Abstract
Small-ruminant lentiviruses (SRLV), which include the caprine arthritis-encephalitis and the maedi-visna virus, cause persistent inflammatory infections in goats and sheep. SRLV are mainly transmitted from mother to offspring through milk. Transmission after prolonged contact between adult animals has also been observed. The observation that certain SRLV subtypes are found in both goats and sheep suggests that interspecies transmission has occurred on several occasions in the past. We investigated seropositive goats and sheep that were kept together in small mixed herds. Phylogenetic analysis of long proviral sequences in gag and pol, combined with epidemiologic information, demonstrated natural sheep-to-goat transmission of the recently identified SRLV subtype A4 in two instances and goat-to-sheep transmission of the same subtype in one instance. In a further mixed cluster, the direction of the interspecies transmission could not be determined. These findings present for the first time direct evidence that natural interspecies transmission of SRLV is ongoing in both directions. The findings are of relevance to virus eradication programs in both species.
Caprine arthritis-encephalitis virus (CAEV) and maedi-visna virus (MVV) are small-ruminant lentiviruses (SRLV) that infect goats and sheep (14, 17, 18, 28). Earlier phylogenetic analysis, mostly based on short sequences, has suggested that these very diverse viruses can be divided into six different sequence clades, I to VI (12, 21, 22, 32). Recent work based on long sequences in gag and pol of more than 100 new isolates from Switzerland and all available database sequences has, however, demonstrated that the SRLV should rather be divided into four principal sequence groups A to D, which differ by 25 to 37% in gag and pol sequences. Sequence groups A and B are further divided into different subtypes that differ from each other by 15 to 27%. Group A contains at least 7 subtypes, A1 to A7, and group B contains two subtypes, B1 and B2 (26). To date, subtypes A1 and A2 have been isolated only from sheep, and subtypes A5, A7, and B1 and groups C and D have been isolated only from goats. In contrast, subtypes A3, A4, A6, and B2 have been isolated from both sheep and goats.
Most transmissions of SRLV among goats or sheep occur through the ingestion of virus-infected colostrum or milk by the newborn (5-8, 13, 23). Less efficient transmission has been associated with prolonged contact with infected animals (6, 8, 23). Contact transmission is seen particularly with the maedi form of MVV infection, in which respiratory exudates may play a role (15). The widespread distribution of these viruses in certain regions and the resulting economic losses have led to segregation-based virus eradication programs for both goats and sheep in several countries (3, 8-10, 19, 23-25).
In Switzerland, a CAEV eradication program in goats was started on a voluntary basis in the early 1980s, and since 1998 the program has been mandatory for all goat holders of the country. The program provides annual serologic testing for all goats. Seropositive animals are to be culled, and a herd in which a seropositive animal is detected is put under a strict quarantine that is maintained until at least three consecutive annual serologic tests of all adult animals yield negative results. When these criteria are met, the farms are certified as CAEV free and may again engage in purchase or sales relations but only with other certified CAEV-free farms. The program has reduced the prevalence of CAEV, which on the herd level was 83% in the early 1980s, to 1.0% in 2002.
Despite the undisputed successes of this eradication program, goat owners, veterinarians, and authorities alike were puzzled time and again by the reemergence of seropositivity, especially in farms that had been CAEV free for many years and had maintained good adherence to the program's guidelines. Since goats in Switzerland are frequently kept together with sheep, the suspicion arose that reinfection of goats from CAEV-free herds might originate from infected sheep. MVV can, indeed, be experimentally transmitted to goats and, vice versa, CAEV can be transmitted to sheep (1). The fact that some SRLV subtypes have been isolated from both sheep and goats furthermore indicates that interspecies transmission must have occurred at least once in each of these subtypes, although the frequency and direction of transmission remain unknown (12, 21, 22, 32). While interspecies transmission from goats to sheep under natural conditions has never been observed (1, 16, 23, 27), we have recently forwarded statistical evidence suggesting that transmission of the newly discovered SRLV subtype A4 from sheep to goats must occur regularly, though rarely (26). Individual events of interspecies transmission, however, have never been described.
