Abstract
5-Aminolevulinic acid (ALA), the committed intermediate of the heme biosynthesis pathway, shows significant promise for cancer treatment. Here, we identified that in addition to hemA and hemL, hemB, hemD, hemF, hemG and hemH are also the major regulatory targets of the heme biosynthesis pathway. Interestingly, up-regulation of hemD and hemF benefited ALA accumulation whereas overexpression of hemB, hemG and hemH diminished ALA accumulation. Accordingly, by combinatorial overexpression of the hemA, hemL, hemD and hemF with different copy-number plasmids, the titer of ALA was improved to 3.25 g l−1. Furthermore, in combination with transcriptional and enzymatic analysis, we demonstrated that ALA dehydratase (HemB) encoded by hemB is feedback inhibited by the downstream intermediate protoporphyrinogen IX. This work has great potential to be scaled-up for microbial production of ALA and provides new important insights into the regulatory mechanism of the heme biosynthesis pathway.
5-aAminolevulinic acid (ALA), a non-protein amino acid, is the first committed intermediate in the common tetrapyrrole pathway (Fig. 1) for synthesis of heme, chlorophyll, cytochrome and vitamin B121. In nature, there exist two known alternate routes by which this committed intermediate is generated2,3. One route is the C4 pathway (Shemin pathway), which involves the condensation of succinyl-CoA and glycine to ALA by ALA synthase (ALAS). The C4 pathway is restricted to mammals, fungi and purple nonsulfur bacteria4. The second route is the C5 pathway5, which involves three enzymatic reactions resulting in the biosynthesis of ALA from glutamate. The C5 pathway is active in most bacteria, all archaea and plants (Fig. 1).
Figure 1. Illustration of the regulation mechanism of the heme pathway in E. coli.
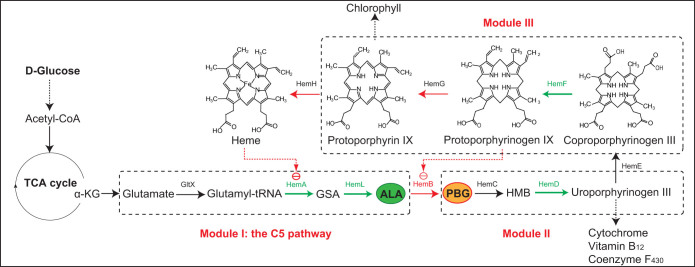
The pathway is divided into three modules (module I, module II and module III in the dotted box). The arrows in green and red represent the enzymes that are positive and negative to ALA accumulation, respectively. Dotted red arrows represent the feedback-inhibition. α-KG: α-ketoglutarate, GSA: glutamate-1-semialdehyde, ALA: 5-aminolevulinic acid, PBG: porphobilinogen, HMB: hydroxymethylbilane, GltX: glutamyl-tRNA synthetase, HemA: glutamyl-tRNA reductase, HemL: glutamate-1-semialdehyde aminotransferase, HemB: 5-aminolevulinic acid dehydratase, HemC: porphobilinogen deaminase, HemD: uroporphyrinogen III synthase, HemE: uroporphyrinogen decarboxylase, HemF: coproporphyrinogen III oxidase, HemG: protoporphyrin oxidase, HemH: ferrochelatase.
ALA has recently attracted much attention for its great potential applications in the fields of medicine (tumor-localizing and cancer photodynamic therapy)6,7,8 and agriculture (biodegradable herbicides, plant growth regulators and insecticides)1,2. Since chemical synthesis of ALA is complicated and low-yielding, studies focusing on alternative microbial production of ALA from renewable and inexpensive sources has received much attention1. In this regard, many native microbes including photosynthetic bacteria (Rhodobacter sphaeroides) have been isolated and randomly mutated to produce ALA2,9,10. Although the titer of ALA has been significantly improved, its application was confined due to many disadvantages, such as the requirement of light illumination and poor cell growth. Consequently, intensive research has been concentrated on construction of recombinant Escherichia coli cell factories for enzyme-transformation of ALA by heterologous expression of different ALAS genes11,12. Notably, by applying a full factorial design model, the production of ALA was significantly increased to 5.20 g l−1 (39 mM)13. In addition, a highest titer of ALA was achieved (7.30 g l−1, 56 mM) after optimization of ALAS expression and cultivation process14,15,16. However, application of the complex medium (LB), artificial addition of the precursors glycine and succinate and the complicated cultivation process would be unbeneficial to industrial production.
Alternatively, from metabolic engineering point of view, Kang et al. recently engineered the C5 pathway by co-overexpression of a mutant hemA17,18,19,20 (encodes glutamyl-tRNA reductase, Salmonella arizona) and hemL (encodes glutamate-1-semialdehyde aminotransferase, E. coli) in E. coli and successfully increased ALA accumulation to 2.01 g l−1. After further overexpression of rhtA (encodes a membrane protein for threonine and homoserine exporting, E. coli) and optimization of minimal medium composition and cultivation process, the titer of ALA was improved to 4.13 g l−1 in batch-fermentation with glucose as the sole carbon source. As an important precursor, the biosynthesis of ALA is tightly regulated by the end product heme. In addition, the regulation mechanism of the heme biosynthesis pathway is more complex than expected21. Although the heme biosynthesis pathway enzymes have been well studied3,22,23, limited information on its regulation mechanism is available3.
