Abstract
Previous reports showed that decreased histone deacetylase activity significantly potentiated the rewarding effects of psychostimulants, and that encoding of the 5-HT3 receptor by the htr3a gene was related to ethanol-seeking behavior. However, the effects of a histone deacetylase inhibitor on ethanol-seeking behavior and epigenetic regulation of htr3a mRNA expression after chronic ethanol exposure are not fully understood. Using quantitative reverse transcription-polymerase chain reaction and chromatin immunoprecipitation analysis, we investigated the effects of chronic ethanol exposure and its interaction with a histone deacetylase inhibitor on histone-acetylation-mediated changes in htr3a mRNA expression in the htr3a promoter region. The conditioned place preference procedure was used to evaluate ethanol-seeking behavior. Chronic exposure to ethanol effectively elicited place conditioning. In the prefrontal cortex, the acetylation of H3K9 and htr3a mRNA expression in the htr3a promoter region were significantly higher in the ethanol group than in the saline group. The histone deacetylase inhibitor sodium butyrate potentiated the effects of ethanol on htr3a mRNA expression and enhanced ethanol-induced conditioned place preferences. These results suggest that ethanol upregulates htr3a levels through mechanisms involving H3K9 acetylation, and that histone acetylation may be a therapeutic target for treating ethanol abuse.
Keywords: Ethanol seeking, chronic ethanol exposure, htr3a, histone deacetylase, histone acetylation, sodium butyrate, neural regeneration
Abbreviations
EtOH, ethanol; PFC: prefrontal cortex; HAT, histone acetyltransferase; HDACs: histone deacetylases; CPP: conditioned place preference; CREB: cAMP-responsive element binding protein; SB, sodium butyrate
INTRODUCTION
Ethanol (EtOH) addiction is a debilitating psychiatric disorder. It is characterized by compulsive EtOH seeking and taking despite severe adverse consequences[1]. It is a complex disorder that involves multiple genes in key reward regions, such as the prefrontal cortex (PFC), ventral tegmental area, and nucleus accumbens. Alterations in gene expression may contribute to the behavioral changes induced by EtOH and other psychoactive substances[1]. For example, the levels of the transcription factor ΔfosB dramatically increase in the nucleus accumbens following chronic cocaine exposure. As chronic exposure to cocaine also increases the sensitivity of the subject to the effects of cocaine, this indicates that ΔfosB may be involved in enhanced sensitivity to the effects of cocaine[2]. Similarly, evidence exists that increased expression of brain-derived neurotrophic factor in the ventral tegmental area and nucleus accumbens induces long-lasting cocaine seeking and a higher risk of relapse during withdrawal[3,4]. However, the underlying mechanisms by which chronic drug exposure promotes changes in gene expression and behavior are not fully understood.
Epigenetics is the study of the heritable changes in gene expression that occur in the absence of DNA sequence alterations[5]. Epigenetic mechanisms alter the genome following exposure to diverse environmental stimuli by altering chromatin structure on specific gene promoters. It can produce robust and often long-lasting changes in gene expression and behavior[6]. Molecular mechanisms of epigenetic regulation include histone modification (acetylation, methylation, and phosphorylation) and DNA methylation[7]. Histone acetylation is an important regulator of gene expression and may contribute to behavioral changes. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) are involved in histone acetylation. HATs can relax the chromatin structure and thereby promote gene expression. HDACs can pack DNA into more condensed chromatin, inhibit access of transcriptional factors, and decrease gene expression. HATs tend to be transcriptional activators, whereas HDACs tend to be transcriptional repressors[8,9].
In the conditioned place preference (CPP) procedure, an animal learns to associate the rewarding effects of drugs with a specific environment. It is an animal model of drug-seeking behavior[10]. In CPP, either systemic or local administration of non-specific HDAC inhibitors significantly potentiates the rewarding effects of cocaine[11]. Consistent with this, mice that are deficient in CREB-binding protein, a HAT, exhibit reduced histone acetylation on the ΔfosB promoter and reduced sensitivity to cocaine[12].