Here we have performed a phylogenetic analysis of SRLV strains isolated from goats and sheep that became infected after they had become newly exposed to SRLV-infected animal(s) of the other species. We provide direct evidence for natural interspecies transmission of SRLV subtype A4 in both directions.
MATERIALS AND METHODS
Animal specimens.
Goats or sheep of epidemiologic interest were identified by the Extension and Health Service for Small Ruminants, a government-supported cooperative involved in the Swiss CAEV eradication program. From all selected animals, 100 ml of EDTA-anticoagulated blood was drawn by venipuncture. Peripheral blood mononuclear cells were isolated by Ficoll density gradient centrifugation, and aliquots of 5 × 106 or 50 × 106 cells were frozen at −70°C as nonviable cell pellets. Milk samples, if available, were also collected from some of the animals and separated into milk plasma and a milk cell pellet by centrifugation.
Amplification of an SRLV sequence by PARRA and molecular cloning of a proviral sequence.
All procedures relating to this work are described in detail elsewhere (11). In short, milk plasma from seropositive goats was tested for reverse transcriptase (RT) activity by the product-enhanced RT (PERT) assay (20), and 1.0 ml of the sample with the highest activity was fractionated on a 7.5 to 60% sucrose density gradient. The fraction with peak RT activity and a density of 1.15 g/ml was used for amplification of the full-length viral RNA sequence by particle-associated retroviral RNA amplification (PARRA), restricting the initial cDNA synthesis in the 5′ rapid amplification of cDNA ends (RACE) step of PARRA to priming by primer pK1 (11). DNA sequence information obtained from PARRA was used to design specific primers for nested PCR. Primers GR-2 (5′-CTTAGTGCAGGCAGGAG-3′) and GU5-1 (5′-GTTATTATCGGGATCCGTC-3′) were used in the first round of long-distance PCR according to the instructions given in the Expand 20 kbPlus PCR system kit (Roche Molecular Biochemicals). After a 2-min denaturation step at 94°C, the temperature profile consisted of five cycles at 93°C for 10 sec, 57°C for 30 sec (with a decrement of 1°C), and 68°C for 8 min, followed by a further 25 cycles at 93°C for 10 sec, 53°C for 30 sec, and 68°C for 8 min, with a time increment of 15 sec for the last 20 cycles. The second round of PCR with 1 μl of the initial PCR product was performed with primers GR-1 (5′-AGATCGCTCTCCAAGAGAC-3′) and GU5-2 (5′-GGATCCGTCACTAATTCTG-3′) under the same conditions as above. The agarose gel-purified 9-kb PCR band was isolated (QIAquick gel extraction kit; QIAGEN) and cloned by TA cloning into the pGemTeasy vector (Promega) according to the instructions of the manufacturer. One full-length clone was selected and sequenced by transposon-mediated insertion sequencing according to kit instructions (Template Generation System F700; Finnzymes).
DNA extraction, sequence amplification, and sequence analysis.
These procedures are also described in detail elsewhere (26). Briefly, DNA was extracted from cell pellets and stored at −70°C prior to use. Proviral sequences from a 1.7-kb sequence in gag-pol and a 1.1-kb sequence in pol were amplified by sequence capture PCR by using published capture probes and amplification primers. Specific PCR products were purified from agarose gel, and both DNA strands were sequenced by means of published amplification and sequencing primers by using dye terminator chemistry and capillary electrophoresis. Nucleic acid sequences were assembled and edited with AutoAssembler 2.1 (Applied Biosystems) and analyzed with MacVector 7.0 (Accelrys, Inc.).
Phylogenetic analysis.
Nucleic acid sequences were aligned by using the ClustalW algorithm of the MacVector software 7.0 (Accelrys, Inc.), and the aligned, length-adjusted sequences were imported as a nexus-file into the software package PAUP* version 4.0 beta 10 (Sinauer Associates, Sunderland, Massachusetts, 2001). All further calculations and tree constructions were carried out with this software.