In the present work, we systematically investigated the effect of overexpression of the downstream genes (hemB, hemC, hemD, hemE, hemF, hemG and hemH) on ALA accumulation. Interestingly, we discovered that in addition to the upstream genes hemA and hemL, hemB, hemD, hemF, hemG and hemH are also the major regulatory targets. Specifically, up-regulation of hemD and hemF was beneficial to ALA accumulation whereas overexpression of hemB, hemG and hemH was adverse to ALA accumulation. Through combinatorial overexpression of hemA, hemL, hemD and hemF, the titer of ALA was increased to 3.25 g l−1. More importantly, in combination with transcriptional and enzymatic analysis, we demonstrated that ALA dehydratase (HemB) encoded by hemB is feedback inhibited by the downstream intermediate protoporphyrinogen IX.
Results
Up-regulation of hemD and hemF increased ALA accumulation
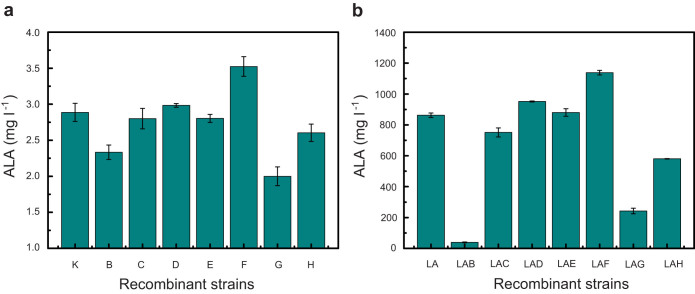
According to previous studies24,25, it has been accepted that the biosynthesis of ALA is the rate-limiting step for heme biosynthesis and is tightly regulated in organisms including E. coli. In order to investigate the effects of downstream genes of the heme synthesis pathway on ALA accumulation, we individually overexpressed hemB, hemC, hemD, hemE, hemF, hemG and hemH with a low-copy number vector pCDFDuet-1 in E. coli. Although the absolute production of ALA was low, distinct changes were observed with single overexpression of the above-mentioned genes (Fig. 2a). Up-regulation of hemD or hemF increased ALA accumulation while reverse results were obtained when overexpressing hemB, hemG or hemH. In contrast, no significant differences were detected after overexpression of hemC or hemE. These findings indicated that up-regulation of the downstream genes hemD or hemF has a positive correlation on ALA accumulation whereas up-regulation of hemB, hemG or hemH has a negative correlation. More importantly, the results also suggested that in addition to hemA, hemL and hemF3, hemB, hemD, hemG and hemH are also the main regulatory points of the heme biosynthesis pathway.
Figure 2. 5-aminolevulinic acid production by recombinant strains.
(a) The production of ALA by recombinant strains that individually overexpress hemB, hemC, hemD, hemE, hemF, hemG and hemH. (b) ALA production of recombinant strains that co-overexpress hemL and hemAs with hemB, hemC, hemD, hemE, hemF, hemG and hemH, respectively.
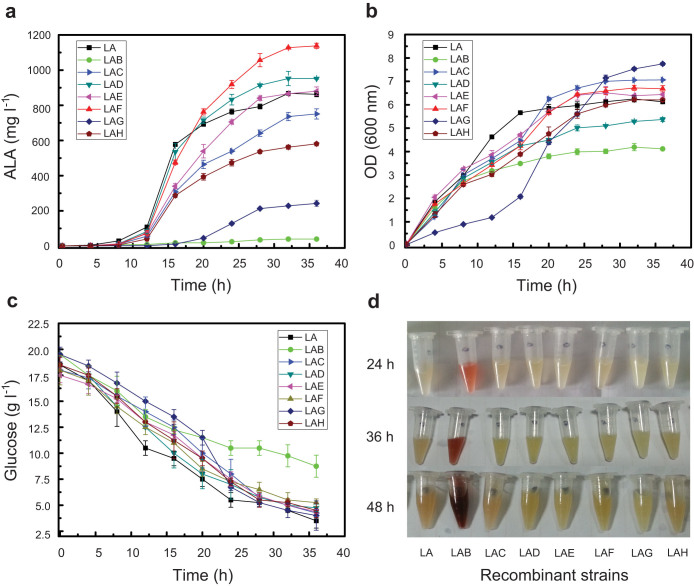
To further validate the above results and in view of ALA biosynthesis is tightly regulated in E. coli17, the upstream metabolic flux towards ALA was increased by co-overexpression of the rate-limiting enzymes HemAs (a variant of HemA from S. arizona) and HemL (E. coli) in the above constructed recombinant strains. Compared to E. coli LA (hemL and hemAs) (862.5 mg l−1), E. coli LAD (hemL, hemAs and hemD) and E. coli LAF (hemL, hemAs and hemF) produced more ALA (951.6 mg l−1 and 1,138.2 mg l−1), respectively (Fig. 2b and Fig. 3a). In contrast, the recombinant strains E. coli LAG (hemL, hemAs and hemG), E. coli LAH (hemL, hemAs and hemH) and especially E. coli LAB (hemL, hemAs and hemB) exhibited remarkably reduced ALA production which were 242.3 mg l−1, 580.6 mg l−1 and 39.2 mg l−1, respectively (Fig. 2b and Fig. 3a). Consistent with the above single expression results, no significant changes were observed when co-overexpressing hemC and hemE. The results confirmed that hemD and hemF are distinct from hemB, hemG and hemH in that they are beneficial to ALA production.