HDAC inhibitors have been found to regulate various genes during drug addiction, playing a major role in drug dependence[11,13]. Administration of sodium butyrate (SB), an HDAC inhibitor, enhanced alcohol-induced locomotor sensitization[14] and significantly potentiated the rewarding effects of cocaine[11]. Consistent with this, reducing histone acetylation by virally overexpressed HDACs in the nucleus accumbens significantly decreased cocaine place conditioning[12]. HDAC knockout mice displayed normal reward responses to initial cocaine exposure, but became hypersensitive if they were previously exposed to a chronic course of cocaine[6]. However, the roles of SB in EtOH-seeking behaviors and related epigenetic mechanism have not been identified.
EtOH-seeking behavior is related to htr3a encoding of the 5-HT3 receptor, which is an ion channel that exhibits strain-dependent and brain-region-specific expression within the central nervous system[15]. Alcohol potentiates 5-HT3-receptor-mediated fast excitatory neurotransmission and hence modulates dopamine release in the reward circuitry[16]. Systemic injection of a 5-HT3 receptor antagonist attenuated foot shock-induced reinstatement of alcohol seeking[17]. In Wistar rats, bilateral microinjection of a 5-HT3 receptor antagonist into the amygdala decreased alcohol drinking[18]. Therefore, histone acetylation may be one of the mechanisms by which EtOH exposure regulates the htr3a gene and EtOH-seeking behavior. To date, no study has examined the effects of HDAC inhibitors on htr3a expression in the PFC, which serves as a major brain region underlying EtOH-seeking behavior[19]. In the present study, we investigated the regulation of htr3a expression in the PFC of Wistar rats after chronic EtOH exposure, and the effects of HDAC inhibition on htr3a expression and EtOH-seeking behavior. We reported that there was a relationship between the level of htr3a expression in PFC and EtOH seeking after exposure to chronic EtOH or EtOH + SB. We also explored the relationship between the level of htr3a expression and H3K9 acetylation, which has been reported to chronically activate most genes, in the htr3a promoter region in the PFC after EtOH or EtOH + SB exposure.
RESULTS
Quantitative analysis of experimental animals
A total of 48 adult male Wistar rats were included in this study. They were equally and randomly divided into four groups according to the exposure conditions: EtOH, ethanol + SB (EtOH + SB), SB and saline groups. The saline group served as the control group. All 48 rats were included in the final analysis.
EtOH-seeking behavior
The rats in each group exhibited comparable baseline data (F(3, 44) = 1.592, P = 0.205). One-way analysis of variance showed that there was a statistically significant CPP in each of the four treatment groups (F(3, 44) = 20.258, P < 0.001). The data presented in Figure 1A show that the CPP scores in the EtOH and EtOH + SB groups were significantly higher than in the saline group (P < 0.01), and the CPP scores in the EtOH + SB group were significantly higher than in the EtOH group (P < 0.05). Moreover, there was no significant difference in CPP scores between the saline and SB groups.
Figure 1.
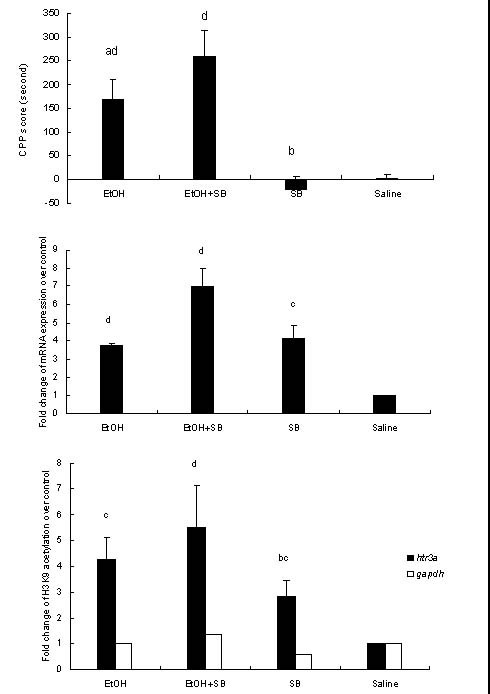
Interactions between the conditioned place preference (CPP) (A), htr3a mRNA expression level (B), and H3K9 acetylation in the htr3a promoter region induced by ethanol (EtOH) (C) and the effects of sodium butyrate (SB).