Pairwise genetic distances were calculated by using the F84 substitution model with default settings, with the exception that all sites with ambiguous codes and gaps were ignored. All unrooted phylogenetic trees shown were constructed by the neighbor-joining method and were based on the distances calculated with the F84 substitution model, again ignoring all sites with ambiguous codes and gaps. Bootstrap values are based on 1,000 repetitions.
Nucleotide sequence accession numbers.
All new nucleotide sequences were deposited in the GenBank database and are available under accession number AY445885 for the nearly full-length sequence and number AY445886 for the 5′ RACE sequence of PARRA; under numbers AY577031 to AY577033 for the 1.7 kb-gag-pol fragments of G7592, S7730, and S7731, respectively; and under numbers AY577034 to AY577039 for the 1.1-kb pol fragments of S7565, S7566, G7569, G7570, G7571, and G7572, respectively.
RESULTS
Goat-to-sheep transmission.
Evidence for goat-to-sheep transmission of SRLV was found in the aftermath of a sheep and goat show that took place in September 1997 in the Swiss town of Interlaken. During this event, 68 (60%) of 114 attending goats, all from certified CAEV-free herds (see introduction), became infected with a single virus strain under unexplained circumstances. Of note, this strain, which was more closely related to MVV than to CAEV, was also isolated from a dairy sheep that had been exhibited at the show (31).
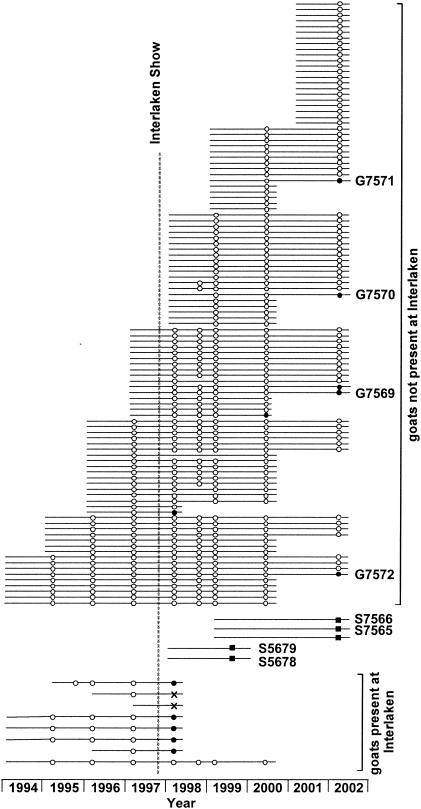
In 2002 we investigated goats from farm A which had been certified CAEV free up to spring 1997 but had become infected through eight goats that were exposed at Interlaken. Although all seropositive or seroindeterminate animals were culled by 1998, the farm has been unable to regain the status of certified CAEV negativity. One goat was found seropositive in the annual testing of 2000, and 5 of the total of 73 goats (7%) were seropositive in 2002. When the 55 dairy sheep of the farm were tested, 15 (27%) were seropositive (Fig. 1).
FIG. 1.
Epidemiology of goats and selected sheep of farm A. Each horizontal line symbolizes an animal. Circles denote goats, and squares denote sheep. Open symbols, documented negative SRLV serology; black symbols, documented positive serology; cross, indeterminate serology. Lines marked with numbers identify the animals from which viral sequences were obtained.