Figure 3. Time course of fermentation by recombinant strains.
(a) ALA production. (b) Cell growth. (c) Glucose consumption. (d) The fermentation broth of the recombinant strains at 24 h, 36 h and 48 h.
Additionally, overexpression of the above genes generated distinct effects on cell growth and glucose consumption. In particular, up-regulation of hemB resulted in reduced biomass (Fig. 3b) and glucose consumption (Fig. 3c), which is likely due to the accumulation of the harmful intermediate porphobilinogen (PBG) and its derivatives12 (Fig. 3d). Interestingly, overexpression of hemG resulted in more biomass coupled with a long lag phase (Fig. 3b and Supplementary Fig. S1), which is likely due to its direct involvement in energy generation26 and the toxic intermediate protoporphyrin IX27.
Modular optimization of the committed enzymes to improve ALA production
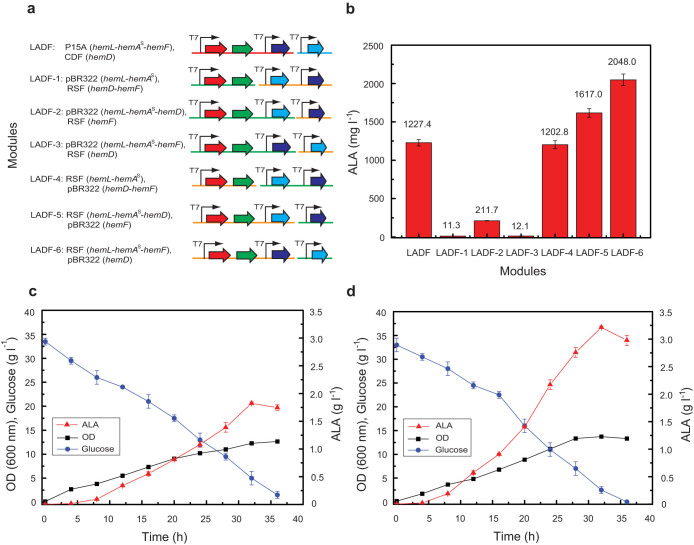
On the base of the above analysis, it could be speculated that simultaneous up-regulation of hemD and hemF would further enhance ALA production. To verify this speculation, the upstream genes (hemAs and hemL) and the downstream genes (hemD and hemF) of the heme biosynthesis pathway were simultaneously overexpressed in E. coli. As expected, the resulting strain E. coli LADF harboring plasmids pACYCDuet-1-hemL-hemAs-hemF and pCDFDuet-1-hemD produced about 1,227.4 mg l−1 of ALA at 36 h (Fig. 4b), which was higher than that of the recombinant strains E. coli LAD (hemL, hemAs and hemD) and E. coli LAF (hemL, hemAs and hemF). To further fine-tune these four committed enzymes and improve the production of ALA, the compatible plasmids pRSFDuet-1 (high-copy number) and pETDuet-1 (medium-copy number) were used to investigate the impact of plasmid copy number on ALA production. According to the above results, the committed genes hemAs, hemL, hemD and hemF were distributed in three modules (Fig. 1). Specifically, due to HemA and HemL forming a tight complex, with a 1:1 ratio, to quickly catalyze glutamyl-tRNA to ALA28, the upstream genes hemL and hemAs were contained within one operon and co-overexpressed with the downstream genes hemD and hemF as described in Fig. 4a. As we expected, the recombinant strains LADF-1, LADF-2 and LADF-3 with comparatively low-level overexpression of hemL and hemAs resulted in dramatically decreased ALA accumulation which were 11.3 mg l−1, 211.7 mg l−1 and 12.1 mg l−1, respectively. In comparison, the recombinants LADF-4, LADF-5 and LADF-6 with high-level overexpression of hemL and hemAs lead to much higher titer of ALA which were 1,202.8 mg l−1, 1,617.0 mg l−1 and 2,048.0 mg l−1, respectively. Furthermore, compared with the recombinant LADF-4 (RSF-hemL-hemAs, pBR322-hemF-hemD), LADF-6 (RSF-hemL-hemAs-hemF, pBR322-hemD) produced more ALA (2,048.0 mg l−1, Fig. 4b). The results suggested that high-level overexpression of the upstream genes hemAs and hemL as well as the downstream gene hemF, and moderate overexpression of the downstream gene hemD are favorable to ALA accumulation.
Figure 4. Modular optimization of ALA production by co-expression of hemAs, hemL, hemD and hemF.
(a) Schematic representation of different combinations. The genes hemL (red arrow), hemAs (green arrow), hemD (light blue arrow) and hemF (purple arrow) are assembled with the plasmids pACYCDuet-1 (red line), pCDFDuet-1 (blue line), pETDuet-1 (green line) and pRSFDuet-1 (orange line), respectively. (b) ALA production of recombinant strains with different gene combinations. (c) Batch fermentation of ALA by the engineered strain E. coli LA in a 3 l bioreactor. (d) Batch fermentation of ALA by the engineered strain LADF-6 in a 3 l bioreactor.
