase gene (pSUPER-p50, pSUPER-calpain 2, pSUPER-thymosin β, or pSUPER-luc, respectively) were transfected in the CRPV-positive rabbit keratinocyte cell line AVS. Transfected cells (105) were plated in the upper chamber of the transwell plate in serum-free medium. The lower chamber contained conditioned medium supplemented with 10% fetal bovine serum. After 48 h, living cells were stained with MTT. Cells collected from the upper or the lower chamber were lysed in DMSO separately, and the extinction was measured at 595 nm. Percent invasion is expressed as the extinction of cells that invaded through the membrane divided by the total extinction. The invasion of the transfected cells is expressed relative to the invasion of cells transfected with the parental pSUPER vector. Error bars indicate standard deviations.
DISCUSSION
To analyze the pathway of papillomavirus-induced carcinogenesis, a number of studies previously investigated the influence of different HPV types on the gene expression of the host cells in tissue culture. By using cDNA microarrays, downregulation of interferon-responsive genes and upregulation of proliferation-responsive genes by HPVs have been identified (13, 35). To study events associated with invasion processes in vivo, we employed the CRPV squamous carcinoma model with New Zealand White rabbits to search for genes involved in malignant progression. Pairs of CRPV-induced benign and malignant biopsies of New Zealand White rabbits were used for suppression subtractive cDNA hybridization and quantitative RT-PCR analysis. These experiments allowed to identify genes, such as those for rh YB-1 protein (21), rh 14-3-3 ζ protein, rh calpain 2, rh NB thymosin β, and rh casein kinase 1 α, that had increased transcript levels in the majority of the carcinomas investigated. Whether these increased transcript levels are due to enhanced transcription or increased mRNA stability cannot at present be differentiated. In contrast, CRPV E7 transcript levels remained rather constant, suggesting that a further increase in the levels of E7 is not necessary for steps following tumor induction, which is in line with earlier observations (52).
With the exception of 14-3-3 ζ, which is identical to phospholipase 1 α and has been described to be downregulated in HPV31-positive human keratinocytes, these genes have so far not been found to be influenced by papillomaviruses (13). However, YB-1, several isoforms of thymosin β, and also 14-3-3 ζ have been demonstrated to be upregulated in several kinds of human cancers (17, 24, 49, 51), suggesting that these genes are involved in the progression rather than the induction of tumors.
One of the most dramatically upregulated genes in carcinomas was the gene encoding the rh for the transcription factor YB-1 (21). Enhanced transcription of YB-1 was previously found to correlate with the outcome of breast cancer (24), which led to the proposal to use YB-1 expression levels as a prognostic marker for this malignant disease (24). More interestingly, it was found that in combination with the transcription factor AP-2 and p53, YB-1 acts to induce transcription and protein secretion of gelatinase A (MMP2) (33), which is a known factor contributing to tumor invasion.
MMP2 was also shown to be induced by the product of the thymosin β gene (6, 27), which is another of the genes found to be upregulated during the progression of CRPV-induced tumors. Several isoforms of the G-actin-sequestering protein thymosin β were found to be upregulated in various transformed cell types and carcinomas (2, 11), which is in line with our findings. Overexpression of thymosin β 15, which shares the highest sequence homology to the thymosin cDNA found to be overexpressed in rabbit carcinomas, correlates with the metastatic potential of mouse lung carcinomas, human breast carcinomas, and prostatic carcinomas and is therefore discussed as a prognostic marker to predict the metastatic potential of these tumors (1, 2). The importance of thymosin β for tumor progression has further been demonstrated by the introduction of antisense constructs of thymosin β in metastatic cell lines, which reduced their metastatic potential (2, 26).
Another gene found to be upregulated in CRPV-induced carcinomas is that for rh calpain 2. It was shown that the expression level of calpain 2 transcripts remains unchanged in healthy prostata tissue and prostata carcinomas. However, calpain 2 cleaves scaffold proteins of focal adhesion plaques, such as talin, paxillin, and the focal adhesion kinase, and also induces expression and secretion of MMP2 and -9, thereby contributing to increased cell motility (12, 14, 38, 39). Inhibition of calpain 2 by introduction of antisense constructs or by using specific calpain 2 inhibitors results in decreased cell invasion of various carcinoma cell lines (31, 38). Interestingly, calpain by itself can be activated by degradation products of collagen type I resulting from the activity of MMP2 (12). These findings are in line with our results that siRNA-mediated interference with gene expression of either mRNP particle major protein p50/YB-1, rh calpain 2, or rh NB thymosin β in a CRPV-positive cell line reduced the invasive potential in matrigel assays.