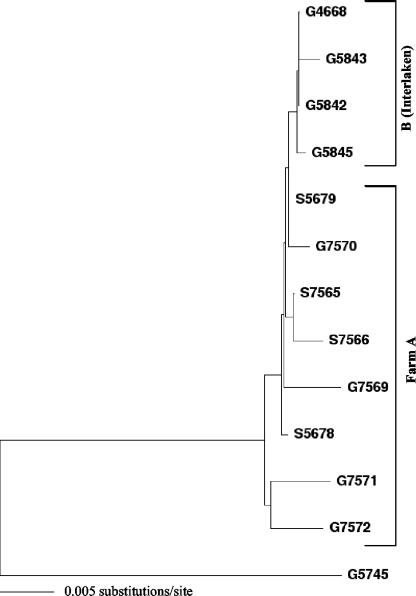
We obtained peripheral blood mononuclear cells from the five seropositive goats and three seropositive sheep and amplified a 1.7-kb fragment comprising sequences in gag and pol (distal two-thirds of capsid, nucleocapsid, and protease and the proximal one-third of RT) by sequence-capture PCR. Provirus amplification was successful in two goats and one sheep; in two other goats and one sheep a 0.7-kb fragment representing the 5′ end of the 1.7-kb sequence was obtained by using alternative primer combinations (data not shown). Amplification of a 1.1-kb sequence located in the deoxyuridine triphosphatase and integrase regions of pol was successful in four goats (G designations; here, G7569 to G7572) and two sheep (S designations; here, S7565 and S7566) but also remained unsuccessful in one goat and one sheep despite the use of alternative primers. Analysis of the pol sequences demonstrated that all six proviruses belonged to SRLV subtype A4 (26) and were closely related, exhibiting 0.0 to 1.0% substitutions between each other (Fig. 2). In comparison, the mean divergence (± standard deviation) between viruses of subtype A4 that were isolated from animals originating from epidemiologically unlinked farms was 6.2% ± 1.2% (26). The viruses isolated from farm A were also closely related to the virus transmitted at Interlaken, as demonstrated by the sequences from five goats from other farms infected at Interlaken, which we had obtained for investigation in 1999 (cluster B). The sequences from farm A and the Interlaken sequences differed by 0.2 to 1.0%. We also discovered that the six sequences from farm A were closely related to sequences that had already been isolated in 1999 from 2 other dairy sheep of the farm, S5678 and S5679 (see Fig. 1), after it had been determined that 3 (23%) out of the 13 dairy sheep the farm kept at that time were seropositive. These two sequences differed by 0.1% from each other and by 0.2% from the most closely related Interlaken sequence, G4668. Phylogenetic analysis of the three 1.7-kb gag-pol sequences and the six 0.7-kb gag sequences available from this cluster confirmed these findings (data not shown). These data thus show that the Interlaken strain had not only been further transmitted to adult goats of farm A that had not been present at Interlaken but had also crossed the species barrier and was now present in the sheep.
FIG. 2.
Phylogenetic analysis of SRLV 1.1-kb pol sequences from goats (G designations) and sheep (S designations) of farm A, together with sequences from the Interlaken goats (cluster B), demonstrating goat-to-sheep transmission. The G5745 sequence used as the outgroup is the most closely related sequence and belongs to SRLV subtype A4. For the position of the G5745 sequence within subtype A4, refer to Fig. 3.
For elucidation of the full-length sequence of this virus, we purified virus particles from the milk of goat G4668, which had exhibited the highest RT activity (327 nU of human immunodeficiency virus type 1 RT equivalent per ml) by sucrose density gradient fractionation, and subjected the viral RNA to PARRA, as described in Materials and Methods. From the DNA extracted from milk cells a 9,023-bp proviral DNA fragment was subsequently amplified, cloned, and sequenced. This first, nearly full-length sequence of SRLV subtype A4 was clearly different throughout the entire genome from all other full-length sequences deposited in the GenBank database and was in all sequence fragments (1,000-bp steps) more closely related to SRLV group A than to groups B or C, thus indicating that it was not a recombinant between viruses of these previously known sequence groups. A comparison with GenBank sequences also revealed identity in positions 7665 to 7842 with a env sequence from the SU5 region (GenBank accession no. AF295075) (2) which had also been isolated from a goat exhibited at Interlaken (G. Bertoni, University of Berne, Switzerland, personal communication).
Sheep-to-goat transmission.