To evaluate the capacity of the recombinant strain LADF-6 for the production of ALA, a scaled-up fermentation experiment was carried out in a 3 l bioreactor with E. coli LA as the control (Fig. 4c). Obviously, compared with E. coli LA, LADF-6 showed much bigger capacity for ALA accumulation (Fig. 4d). At 32 h, the titer of ALA was increased to 3.25 g l−1 with a final productivity of 0.102 g l−1 h−1 which was 1.79-fold of that of E. coli LA (1.82 g l−1). The results further demonstrated that up-regulation hemD and hemF is beneficial to ALA accumulation and rational up-regulation of the upstream genes (hemA and hemL) and the downstream genes (hemD and hemF) is a good strategy for efficient synthesis of ALA. Moreover, the results also confirmed that the regulatory mechanism of the heme biosynthesis pathway is complicated and needs to be uncovered.
The heme biosynthesis pathway is tightly regulated at the transcriptional level
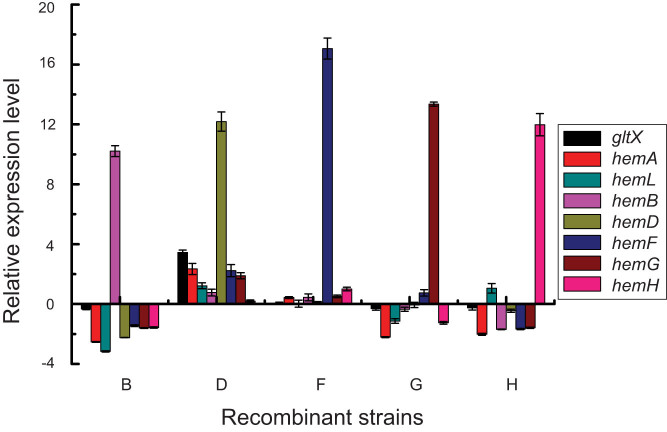
To understand the underlying biology behind the different effects of overexpressing the above mentioned five key genes hemB, hemD, hemF, hemG and hemH on the heme biosynthesis pathway, we quantified the mRNA expression of these genes. As expected, single overexpression of these genes substantially increased the level of each respective mRNA transcript. Moreover, we observed that overexpression of hemB, hemG or hemH caused reduction of most of the pathway genes while overexpression of hemD resulted in significant increase of the all pathway genes (especially gltX and hemA) (Fig. 5), which are in agreement with ALA production (Fig. 2) and the results from the C4 pathway29. These findings suggested that hemB, hemG, hemH and hemD have central regulatory roles in the heme biosynthesis pathway and their transcriptional levels are tightly regulated.
Figure 5. Alteration in pathway gene expression in the recombinant strains.
Relative gene expression in recombinant strains was compared using gapA transcript levels for normalization. The control was the E. coli BL21 (DE3) harboring plasmid pCDFDuet-1. The results are on a logarithmic scale and the error bars represent one standard deviation of three independent experiments.
Although ALA accumulation was significantly increased with overexpression of hemF, no obvious transcriptional alterations of the pathway genes were detected (Fig. 5), suggesting that up-regulation of hemF resulted in other effects but not transcriptional changes in pathway genes. Compared to E. coli LA (hemL, hemAs) and LAB (hemL, hemAs and hemB), E. coli LAF (hemL, hemAs and hemF) showed lighter color even with accumulation of more ALA (Fig. 3d). In addition, although up-regulation of hemG significantly decreased the transcriptional level of the genomic hemA gene, the recombinant strain E. coli LAG (hemL, hemAs and hemG) harboring the plasmid-borne genes hemL and hemAs still accumulated much less ALA compared with E. coli LA (hemL, hemAs). Taken together, we concluded that HemB is a key control node and might be feedback inhibited by the intermediate protoporphyrinogen IX which catalyzed from coproporphyrinogen III by coproporphyrinogen III oxidase (HemF, encoded by hemF) (Fig. 1).
HemB is a feedback inhibitory node of the heme biosynthesis pathway
To further validate our speculation, the HemB activity of the recombinant strains E. coli LA (hemL, hemAs), E. coli LAF (hemL, hemAs and hemF), E. coli LAG (hemL, hemAs and hemG) and E. coli LAB (hemL, hemAs and hemB) were directly examined (Table 1). Compared with E. coli LAB (52.04 U mg−1), E. coli LA exhibited much lower activity (6.97 U mg−1) which further confirmed that expression of HemB is tightly regulated to maintain a relatively low level in E. coli. Notably, E. coli LAF with overexpression of hemF showed an obvious decline of the HemB activity (3.34 U mg−1), which supported our speculation that HemB is feedback inhibited by the intermediate protoporphyrinogen IX (Fig. 1). Furthermore, E. coli LAG with overexpression of hemG gave rise to a substantial increase in HemB activity (22.21 U mg−1), which might be attributed to the direct activation by protoporphyrin IX or the weakened inhibition that caused by decreased protoporphyrinogen IX (Fig. 1).
Table 1. HemB enzyme activity in different recombinant strains.
| Recombinants | HemB activity (U mg−1)a | Protoporphyrin IX (mg l−1)b |
|---|---|---|
| E. coli LA | 6.97 ± 0.26 | - |
| E. coli LAF | 3.34 ± 0.05 | - |
| E. coli LAG | 22.21 ± 0.42 | - |
| E. coli LAB | 52.04 ± 1.47 | - |
| E. coli LAB | 51.78 ± 1.50 | 5.0 |
| E. coli LAB | 51.70 ± 1.53 | 10.0 |
aData are the means ± standard deviation (SD) from three parallel experiments.
bIntermediate protoporphyrin IX was added into the reaction system to study the effect on HemB activity.