Both mouse thymosin β and calpain 2 can be regulated in their expression by the ERK/MAPK signal cascade (22, 29). Interestingly, the 14-3-3 ζ gene, whose rh was found to be constantly overexpressed in CRPV-induced carcinomas, is capable of binding and activating c-Raf, leading to the subsequent activation of the downstream effectors of the ERK/MAPK pathway (28, 34, 48). It is therefore conceivable that upregulation of 14-3-3 ζ may be responsible for the observed overexpression of thymosin β and calpain 2. Another intriguing ability with regard to enhanced cell motility is the capacity of 14-3-3 ζ to rearrange the actin cytoskeleton by direct interaction with the actin-dephosphorylating factor cofilin and its regulating kinase (5). In context with these results, the transcript for casein kinase 1 α, a serine/threonine kinase which is able to phosphorylate 14-3-3 ζ (19) and β-catenin (30) in vivo and in vitro, was also found to be upregulated in seven of eight CRPV-induced carcinomas investigated. At present, however, the functional consequence of the phosphorylation of 14-3-3 ζ is unknown (47).
In summary, this study identified a number of cellular genes involved in cell motility and invasion, some of which are already known from studies with humans to contribute to tumor invasion, that showed upregulated transcription during the progression of CRPV-induced tumors. Functional analysis by siRNA-mediated interference with the expression of the genes for YB-1, thymosin β, and calpain 2, which were the most upregulated in CRPV-induced carcinomas, revealed their direct role in tissue invasion.
Acknowledgments
We thank Felix O. Wettstein for providing the CRPV virus stock and biopsy material. We further thank Raffael Kurek for use of the laser capture microdissection system, Andreas Blum and Karl Sotlar for help with the histopathological diagnosis, Andreas Behren for help with the matrigel invasion assays, and R. Agami for providing pSUPER plasmid.
This work was supported by a grant (no. 99.111.1) from the Wilhelm Sander-Stiftung to T.I.
REFERENCES
- 1.Bao, L., M. Loda, and B. R. Zetter. 1998. Thymosin beta15 expression in tumor cell lines with varying metastatic potential. Clin. Exp. Metastasis 16:227-233. [DOI] [PubMed] [Google Scholar]
- 2.Bao, L., M. Loda, P. A. Janmey, R. Stewart, B. Anand-Apte, and B. R. Zetter. 1996. Thymosin beta 15: a novel regulator of tumor cell motility upregulated in metastatic prostate cancer. Nat. Med. 2:1322-1328. [DOI] [PubMed] [Google Scholar]
- 3.Berkhout, R. J., J. N. Bouwes Bavinck, and J. ter Schegget. 2000. Persistence of human papillomavirus DNA in benign and (pre)malignant skin lesions from renal transplant recipients. J. Clin. Microbiol. 38:2087-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout, R. J., L. M. Tieben, H. L. Smits, J. N. Bavinck, B. J. Vermeer, and J. ter Schegget. 1995. Nested PCR approach for detection and typing of epidermodysplasia verruciformis-associated human papillomavirus types in cutaneous cancers from renal transplant recipients. J. Clin. Microbiol. 33:690-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birkenfeld, J., H. Betz, and D. Roth. 2003. Identification of cofilin and LIM-domain-containing protein kinase 1 as novel interaction partners of14-3-3 zeta. Biochem J. 369:45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blain, E. J., D. J. Mason, and V. C. Duance. 2002. The effect of thymosin beta4 on articular cartilage chondrocyte matrix metalloproteinase expression. Biochem. Soc. Trans. 30:879-882. [DOI] [PubMed] [Google Scholar]