Data are expressed as mean ± SEM (n = 12 rats per group).
Significant differences among multiple groups were analyzed by one-way analysis of variance followed by least significance difference or Dunnett's post hoc test. aP < 0.05, bP < 0.01, vs. EtOH + SB group; cP < 0.05, dP < 0.01, vs. the saline group.
htr3a mRNA expression
One-way analysis of variance showed that there were significant differences in htr3a mRNA expression across the four treatment groups (F(3, 44) = 19.752, P < 0.001). As shown in Figure 1B, htr3a mRNA expression levels in the EtOH, EtOH + SB, and SB groups were significantly higher than in the saline group (P < 0.05 or 0.01). Although there was no significant difference in htr3a mRNA expression among the EtOH, EtOH + SB and SB groups, htr3a mRNA expression in the EtOH + SB group was higher than in the EtOH and SB groups.
Histone H3K9 acetylation in the htr3a and gapdh promoter regions
One-way analysis of variance showed that chronic EtOH, EtOH + SB, and SB exposure generally elicited significant increases in H3K9 acetylation in the htr3a promoter region (F(3, 44) = 5.618, P < 0.01). As shown in Figure 1C, H3K9 acetylation in the htr3a promoter region in the EtOH, EtOH + SB, SB groups was significantly higher than in the saline group (P < 0.05 or 0.01).
H3K9 acetylation in the htr3a promoter region was significantly higher in the EtOH + SB group than in the SB and saline groups (P < 0.01). The EtOH + SB treatment led to a greater increase of H3K9 acetylation than EtOH treatment alone, but this difference did not reach statistical significance. There was no significant difference in H3K9 acetylation in the gapdh promoter region among the four groups. This suggests that EtOH, EtOH + SB, and SB treatments specifically affect H3K9 acetylation in htr3a promoter region.
Correlation analyses between CPP scores and htr3a mRNA expression levels
Pearson's correlation test showed that htr3a mRNA expression levels in the PFC in the EtOH group and EtOH + SB group were positively correlated with CPP scores (r = 0.347, P = 0.033; Figure 2).
Figure 2.
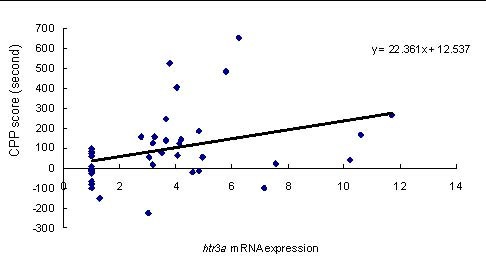
Correlation between conditioned place preference (CPP) scores and htr3a mRNA expression levels in both the ethanol (EtOH) and EtOH + sodium butyrate (SB) groups as assessed by Pearson's correlation test (n = 12 rats per group).
Correlation analyses between htr3a mRNA expression levels and H3K9 acetylation in htr3a promoter region.
Pearson's correlation test showed that htr3a mRNA expression levels in the PFC after any of the treatments were positively correlated with H3K9 acetylation in the htr3a promoter region (r = 0.434, P = 0.009; Figure 3).
Figure 3.
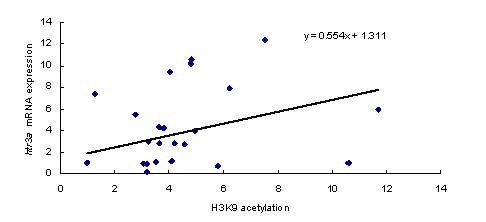
Correlation between htr3a mRNA expression levels and H3K9 acetylation in the htr3a promoter region as assessed by Pearson's correlation test. The pooled data from the four treatment groups were used for this analysis (n = 12 rats per group).