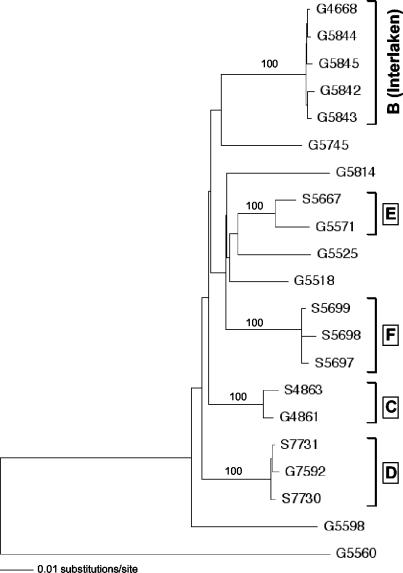
Evidence for interspecies transmission involving SRLV subtype A4 was also found when other clusters of seropositive goats kept together with sheep were investigated (Fig. 3, clusters C, D, and E). The strains isolated from goats and sheep within each of these clusters were closely related, with strains in cluster D related as closely as strains in the Interlaken cluster (B). Clusters C and D represent particularly well documented cases of sheep-to-goat transmission. Farm C kept two sheep and then bought two young goats from a certified CAEV-free farm, which has maintained this status to the present date. One of the newly acquired goats subsequently seroconverted. The close relatedness of the viruses isolated from this goat (G4861) and from one of the sheep (S4863) demonstrates beyond reasonable doubt that the goat, which was initially seronegative and had originated from a herd that was and still is certified CAEV negative, must have been infected by one of the sheep. Analysis of the 1.1-kb pol sequence confirmed the close relationship (data not shown).
FIG. 3.
Phylogenetic analysis of SRLV 1.7-kb gag-pol sequences from epidemiologically linked clusters of goats (G designations) and/or sheep (S designations). Boxed letters denote the different clusters, as described in the text. The G5560 sequence used as the outgroup is the most closely related sequence and belongs to SRLV subtype A5 (26).
Farm D kept 10 sheep and a single old goat born in 1992 (G7592). Through all the years this goat and all of its offspring had been CAEV negative, but in 2002 it was suddenly found to be seropositive, together with its kid born in 2001 (not shown in the figure since the 1.7-kb gag-pol sequence could not be amplified; analysis of the shortened 0.7-kb gag fragment showed, however, that the two sequences differed by only 0.48%). Since two of the sheep, which, in contrast to the old goat, had never before been tested, were found infected and carried the same virus strain, infection of the goat must have come from one of the sheep.
Sequences from another mixed cluster, E, were also closely related to each other, but epidemiologic information permitting elucidation of the direction of the interspecies transmission is not available. Cluster F finally presents intraspecies transmission of a subtype A4 strain in a small flock of seropositive sheep and is shown for reference only.
DISCUSSION
Although some subtypes of the large and diverse SRLV group are found among both goats and sheep, thus presenting evidence for more than one event of interspecies transmission in the past (12, 26, 32), and although interspecies transmission has been achieved experimentally (1), the natural transmission of these viruses across the species barrier has never been documented directly. Here we have phylogenetically analyzed viruses isolated from goats or sheep that seroconverted after they were newly exposed to SRLV-infected animals of the other species. We demonstrate a much closer genetic relationship among these epidemiologically linked sequences, which differ by less than 1% from each other, than that found among viruses from epidemiologically unlinked animals, which differ by 4 to 8%, as is evident from Fig. 3 and has been reported previously (12, 26, 32). We thus present the first direct evidence that the recently identified SRLV subtype A4 is transmitted across the species barrier in both directions by the natural horizontal infection of adult animals. While the direction of transmission is unclear in cluster E, cluster A demonstrates goat-to-sheep transmission (Fig. 1 and 2) and clusters C and D represent sheep-to-goat transmissions (Fig. 3).
With regard to the goat-to-sheep transmission, the fact that farm A was always certified CAEV free prior to Interlaken and that five goats exposed at Interlaken subsequently became seropositive suggests strongly that the virus was newly introduced into the farm by these five goats and that it was later transmitted to the sheep, as first documented by the two seropositive sheep, S5678 and S5679, which we investigated in 1999. There is a theoretical, alternative explanation: that the sheep of farm A already harbored this virus strain prior to Interlaken, that they transmitted it to at least one of the goats brought to the show shortly after, and that these goats in their early state of infection actually represented the source of the virus distributed at the show. This explanation is rendered highly unlikely by the fact that the founder animals of the herd, which was started in 1996, were purchased as lambs from a farm that had tested MVV negative in 1995 and remained negative in 1997 and 1999. The rams of farm A were also always selected from MVV-free farms, thus minimizing the risk of introducing the virus into the expanding young herd. There is, thus, no indication that infection of the sheep of farm A by the Interlaken strain originated from a source other than an infected goat.