- 7.Bosch, F. X., A. Lorincz, N. Munoz, C. J. Meijer, and K. V. Shah. 2002. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 55:244-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouwes Bavinck, J. N., D. R. Hardie, A. Green, S. Cutmore, A. MacNaught, B. O'Sullivan, V. Siskind, F. J. Van Der Woude, and I. R. Hardie. 1996. The risk of skin cancer in renal transplant recipients in Queensland, Australia. A follow-up study. Transplantation 15:715-721. [DOI] [PubMed] [Google Scholar]
- 9.Brandsma, J. L., Z. H. Yang, S. W. Barthold, and E. A. Johnson. 1991. Use of a rapid, efficient inoculation method to induce papillomas by cottontail rabbit papillomavirus DNA shows that the E7 gene is required. Proc. Natl. Acad. Sci. USA 88:4816-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2:243-247. [DOI] [PubMed] [Google Scholar]
- 11.Califano, D., C. Monaco, G. Santelli, A. Giuliano, M. L. Veronese, M. T. Berlingieri, V. de Franciscis, N. Berger, F. Trapasso, M. Santoro, G. Viglietto, and A. Fusco. 1998. Thymosin beta-10 gene overexpression correlated with the highly malignant neoplastic phenotype of transformed thyroid cells in vivo and in vitro. Cancer Res. 58:823-828. [PubMed] [Google Scholar]
- 12.Carragher, N. O., B. Levkau, R. Ross, and E. W. Raines. 1999. Degraded collagen fragments promote rapid disassembly of smooth muscle focal adhesions that correlates with cleavage of pp125(FAK), paxillin, and talin. J. Cell Biol. 147:619-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, Y. E., and L. A. Laimins. 2000. Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J. Virol. 74:4174-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooray, P., Y. Yuan, S. M. Schoenwaelder, C. 8A. Mitchell, H. H. Salem, and S. P. Jackson. 1996. Focal adhesion kinase (pp125FAK) cleavage and regulation by calpain. Biochem. J. 318:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox, K. H., D. V. DeLeon, L. M. Angerer, and R. C. Angerer. 1984. Detection of mRNAs in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev. Biol. 101:485-502. [DOI] [PubMed] [Google Scholar]
- 16.de Jong-Tieben L. M., R. J. Berkhout, H. L. Smits, J. N. Bouwes Bavinck, B. J. Vermeer, F. J. van der Woude, and J. ter Schegget. 1995. High frequency of detection of epidermodysplasia verruciformis-associated human papillomavirus DNA in biopsies from malignant and premalignant skin lesions from renal transplant recipients. J. Investig. Dermatol. 105:367-371. [DOI] [PubMed] [Google Scholar]
- 17.Diamond, D. L., Y. Zhang, A. Gaiger, M. Smithgall, T. S. Vedvick, and D. Carter. 2003. Use of ProteinChip array surface enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF MS) to identify thymosin beta-4, a differentially secreted protein from lymphoblastoid cell lines. J. Am. Soc. Mass Spectrom. 14:760-765. [DOI] [PubMed] [Google Scholar]
- 18.Diatchenko, L., Y. F. Lau, A. P. Campbell, A. Chenchik, F. Moqadam, B. Huang, S. Lukyanov, K. Lukyanov, N. Gurskaya, E. D. Sverdlov, and P. D. Siebert. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubois, T., C. Rommel, S. Howell, U. Steinhussen, Y. Soneji, N. Morrice, K. Moelling, and A. Aitken. 1997. 14-3-3 is phosphorylated by casein kinase I on residue 233. Phosphorylation at this site in vivo regulates Raf/14-3-3 interaction. J. Biol. Chem. 272:28882-28888. [DOI] [PubMed] [Google Scholar]
- 20.Elbashir, S. M., J. Harborth, K. Weber, and T. Tuschl. 2002. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26:199-213. [DOI] [PubMed] [Google Scholar]
- 21.Evdokimova, V. M., C. L. Wei, A. S. Sitikov, P. N. Simonenko, O. A. Lazarev, K. S. Vasilenko, V. A. Ustinov, J. W. Hershey, and L. P. Ovchinnikov. 1995. The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. J. Biol. Chem. 270:3186-3192. [DOI] [PubMed] [Google Scholar]