DISCUSSION
EtOH-induced histone acetylation
In the present study, we found that H3K9 acetylation was one of the mechanisms by which chronic EtOH exposure regulates htr3a expression in the PFC of Wister rats. A very consistent pattern was observed for EtOH, in which it induced htr3a mRNA expression and increased H3K9 acetylation in the htr3a gene promoter region, and these effects were synergistic with those of SB. There were no consistent results concerning EtOH-induced histone acetylation. Pandey et al[20] found increases in the acetylation of H3 and H4, the levels of cAMP-responsive element binding protein (CREB), and neuropeptide Y expression and a decrease in HDAC activity in amygdaloid brain regions of rats following acute EtOH exposure (1 g/kg, i.p.). Moreover, these effects were associated with anxiolytic effects. Furthermore, during withdrawal after chronic EtOH exposure, HDAC activity increased and acetylation of H3 and H4, the level CREB, and neuropeptide Y expression decreased in the amygdala. However, Renthal and Nestler[21] suggested that acute EtOH exposure may increase HDAC activity and thereby reduce histone acetylation, whereas withdrawal from chronic EtOH treatment may reduce HDAC activity and increase histone acetylation. Kim and Shukla[22] reported that alcohol had no significant effect on acetylation of H3 histone in the cerebral hemispheres. Another study showed that intermittent treatment with alcohol in adolescent rats significantly increased histone H3 and H4 acetylation in the frontal cortex[23]. These inconsistencies may be partially attributable to differences in animals with differential genetic vulnerabilities, differences in histone acetylation across different brain regions, or procedural differences across studies.
The cAMP-response element (CRE) sites are found within the regulatory (promoter or enhancer) regions of numerous genes[24]. CREB is ubiquitously expressed in brain cells and has shown to be involved in many aspects of central nervous system function including addiction, long-term memory formation, and synaptic plasticity[25,26]. In addition, CREB is activated by phosphorylation at serine-133 via calcium/calmodulin-dependent protein kinase II and IV, cAMP-dependent protein kinase A, mitogen-activated protein kinase[27,28], neurotransmitters, hormones, membrane depolarization, and growth and neurotrophic factors[29]. Activated CREB can recruit CREB binding protein[8,30], which functions both as a platform for recruiting other required components of the transcriptional machinery and as a HAT that alters chromatin structure, activates transcription, and finally promotes gene expression[30]. No single process that can account for the effects of EtOH. Instead, the effects of EtOH use many different mechanisms. EtOH has a complex pharmacological profile that includes effects such as modulation of 5-HT3 receptor currents to induce membrane depolarization[31] and activation of various signaling systems such as cAMP-dependent protein kinase A and calcium/calmodulin-dependent protein kinase II, which activate CREB[28]. Therefore, after EtOH exposure, activated CREB recruits CREB binding protein and increase histone acetylation. Genome-wide studies have shown that hyperacetylation in promoter regions is strongly associated with gene activation[9]. Taken together, these findings suggest that histone acetylation is associated with the mechanism through which EtOH upregulates htr3a expression.
HDAC modulates the behavioral effects of EtOH
In the present study, the HDAC inhibitor increased acetylation of histones that activate gene transcription. Earlier studies showed that manipulations that increased gene transcription generally promoted behavioral sensitivity to drugs of abuse[21]. CPP is widely used to assess EtOH-seeking behavior. To our knowledge, no previous report examined the effects of HDAC inhibitors on EtOH-induced CPP. Our results confirm and extend earlier reports that HDAC inhibitors modify the behavioral effects of EtOH. In the present study, we provided the first evidence that the HDAC inhibitor SB enhanced EtOH-induced CPP, whereas SB treatment alone did not affect this CPP. Although the effects of EtOH and EtOH + SB treatment on htr3a expression were not significantly different, there was a trend for EtOH + SB to synergistically facilitate htr3a expression. Our correlation analyses yield clear evidence for the behavioral effects of SB on EtOH-induced seeking behavior as SB administration. Taken together, these findings indicate that histone acetylation in the htr3a promoter region plays an important role in EtOH-seeking behavior.