Sheep-to-goat transmission is clearly demonstrated by clusters C and D, in which the sheep kept on the respective farms represented the only possible source of infection. That sheep-to-goat transmission may not be exceptional is suggested by a previously reported finding that the infection of goats by viruses of subtypes A3 and A4, which we have found to be present among sheep in Switzerland, is significantly associated with both a previously CAEV-free herd status and documented exposure to sheep (26). Although such events may be infrequent, they must occur regularly since truly exceptional events would not be amenable to statistics.
In combination, these findings demonstrate that sheep may function as reservoirs for SRLV subtypes that can be naturally transmitted to goats by horizontal infection. Conversely, goats may function as reservoirs for SRLV infection in sheep. These new facts should be considered in all SRLV eradication programs. Whether subtype A4 may be particularly prone to interspecies transmission is unknown, but the fact that subtypes A3, A6, and B2 can be found in both sheep and goats is as compatible with ongoing interspecies transmission as with a single event in the past (12, 26, 32).
Little information is available as to the route of infection in these interspecies transmissions. On farm A, the goats and dairy sheep were housed in separated areas of a large barn but shared a common milking berth with a large feeding trough, and they grazed together on the same pastures, where they used the same water source and frequented the same salt lick. In the smaller herds of the farms in which we demonstrated sheep-to-goat transmission, the two species probably had even closer contact. It is thus conceivable that these transmissions occurred by the oral route. Contact transmission among sheep is seen particularly with the pulmonary manifestation form of MVV infection, suggesting that respiratory exudates may be involved (15). It has also been shown that coinfection, e.g., with the pulmonary adenomatosis (Jaagsiekte) retrovirus, which is also enzootic in Switzerland (29, 30), increases the pulmonary MVV load, thus leading to more efficient contact transmission among sheep (4, 15). Since SRLV infection of goats may also manifest itself as a chronic progressive interstitial pneumonia, it is conceivable that a similar mechanism may be involved in horizontal transmission originating from goats, e.g., via commonly frequented salt licks or feeding troughs. Further research is needed to elucidate these possibilities.
Acknowledgments
We thank Antonietta Baumgartner and Lucia Bertodatto for excellent technical assistance and Giuseppe Bertoni, Ernst Peterhans, Hans-Rudolf Vogt, Reto Zanoni, and Alfred Zaugg for valuable discussions.
This work was partially financed by the Extension and Health Service for Small Ruminants, Herzogenbuchsee, Switzerland, and supported by grants from the Swiss Federal Office of Veterinary Health, Berne, and Schweizerischer Vorort, Viehandelskonkordat/Concordat Sur Le Commerce Du Bétail, Aarau, Switzerland.