- 22.Frame, M. C., V. J. Fincham, N. O. Carragher, and J. A. Wyke. 2002. v-Src's hold over actin and cell adhesions. Nat. Rev. Mol. Cell. Biol. 3:233-245. [DOI] [PubMed] [Google Scholar]
- 23.Harwood, C. A., T. Surentheran, J. M. McGregor, P. J. Spink, I. M. Leigh, J. Breuer, and C. M. Proby. 2000. Human papillomavirus infection and non-melanoma skin cancer in immunosuppressed and immunocompetent individuals. J. Med. Virol. 61:289-297. [DOI] [PubMed] [Google Scholar]
- 24.Janz, M., N. Harbeck, P. Dettmar, U. Berger, A. Schmidt, K. Jurchott, M. Schmitt, and H. D. Royer. 2002. Y-box factor YB-1 predicts drug resistance and patient outcome in breast cancer independent of clinically relevant tumor biologic factors HER2, uPA and PAI-1. Int. J. Cancer 97:278-282. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi, H., H. Ohi, M. Sugimura, H. Shinohara, T. Fujii, and T. Terao. 1992. Inhibition of in vitro ovarian cancer cell invasion by modulation of urokinase-type plasminogen activator and cathepsin B. Cancer Res. 52:3610-3614. [PubMed] [Google Scholar]
- 26.Kobayashi, T., F. Okada, N. Fujii, N. Tomita, S. Ito, H. Tazawa, T. Aoyama, S. K. Choi, T. Shibata, H. Fujita, and M. Hosokawa. 2002. Thymosin-beta4 regulates motility and metastasis of malignant mouse fibrosarcoma cells. Am. J. Pathol. 160:869-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Q. Y., P. L. Jones, R. P. Lafferty, D. Safer, and R. J. Levy. 2002. Thymosin beta4 regulation, expression and function in aortic valve interstitial cells. J. Heart Valve Dis. 11:726-735. [PubMed] [Google Scholar]
- 28.Li, W., E. M. Skoulakis, R. L. Davis, and N. Perrimon. 1997. The Drosophila 14-3-3 protein Leonardo enhances Torso signaling through D-Raf in a Ras 1-dependent manner. Development 124:4163-4171. [DOI] [PubMed] [Google Scholar]
- 29.Li, X., A. Zimmerman, N. G. Copeland, D. J. Gilbert, N. A. Jenkins, and H. L. Yin. 1995. The mouse thymosin beta 4 gene: structure, promoter identification, and chromosome localization. Genomics 32:388-394. [DOI] [PubMed] [Google Scholar]
- 30.Liu, C., Y. Li, M. Semenov, C. Han, G. H. Baeg, Y. Tan, Z. Zhang, X. Lin, and X. He. 2002. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108:837-847. [DOI] [PubMed] [Google Scholar]
- 31.Mamoune, A., J. H. Luo, D. A. Lauffenburger, and A. Wells. 2003. Calpain-2 as a target for limiting prostate cancer invasion. Cancer Res. 63:4632-4640. [PubMed] [Google Scholar]
- 32.Melchiori, A., A. Albini, J. M. Ray, and W. G. Stetler-Stevenson. 1992. Inhibition of tumor cell invasion by a highly conserved peptide sequence from the matrix metalloproteinase enzyme prosegment. Cancer Res. 52:2353-2356. [PubMed] [Google Scholar]
- 33.Mertens, P. R., K. Steinmann, M. A. Alfonso-Jaume, A. En-Nia, Y. Sun, and D. H. Lovett. 2002. Combinatorial interactions of p53, activating protein-2, and YB-1 with a single enhancer element regulate gelatinase A expression in neoplastic cells. J. Biol. Chem. 277:24875-24882. [DOI] [PubMed] [Google Scholar]
- 34.Muslin, A. J., J. W. Tanner, P. M. Allen, and A. S. Shaw. 1996. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84:889-897. [DOI] [PubMed] [Google Scholar]
- 35.Nees, M., J. M. Geoghegan, T. Hyman, S. Frank, L. Miller, and C. D. Woodworth. 2001. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-κB-responsive genes in cervical keratinocytes. J. Virol. 75:4283-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orth, G. 1987. The Papovaviridae, p. 199-243. In N. P. Salzman and P. M. Howley (ed.), The papillomaviruses. Plenum Press, New York, N.Y.