Previous studies have demonstrated that SB administration enhanced EtOH-induced locomotor sensitization as well as cocaine- and morphine-induced locomotor sensitization and CPP[11,14]. Consistent with these results, reducing histone acetylation in the nucleus accumbens via virally overexpressing HDAC5 in the nucleus accumbens significantly decreased cocaine-induced CPP[6]. Mice that were deficient in CREB binding protein exhibited reduced histone acetylation and reduced sensitivity to cocaine[12]. Interestingly, some studies, as described below, suggested that treatment with the HDAC inhibitor trichostatin A reversed alcohol-related brain damage[32] and the effects of EtOH withdrawal on chromatin remodeling, neuropeptide Y expression, and anxiety-like behaviors[20], and it displayed a neuroprotective role[33]. Other investigators reported that SB facilitated extinction of either cocaine-[34] or morphine-induced[35] CPP. Different genes, different behaviors related to EtOH addiction, and different effects of various drugs of abuse may account for these inconsistencies. Despite these inconsistencies, histone acetylation provided a substrate for the long-term changes in gene expression underlying behavior changes. Accordingly, a better understanding of histone acetylation associated with EtOH exposure may provide insights into EtOH dependence and more effective treatments and preventions.
Our behavioral, gene expression and histone acetylation studies support the hypothesis that EtOH-induced histone acetylation underlies EtOH-seeking behavior. The results provide the first characterization of the alterations in htr3a mRNA expression and histone acetylation in the promoter region of htr3a in the PFC of Wistar rats after chronic EtOH exposure. We explored the effects of histone acetylation and the HDAC inhibitor SB on EtOH-seeking behavior. Although our understanding of the molecular mechanism underlying EtOH dependence remains incomplete, the identification of histone acetylation as an important mediator of EtOH-induced transcriptional and behavioral changes represents an exciting new area of research with potential therapeutic implications. The ability to reverse epigenetic alterations offers an approach that may profit the development of more effective treatments for EtOH dependence, through both direct targeting of aberrant chromatin acetylation and the identification of genes involved in EtOH dependence.
In conclusion, the results illustrate that EtOH upregulates htr3a mRNA expression through mechanisms that involve H3K9 acetylation, which indicates that histone acetylation may be a therapeutic target for EtOH abuse.
MATERIALS AND METHODS
Design
A randomized, controlled animal experiment.
Time and setting
This study was performed at the Key Psychiatric Laboratory of the Second Affiliated Hospital of Xinxiang Medical University, China between April 2010 and February 2011.
Materials
We examined 48 male adult inbred Wistar rats weighing 180–220 g. These animals were provided by the Animal Center of the Second Affiliated Hospital of Xinxiang Medical University, China (license No. SYXK (Yu) 2008-0105). All rats were group-housed with free access to water and food in the vivarium, which had a 12-hour light/dark cycle and a thermoregulated environment.
Methods
CPP, chronic EtOH exposure, and SB administration
The CPP apparatus consisted of a black and white chamber (60 cm × 30 cm × 30 cm) with a guillotine door. There was a smooth floor in the black chamber and a rough floor in the white chamber. The time a rat spent in each chamber was recorded by a video-tracking system (Anilab, Ningbo, China) through which the researchers could observe the animal's behaviors. All rats were allowed to adapt to the CPP apparatus for 60 minutes per day for 3 days. On day 4, the rats were placed in the center of the apparatus with the guillotine doors open. They were allowed free access to the entire apparatus for 15 minutes, and the time spent in each chamber was recorded to identify any innate preferences for one of the chambers (≥ 450 seconds). The less preferred chamber was used for the drug pairing, and the time spent in that chamber (< 450 seconds) was set as the baseline.
Similar to previous studies[36,37], experimental groups were treated with EtOH (0.5 g/kg per day, 10% w/v, Tianjin, China), EtOH + SB (200 mg/kg per day, Tianjin, China) (EtOH first and SB later), SB (200 mg/kg per day) or saline (10 mg/kg per day) via intraperitoneal injections. Each treatment was administered four times per day at 3-hour intervals from 08:00 to 17:00 for 15 days. Animals received EtOH, EtOH + SB, or SB in the morning and saline in the afternoon or saline in the morning and EtOH, EtOH + SB, or SB in the afternoon on alternating days. Rats receiving chronic EtOH, EtOH + SB, or SB treatments were confined in the drug-paired compartment for 60 minutes immediately after the injections, and in the saline-paired compartment immediately after the saline injection. The saline-treated rats received saline injections four times per day at 3-hour intervals from 08:00 to 17:00 for 15 days and were placed in the compartments randomly. During these sessions, the removable wall was inserted to separate the two compartments. The CPP test began 24 hours later after the last conditioning trial for all animals and lasted 15 minutes. The time spent in the drug-paired chamber during the CPP test was defined as the CPPtest and the CPP baseline was defined as the CPPbaseline. The CPP score was calculated as the CPPtest minus the CPPbaseline. A positive score was interpreted as a result of the rewarding effects of EtOH on associative learning[38], and it was used to evaluate EtOH seeking after chronic EtOH exposure.