REFERENCES
- 1.Banks, K. L., D. S. Adams, T. C. McGuire, and J. Carlson. 1983. Experimental infection of sheep by caprine arthritis-encephalitis virus and goats by progressive pneumonia virus. Am. J. Vet. Res. 44:2307-2311. [PubMed] [Google Scholar]
- 2.Bertoni, G., C. Hertig, M. L. Zahno, H. R. Vogt, S. Dufour, P. Cordano, E. Peterhans, W. P. Cheevers, P. Sonigo, and G. Pancino. 2000. B-cell epitopes of the envelope glycoprotein of caprine arthritis-encephalitis virus and antibody response in infected goats. J. Gen. Virol. 12:2929-2940. [DOI] [PubMed] [Google Scholar]
- 3.Cutlip, R. C., and H. D. Lehmkuhl. 1986. Eradication of ovine progressive pneumonia from sheep flocks. J. Am. Vet. Med. Assoc. 188:1026-1027. [PubMed] [Google Scholar]
- 4.Dawson, M., S. H. Done, C. Venables, and C. E. Jenkins. 1990. Maedi-visna and sheep pulmonary adenomatosis: a study of concurrent infection. Br. Vet. J. 146:531-538. [DOI] [PubMed] [Google Scholar]
- 5.De Boer, G. F., C. Terpstra, D. J. Houwers, and J. Hendriks. 1979. Studies in epidemiology of maedi/visna in sheep. Res. Vet. Sci. 26:202-208. [PubMed] [Google Scholar]
- 6.East, N. E., J. D. Rowe, B. R. Madewell, and K. Floyd. 1987. Serologic prevalence of caprine arthritis-encephalitis virus in California goat dairies. J. Am. Vet. Med. Assoc. 190:182-186. [PubMed] [Google Scholar]
- 7.Ellis, T., W. Robinson, and G. Wilcox. 1983. Effect of colostrum deprivation of goat kids on the natural transmission of caprine retrovirus infection. Aust. Vet. J. 60:326-329. [DOI] [PubMed] [Google Scholar]
- 8.Greenwood, P. L., R. N. North, and P. D. Kirkland. 1995. Prevalence, spread and control of caprine arthritis-encephalitis virus in dairy goat herds in New South Wales. Aust. Vet. J. 72:341-345. [DOI] [PubMed] [Google Scholar]
- 9.Houwers, D. J. 1980. Maedi and maedi control [author's translation]. Tijdschr. Diergeneeskd. 105:661-664. (In Dutch.) [PubMed] [Google Scholar]
- 10.Houwers, D. J., C. D. Konig, J. Bakker, M. J. de Boer, J. J. Pekelder, J. Sol, P. Vellema, and G. de Vries. 1987. Maedi-visna control in sheep. III: results and evaluation of a voluntary control program in The Netherlands over a period of four years. Vet. Q. 9:29S-36S. [PubMed] [Google Scholar]
- 11.Huder, J. B., J. Boni, J. M. Hatt, G. Soldati, H. Lutz, and J. Schupbach. 2002. Identification and characterization of two closely related unclassifiable endogenous retroviruses in pythons (Python molurus and Python curtus). J. Virol. 76:7607-7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leroux, C., J. Chastang, T. Greenland, and J. F. Mornex. 1997. Genomic heterogeneity of small-ruminant lentiviruses: existence of heterogeneous populations in sheep and of the same lentiviral genotypes in sheep and goats. Arch. Virol. 142:1125-1137. [DOI] [PubMed] [Google Scholar]
- 13.McGuire, T. C., K. I. O'Rourke, D. P. Knowles, and W. P. Cheevers. 1990. Caprine arthritis encephalitis lentivirus transmission and disease. Curr. Top. Microbiol. Immunol. 160:61-75. [DOI] [PubMed] [Google Scholar]
- 14.Narayan, O., J. E. Clements, J. D. Strandberg, L. C. Cork, and D. E. Griffin. 1980. Biological characterization of the virus causing leukoencephalitis and arthritis in goats. J. Gen. Virol. 50:69-79. [DOI] [PubMed] [Google Scholar]
- 15.Narayan, O., and L. C. Cork. 1985. Lentiviral diseases of sheep and goats: chronic pneumonia leukoencephalomyelitis and arthritis. Rev. Infect. Dis. 7:89-98. [DOI] [PubMed] [Google Scholar]