To further confirm the above speculation and due to the unavailability of the intermediate protoporphyrinogen IX, we quantified HemB activity of the E. coli LAB strain extracts with addition of protoporphyrin IX in vitro to indirectly determine which of the two possibilities is correct. As shown in Table 1, activation of HemB following the addition of protoporphyrin IX was not detected. In combination with the above results from E. coli LAF, our speculation that HemB is a key regulatory node of the heme biosynthesis pathway and feedback inhibited by the intermediate protoporphyrinogen IX (Fig. 1) was demonstrated, which in turn provided the reason why high-level overexpression of hemF enhanced the production of ALA.
Discussion
Because of its numerous potential applications in medicine and agriculture, market demand of the valuable compound ALA is rapidly rising. Over the past several years, much research has been dedicated to optimizing whole-cell catalytic synthesis of ALA1 by overexpression of ALAS. Although metabolic engineering has been widely used as a powerful tool for construction and optimization of the target pathways towards various valuable compounds30,31,32,33,34, few related studies have made significant contributions towards elucidating the regulatory mechanism of the heme biosynthesis pathway1,2,35,36 (Fig. 1), while the structure-function relationship for all the heme biosynthesis enzymes have been well understood23. Therefore, exploration of the heme synthesis pathway would not only be beneficial to rational engineering towards ALA production but also invaluable for uncovering regulatory mechanism of this highly conserved pathway.
Although the transcriptional model of hemA and hemF has been previously examined under different conditions3, the overall regulatory mechanism of heme biosynthesis pathway genes is still vague. In this study, we discovered that in addition to HemA, HemL and HemF3,35, HemB, HemD, HemG and HemH are also the key regulatory targets of the heme biosynthesis pathway. Overexpression of these genes showed significant effects on cell growth and metabolism (Fig. 3b and 3c) suggesting that they are tightly controlled under normal conditions. Recent studies have reported that hemB and hemH are negatively regulated by RyhB (an iron-associated small non-coding RNA) at the post-transcriptional level37. Moreover, by investigation at transcriptional and protein levels, we demonstrated that HemB is feedback inhibited by the intermediate protoporphyrinogen IX (Fig. 1), which suggest that regulation of heme biosynthesis pathway genes is far more complicated than imagined. In addition, Kang et al. previously discovered that overexpression of the native gltX gene (glutamyl-tRNA synthetase, GluRS) resulted in dramatic decrease of ALA17. Here, we also confirmed that overexpression of gltX and artificial addition of glutamate lead to reduced ALA accumulation and cell growth (Supplementary Fig. S2). The reason might be attributed to the synthesis of glutamyl-tRNAGln (harmful to cell growth) that catalyzed by the nonspecific GluRS35,38.
In the field of metabolic engineering, it was generally accepted that increase and balance of the objective synthetic pathways are the most critical parameters for high-yield production of the end-product34,39,40. For this purpose, many combinatorial optimization strategies have been developed and applied to assemble and regulate expression of the key genes41,42,43,44,45. In this study, after investigation and identification of the four positive key genes towards ALA, the heme biosynthesis pathway was further optimized with two compatible plasmids pRSFDuet-1 (high-copy number) and pETDuet-1 (medium-copy number) according to previous results. As we expected, the production of ALA was significantly increased (Fig. 4d) with an optimized combination of the increased flux towards ALA. One side, the results demonstrated that moderate expression of HemD is crucial to ALA accumulation since overexpression of HemD not only up-regulates the upstream genes (Fig. 5) but also draws more flux to downstream reactions from ALA (Fig. 1). Moreover, the results were consistent with our finding that HemB is a key regulatory point of the heme biosynthesis pathway and feedback inhibited by protoporphyrinogen IX. Previously, it has been reported that a hemB mutant E. coli strain failed to increase the production of ALA46 indicating that low activity of HemB is essential for cell growth and ALA accumulation. In comparison, the strategy of down-regulating HemB with overexpression of HemF was an alternative to improve ALA production.
In conclusion, through investigation of the heme biosynthesis pathway genes we discovered that hemB, hemD, hemF, hemG and hemH are also the main regulatory targets of the heme biosynthesis pathway. More importantly, overexpression of hemD and hemF increased the accumulation of the upstream intermediate ALA. By combinatorial overexpression of hemA, hemL, hemD and hemF with different copy-number plasmids, the titer of ALA was increased from 862.5 mg l−1 to 3.25 g l−1. Furthermore, in combination with transcriptional and enzymatic analysis, we demonstrated that HemB is feedback inhibited by the downstream intermediate protoporphyrinogen IX, which provides new important insights into the regulatory mechanism of the heme biosynthesis pathway. To further increase the production of ALA, comparative investigation of different E. coli hosts and overexpression of the ALA transporter RhtA17 should be available. In addition, dynamic regulation of HemB with synthetic regulatory elements or circuits40,47,48 should also be desirable for efficient production of ALA.
Methods
Strains and plasmids
Bacterial strains and plasmids used in this study are described in Supplementary Table S1 and Table S2, respectively. Specifically, E. coli BL21 (DE3) was used as host for gene expression due to the T7 RNA polymerase. E. coli JM109 was used for DNA manipulations and plasmids construction.