- 37.Pfister, H. 1992. Human papillomaviruses and skin cancer. Semin. Cancer Biol. 3:263-271. [PubMed] [Google Scholar]
- 38.Popp, O., M. Heidinger, L. Ruiz-Heinrich, C. Ries, M. Jochum, and S. Gil-Parrado. 2003. The calpastatin-derived calpain inhibitor CP1B reduces mRNA expression of matrix metalloproteinase-2 and -9 and invasion by leukemic THP-1 cells. Biol. Chem. 384:951-958. [DOI] [PubMed] [Google Scholar]
- 39.Postovit, L. M., P. Dutt, N. Dourdin, M. Park, P. A. Greer, C. H. Graham, and J. S Elce. 2002. Calpain is required for MMP-2 and u-PA expression in SV40 large T-antigen-immortalized cells. Biochem. Biophys. Res. Commun. 297:294-301. [DOI] [PubMed] [Google Scholar]
- 40.Rous, P., and J. W. Beard. 1935. The progression to carcinoma of virus-induced rabbit papillomas (Shope). J. Exp. Med. 62:523-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruesch, M. N., F. Stubenrauch, and L. A. Laimins. 1998. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J. Virol. 72:5016-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitt, A., A. Rochat, R. Zeltner, L. Borenstein, Y. Barrandon, F. O. Wettstein, and T. Iftner. 1996. The primary target cells of the high-risk cottontail rabbit papillomavirus colocalize with hair follicle stem cells. J. Virol. 70:1912-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shamanin, V., H. zur Hausen, D. Lavergne, C. M. Proby, I. M. Leigh, C. Neumann, H. Hamm, M. Goos, U. F. Haustein, E. G. Jung, G. Plewig, H. Wolff, and E. M. de Villiers. 1996. Human papillomavirus infections in nonmelanoma skin cancers from renal transplant recipients and nonimmunosuppressed patients. J. Natl. Cancer Inst. 88:802-811. [DOI] [PubMed] [Google Scholar]
- 44.Simon, C., M. Simon, G. Vucelic, M. J. Hicks, P. K. Plinkert, A. Koitschev, and H. P. Zenner. 2001. The p38 SAPK pathway regulates the expression of the MMP-9 collagenase via AP-1-dependent promoter activation. Exp. Cell Res. 271:344-355. [DOI] [PubMed] [Google Scholar]
- 45.Syverton, J. T. 1952. The pathogenesis of the rabbit papilloma-to-carcinoma sequence. Ann. N. Y. Acad. Sci. 54:1126-1140. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, E. W., S. Nakamura, T. B. Shima, A. Melchiori, G. R. Martin, S. Z. Salahuddin, R. C. Gallo, and A. Albini. 1991. Supernatants of acquired immunodeficiency syndrome-related Kaposi's sarcoma cells induce endothelial cell chemotaxis and invasiveness. Cancer Res. 51:2670-2676. [PubMed] [Google Scholar]
- 47.Tzivion, G., and J. Avruch. 2001. 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem. 277:3061-3064. [DOI] [PubMed] [Google Scholar]
- 48.Tzivion, G., Y. H. Shen, and J. Zhu. 2001. 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene 20:6331-6338. [DOI] [PubMed] [Google Scholar]
- 49.Wang, W. S., P. M. Chen, H. L. Hsiao, S. Y. Ju, and Y. Su. 2003. Overexpression of the thymosin beta-4 gene is associated with malignant progression of SW480 colon cancer cells. Oncogene 22:3297-3306. [DOI] [PubMed] [Google Scholar]
- 50.Wettstein, F. O., M. S. Barbosa, and M. Nasseri. 1987. Identification of the major cottontail rabbit papillomavirus late RNA cap site and mapping and quantitation of an E2 and minor E6 coding mRNA in papillomas and carcinomas. Virology 159:321-328. [DOI] [PubMed] [Google Scholar]
- 51.Wulfkuhle, J. D., D. C. Sgroi, H. Krutzsch, K. McLean, K. McGarvey, M. Knowlton, S. Chen, H. Shu, A. Sahin, R. Kurek, D. Wallwiener, M. J. Merino, E. F. Petricoin III, Y. Zhao, and P. S. Steeg. 2002. Proteomics of human breast ductal carcinoma in situ. Cancer Res. 62:6740-6749. [PubMed] [Google Scholar]
- 52.Zeltner, R., L. A. Borenstein, F. O. Wettstein, and T. Iftner. 1994. Changes in RNA expression pattern during the malignant progression of cottontail rabbit papillomavirus-induced tumors in rabbits. J. Virol. 68:3620-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]