- 16.Oliver, R., A. Cathcart, R. McNiven, W. Poole, and G. Robati. 1985. Infection of lambs with caprine arthritis encephalitis virus by feeding milk from infected goats. Vet. Rec. 116:83. [DOI] [PubMed] [Google Scholar]
- 17.Pasick, J. 1998. Maedi-visna virus and caprine arthritis-encephalitis virus: distinct species or quasispecies and its implications for laboratory diagnosis. Can. J. Vet. Res. 62:241-244. [PMC free article] [PubMed] [Google Scholar]
- 18.Pepin, M., C. Vitu, P. Russo, J. F. Mornex, and E. Peterhans. 1998. Maedi-visna virus infection in sheep: a review. Vet. Res. 29:341-367. [PubMed] [Google Scholar]
- 19.Peretz, G., F. Bugnard, and D. Calavas. 1994. Study of a prevention programme for caprine arthritis-encephalitis. Vet. Res. 25:322-326. [PubMed] [Google Scholar]
- 20.Pyra, H., J. Böni, and J. Schüpbach. 1994. Ultrasensitive retrovirus detection by a reverse transcriptase assay based on product enhancement. Proc. Natl. Acad. Sci. USA 91:1544-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravazzolo, A. P., D. Reischak, E. Peterhans, and R. Zanoni. 2001. Phylogenetic analysis of small ruminant lentiviruses from Southern Brazil. Virus Res. 79:117-123. [DOI] [PubMed] [Google Scholar]
- 22.Rolland, M., J. Mooney, S. Valas, G. Perrin, and R. Z. Mamoun. 2002. Characterisation of an Irish caprine lentivirus strain-SRLV phylogeny revisited. Virus Res. 85:29-39. [DOI] [PubMed] [Google Scholar]
- 23.Rowe, J. D., and N. E. East. 1997. Risk factors for transmission and methods for control of caprine arthritis-encephalitis virus infection. Vet. Clin. North Am.Food. Anim. Pract. 13:35-53. [DOI] [PubMed] [Google Scholar]
- 24.Rowe, J. D., N. E. East, M. C. Thurmond, C. E. Franti, and N. C. Pedersen. 1992. Cohort study of natural transmission and two methods for control of caprine arthritis-encephalitis virus infection in goats on a California dairy. Am. J. Vet. Res. 53:2386-2395. [PubMed] [Google Scholar]
- 25.Scheer-Czechowski, P., H. R. Vogt, A. Tontis, E. Peterhans, and R. Zanoni. 2000. Pilot project for eradicating maedi-visna in Walliser blacknose sheep. Schweiz. Arch. Tierheilkd. 142:155-164. [PubMed] [Google Scholar]
- 26.Shah, C. A., J. Böni, J. B. Huder, H. R. Vogt, J. Mühlherr, R. Zanoni, R. Miserez, H. Lutz, and J. Schüpbach. 2004. Phylogenetic analysis and reclassification of caprine and ovine lentiviruses based on 104 new isolates: evidence for regular sheep-to-goat transmission and world-wide propagation through livestock trade. Virology 319:12-26. [DOI] [PubMed] [Google Scholar]
- 27.Smith, V. W., J. Dickson, W. Coackley, and H. Carman. 1985. Response of merino sheep to inoculation with a caprine retrovirus. Vet. Rec. 117:61-63. [DOI] [PubMed] [Google Scholar]
- 28.Sundquist, B. 1981. Goat visna virus: isolation of a retrovirus related to visna virus of sheep. Arch. Virol. 68:115-127. [DOI] [PubMed] [Google Scholar]
- 29.Tontis, A., G. Bestetti, H. Konig, and H. Luginbuhl. 1979. Enzootic occurrence of pulmonary adenomatosis in 13 sheep near Bern. Schweiz. Arch. Tierheilkd. 121:251-262. [PubMed] [Google Scholar]
- 30.Tontis, A., and R. Zwahlen. 1984. Further cases of respiratory slow virus diseases (pulmonary adenomatosis and maedi) in sheep in the Bern area. Schweiz. Arch. Tierheilkd. 126:305-311. [PubMed] [Google Scholar]
- 31.Vogt, H. R. 2000. Caprine arthritis encephalitis: analysis of an outbreak associated with a goat and sheep market. D. V. M. dissertation. University of Berne, Berne, Switzerland.
- 32.Zanoni, R. G. 1998. Phylogenetic analysis of small ruminant lentiviruses. J. Gen. Virol. 79:1951-1961. [DOI] [PubMed] [Google Scholar]