Media and culture conditions
Luria-Bertani (LB) medium (g l−1) composed of tryptone 10.0, yeast extract 5.0 and NaCl 10.0 was used for the DNA manipulation process and seed cultures. Agar (2.0%) was added when a solid medium was required. The modified minimal medium (g l−1), which contains glucose 20.0, yeast extract 2.0, (NH4)2SO4 16.0, KH2PO4 3.0, Na2HPO4·12H2O 16.0, MgSO4·7H2O 1.0, MnSO4·H2O 0.01 was used for microbial production of ALA17. All recombinant strains were cultured at 37°C, 200 r min−1 and isopropyl-β-D-thiogalactopyranoside (IPTG) was initially added into the medium with a final concentration of 0.1 mM to induce the genes expression. Ampicillin (100 μg ml−1), chloramphenicol (20 μg ml−1), streptomycin (50 μg ml−1) or kanamycin (50 μg ml−1) was added to the medium for selection when necessary based on the harboring vectors.
The batch culture was performed in a 3 l fermentor (BilFlo 115, New Brunswick Scientific Co., Edison, NJ, USA). 2.0% inoculation of the seed culture was transferred into the fermentor with approximate 1.5 l medium after being cultured at 37°C for about 12 h. Chloramphenicol (20 μg ml−1) and streptomycin (50 μg ml−1) or ampicillin (100 μg ml−1) and kanamycin (50 μg ml−1), glucose (35.0 g l−1) and IPTG with a final concentration of 0.1 mM to induce the expression of genes were initially added to the medium. Agitation speed was 400 r min−1 and aeration rate was 1.0 vvm. The cultures were incubated at 37°C and pH was maintained at approximate 6.5 by adding 4.0 mol l−1 NaOH.
Analytical procedures
All engineered E. coli strains were cultured in the modified minimal medium containing appropriate antibiotics. For growth studies, optical densities (OD) of the cell were measured at a wavelength of 600 nm with a UV-1,700 spectrophotometer (Shimadzu, Kyoto, Japan). Glucose concentration in the supernatant was detected using a glucose-glutamate analyzer SBA-40C (Biology Institute of Shandong Academy of Sciences, Jinan, China). The production of ALA was analyzed using the Modified Ehrlich's Reagent after the cultures were centrifuged49.
Enzyme assays of HemB
For detecting HemB activity, cells were harvested after addition of IPTG following 8 h of culture by centrifugation for 10 min (10,000 r min−1, 4°C). Cells were washed twice with disodium hydrogen phosphate-citric acid buffer (pH 6.7), then they were resuspended in the above buffer and disrupted for 5 min using an ultrasonic oscillation (Sonics VCX750, amplitude 25%). After removing the cellular debris by centrifugation (10,000 r min−1, 4°C), the supernatant was analyzed for enzyme activity. Enzyme activity was quantified using the method described previously without the addition of dithiothreitol (DTT) to the reaction mixture50. HemB enzyme activity of one unit was defined as the amount of enzyme required to consume 1 pmol of ALA per min.
Quantitative real-time PCR (qRT-PCR) analysis
Cultures used for RNA extraction were cultivated for approximately 5 h after the addition of 0.1 mM IPTG. Cell quantities corresponding to approximately 1.5–2.0 OD600 nm were harvested and frozen immediately at −80°C. Total RNA of all E. coli strains was extracted using the RNA Extraction Kit (Takara, Dalian, China) according to the manufacturer's instructions. The quantity and purity of the RNA was determined using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) by optical density measurements at 260 and 280 nm.
The RNA level was measured by qRT-PCR. Genes and their respective primer sequences that were used for qRT-PCR studies are listed in Supplementary Table S3. gapA encoding D-glyceraldehyde-3-phosphate dehydrogenase was selected as internal standard. The cDNA templates used for qRT-PCR were obtained by reverse transcribing mRNA transcripts using PrimeScriptTM RT-PCR Kit (Takara, Dalian, China). Gene expression analysis via qRT-PCR was carried out in a 96-well plate with a total reaction volume of approximately 20 μl using a SYBR® Premix Ex TaqTM (Takara, Dalian, China) according to manufacturer's specifications. Reactions were performed with a LightCycler 480 II Real-time PCR instrument (Roche Applied Science, Mannheim, Germany).
Author Contributions
Z.K. and J.Z. designed research; J.Z. performed research; Z.K. and J.Z. analyzed data; Z.K., J.Z., G.D. and J.C. wrote the paper; all authors reviewed the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Professor Jens Nielsen (Chalmers University of Technology) for meaningful discussion. This work was financially supported by the National Natural Science Foundation of China (31200020), the Major State Basic Research Development Program of China (973 Program, 2013CB733602, 2014CB745103), the Program for Changjiang Scholars and Innovative Research Team in University (No. IRT1135) and the 111 Project.
References
- Kang Z. et al. Recent advances in microbial production of δ-aminolevulinic acid and vitamin B12. Biotechnol Adv 30, 1533–1542 (2012). [DOI] [PubMed] [Google Scholar]
- Sasaki K., Watanabe M. & Tanaka T. Biosynthesis, biotechnological production and applications of 5-aminolevulinic acid. Appl Microbiol Biotechnol 58, 23–29 (2002). [DOI] [PubMed] [Google Scholar]
- Schobert M. & Jahn D. Regulation of heme biosynthesis in non-phototrophic bacteria. J Mol Microbiol Biotechnol 4, 287–294 (2002). [PubMed] [Google Scholar]
- Burnham B. F. δ-Aminolevulinic acid synthase (Rhodopseudomonas sphaeroides). Methods Enzymol 17A, 195–204 (1970). [Google Scholar]
- Woodard S. I. & Dailey H. A. Regulation of heme biosynthesis in Escherichia coli. Arch Biochem Biophys 316, 110–115 (1995). [DOI] [PubMed] [Google Scholar]
- Bhowmick R. & Girotti A. W. Cytoprotective induction of nitric oxide synthase in a cellular model of 5-aminolevulinic acid-based photodynamic therapy. Free Radic Biol Med 48, 1296–1301 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolajewska P. et al. Microneedle pre-treatment of human skin improves 5-aminolevulininc acid (ALA)- and 5-aminolevulinic acid methyl ester (MAL)-induced PpIX production for topical photodynamic therapy without increase in pain or erythema. Pharm Res 27, 2213–2220 (2010). [DOI] [PubMed] [Google Scholar]
- Sakamoto F. H., Torezan L. & Anderson R. R. Photodynamic therapy for acne vulgaris: a critical review from basics to clinical practice: part II. Understanding parameters for acne treatment with photodynamic therapy. J Am Acad Dermatol 63, 195–211 (2010). [DOI] [PubMed] [Google Scholar]
- Yubisui Y. & Yoneyama Y. 5-Aminolevulinic acid synthase Rhodobacter sphaeroides: purification and properties of the enzyme. Arch Biochem Biophys 150, 77–85 (1972). [DOI] [PubMed] [Google Scholar]
- Nishikawa S. et al. Rhodobacter sphaeroides mutants which accumulate 5-aminolevulinic acid under aerobic and dark conditions. J Biosci Bioeng 87, 798–804 (1999). [DOI] [PubMed] [Google Scholar]
- Neidle E. L. & Kaplan S. Expression of the Rhodobacter sphaeroides hemA and hemT genes, encoding two 5-aminolevulinic acid synthase isozymes. J Bacteriol 175, 2292–2303 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Werf M. J. & Zeikus J. G. 5-Aminolevulinate production by Escherichia coli containing the Rhodobacter sphaeroides hemA gene. Appl Environ Microbiol 62, 3560–3566 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Hall D., Eiteman M. A. & Altman E. Optimization of recombinant aminolevulinate synthase production in Escherichia coli using factorial design. Appl Microbiol Biotechnol 63, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- Qin G., Lin J., Liu X. & Cen P. Effects of medium composition on production of 5-aminolevulinic acid by recombinant Escherichia coli. J Biosci Bioeng 102, 316–322 (2006). [DOI] [PubMed] [Google Scholar]
- Fu W., Lin J. & Cen P. Enhancement of 5-aminolevulinate production with recombinant Escherichia coli using batch and fed-batch culture system. Bioresour Technol 99, 4864–4870 (2008). [DOI] [PubMed] [Google Scholar]
- Lin J., Fu W. & Cen P. Characterization of 5-aminolevulinate synthase from Agrobacterium radiobacter, screening new inhibitors for 5-aminolevulinate dehydratase from Escherichia coli and their potential use for high 5-aminolevulinate production. Bioresour Technol 100, 2293–2297 (2009). [DOI] [PubMed] [Google Scholar]
- Kang Z., Wang Y., Gu P., Wang Q. & Qi Q. Engineering Escherichia coli for efficient production of 5-aminolevulinic acid from glucose. Metab Eng 13, 492–498 (2011). [DOI] [PubMed] [Google Scholar]
- Wang L., Wilson S. & Elliott T. A mutant HemA protein with positive charge close to the N terminus is stabilized against heme-regulated proteolysis in Salmonella typhimurium. J Bacteriol 181, 6033–6041 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Elliott M. & Elliott T. Conditional stability of the HemA protein (glutamyl-tRNA reductase) regulates heme biosynthesis in Salmonella typhimurium. J Bacteriol 181, 1211–1219 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. M. & Elliott T. A purified mutant HemA protein from Salmonella enterica serovar Typhimurium lacks bound heme and is defective for heme-mediated regulation in vivo. FEMS Microbiol Lett 307, 41–47 (2010). [DOI] [PubMed] [Google Scholar]
- Wang L., Brown L., Elliott M. & Elliott T. Regulation of heme biosynthesis in Salmonella typhimurium: activity of glutamyl-tRNA reductase (HemA) is greatly elevated during heme limitation by a mechanism which increases abundance of the protein. J Bacteriol 179, 2907–2914 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann I. U., Jahn M. & Jahn D. The biochemistry of heme biosynthesis. Arch Biochem Biophys 474, 238–251 (2008). [DOI] [PubMed] [Google Scholar]
- Layer G., Reichelt J., Jahn D. & Heinz D. W. Structure and function of enzymes in heme biosynthesis. Protein Sci 19, 1137–1161 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Russell C. S., Murooka Y. & Cosloy S. D. 5-Aminolevulinic acid synthesis in Escherichia coli requires expression of hemA. J Bacteriol 176, 2743–2746 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verderber E. et al. Role of the hemA gene product and δ-aminolevulinic acid in regulation of Escherichia coli heme synthesis. J Bacteriol 179, 4583–4590 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möbius K. et al. Heme biosynthesis is coupled to electron transport chains for energy generation. Proc Natl Acad Sci USA 107, 10436–10441 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I., Kruse E., Mock H. P. & Grimm B. Cloning and characterization of a plastidal and a mitochondrial isoform of tobacco protoporphyrinogen IX oxidase. Proc Natl Acad Sci USA 94, 8895–8900 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luer C. et al. Complex formation between glutamyl-tRNA reductase and glutamate-1-semialdehyde 2,1-aminomutase in Escherichia coli during the initial reactions of porphyrin biosynthesis. J Biol Chem 280, 18568–18572 (2005). [DOI] [PubMed] [Google Scholar]
- Lee M. J. et al. Effect of gene amplifications in porphyrin pathway on heme biosynthesis in a recombinant Escherichia coli. J Microbiol Biotechnol 23, 668–673 (2013). [DOI] [PubMed] [Google Scholar]
- Nielsen J., Larsson C., van Maris A. & Pronk J. Metabolic engineering of yeast for production of fuels and chemicals. Curr Opin Biotechnol 24, 398–404 (2013). [DOI] [PubMed] [Google Scholar]
- Stephanopoulos G. Synthetic biology and metabolic engineering. ACS Synth Biol 1, 514–525 (2012). [DOI] [PubMed] [Google Scholar]
- Keasling J. D. Synthetic biology and the development of tools for metabolic engineering. Metab Eng 14, 189–195 (2012). [DOI] [PubMed] [Google Scholar]
- Lee S. Y. Metabolic engineering and synthetic biology in strain development. ACS Synth Biol 1, 491–492 (2012). [DOI] [PubMed] [Google Scholar]
- Chen Y. & Nielsen J. Advances in metabolic pathway and strain engineering paving the way for sustainable production of chemical building blocks. Curr Opin Biotechnol 24, 965–972 (2013). [DOI] [PubMed] [Google Scholar]
- Levicán G., Katz A., de Armas M., Núñez H. & Orellana O. Regulation of a glutamyl-tRNA synthetase by the heme status. Proc Natl Acad Sci USA 104, 3135–3140 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNicholas P. M., Javor G., Darie S. & Gunsalus R. P. Expression of the heme biosynthetic pathway genes hemCD, hemH, hemM and hemA of Escherichia coli. FEMS Microbiol Lett 146, 143–148 (1997). [DOI] [PubMed] [Google Scholar]
- Li F. et al. Constitutive expression of RyhB regulates the heme biosynthesis pathway and increases the 5-aminolevulinic acid accumulation in Escherichia coli. FEMS Microbiol Lett 350, 209–215 (2014). [DOI] [PubMed] [Google Scholar]
- Núñez H., Lefimil C., Min B., Söll D. & Orellana O. In vivo formation of glutamyl-tRNAGln in Escherichia coli by heterologous glutamyl-tRNA synthetases. FEBS Lett 557, 133–135 (2004). [DOI] [PubMed] [Google Scholar]
- Zamboni N. et al. Transient expression and flux changes during a shift from high to low riboflavin production in continuous cultures of Bacillus subtilis. Biotechnology and Bioengineering 89, 219–232 (2005). [DOI] [PubMed] [Google Scholar]
- Liu L., Martinez J. L., Liu Z., Petranovic D. & Nielsen J. Balanced globin protein expression and heme biosynthesis improve production of human hemoglobin in Saccharomyces cerevisiae. Metab Eng 21, 9–16 (2014). [DOI] [PubMed] [Google Scholar]
- Xu P. et al. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat Commun 4, 1409–1416 (2013). [DOI] [PubMed] [Google Scholar]
- Ajikumar P. K. et al. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science 330, 70–74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juminaga D. et al. Modular engineering of L-tyrosine production in Escherichia coli. Appl Environ Microbiol 78, 89–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos C. N., Xiao W. H. & Stephanopoulos G. Rational, combinatorial, and genomic approaches for engineering L-tyrosine production in Escherichia coli. Proc Natl Acad Sci USA 109, 13538–13543 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger B. F., Pitera D. J., Smolke C. D. & Keasling J. D. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat biotechnol 24, 1027–1032 (2006). [DOI] [PubMed] [Google Scholar]
- Xie L., Eiteman M. A. & Altman E. Production of 5-aminolevulinic acid by an Escherichia coli aminolevulinate dehydratase mutant that overproduces Rhodobacter sphaeroides aminolevulinate synthase. Biotechnol Lett 25, 1751–1755 (2003). [DOI] [PubMed] [Google Scholar]
- Kang Z. et al. Small RNA regulators in bacteria: powerful tools for metabolic engineering and synthetic biology. Appl Microbiol Biotechnol 98, 3413–3424 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang F., Carothers J. M. & Keasling J. D. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol 30, 354–359 (2012). [DOI] [PubMed] [Google Scholar]
- Mauzerall D. & Granick S. The occurrence and determination of δ-aminolevulinic acid and porphobilinogen in urine. J Biol Chem 219, 435–446 (1956). [PubMed] [Google Scholar]
- Sassa S. Delta-aminolevulinic acid dehydratase assay. Enzyme 28, 133–145 (1982). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information