Quantitative reverse transcription-PCR (qRT-PCR) for htr3a mRNA expression
After the CPP test, the rats were anaesthetized with napental (30–40 mg/kg, i.p.), and the PFC was removed following decapitation by gross dissection and washed with ice-cold PBS. RNA was extracted using a Trizol reagent purchased from Invitrogen (Shanghai, China) and precipitated with isopropanol. The purified RNA was DNase treated. The mRNA was reverse transcribed to cDNA using first-strand synthesis kits (Invitrogen). The following primers were employed to amplify specific cDNA regions of the transcriptions of interest: htr3a: 5’-GCT GGT GAC CGC CTG TAG CC-3’ and 5’-GCC GGC GGA TGA CCA CGT AG-3’, glyceraldehyde-3-phosphate dehydrogenase (gapdh) 5’-ACC ACA GTC CAT GCC ATC AC-3’ and 5’-TCC ACC ACC CTG TTG CTG TA-3’. The gapdh quantification was used as an internal control for normalization. The PCR cycling conditions were 95°C for 5 minutes, 35 cycles of 95°C for 30 seconds, 60°C for 45 seconds, 72°C for 45 seconds, and 72°C for 5 minutes. The products were stored at 4°C. The cDNA amplification reactions were run in triplicate in the presence of SYBR-Green. The CT values of each sample were recorded using FTC-2000 software (Shanghai, China). The fold change in mRNA levels over control values were calculated using the delta-delta method[39] for comparing relative expression results between different treatments.
The fold change of mRNA level over control was calculated as 2-ΔΔCT, whereinΔCT = CThtr3a–CTgapdh, ΔΔCT = ΔCTtreatment–ΔCTcontrol
Chromatin immunoprecipitation (ChIP) and qRT-PCR for H3K9 acetylation
The ChIP procedure was performed using the instructions provided with the ChIP kit (number 17-371, Millipore, Bedford, MA, USA) with a few modifications. The frozen PFC was sectioned and immediately cross-linked in formaldehyde for 10 minutes. Glycine was added to quench the cross-linked reaction. Subsequently, the cells were rinsed twice in ice-cold PBS containing a proteinase inhibitor, scraped, and collected by centrifugation. Cells were then resuspended in SDS lysis buffer. With the optimal conditions for sonication, the chromatin was sheared to 200–1 000 bp concentrating on 400–500 bp. The homogenates were centrifuged, and the supernatants were placed in fresh microfuge tubes and defined as the positive control (anti-RNA Polymerase II), the negative control (Normal Mouse IgG), and the anti-acetylation. ChIP dilution buffer containing a protease inhibitor was added to each tube to dilute the chromatin lysate. The chromatin solution was pre-cleared with salmon sperm DNA/protein G agarose, briefly centrifuged, and 10 μL of the supernatant was removed as the input for later normalization. The remaining supernatant was collected, and it was immunoprecipitated overnight at 4°C with an antibody directed against H3 acetylation on Lys9 (kit number 9671, Cell Signaling Technology, MA, USA), anti-RNA polymerase II, and normal mouse IgG. After immunoprecipitation, the DNA-histone complex with salmon sperm DNA/protein G agarose beads was collected. The beads were washed with low-salt buffer, high-salt buffer, LiCl buffer and TE buffer. The DNA-histone complex was eluted from the beads with elution buffer in fresh tubes. All tubes including the inputs and immunoprecipitates were incubated at 65°C for 5 hours. After addition of RNase A, the mixtures were incubated at 37°C for 30 minutes. After addition of proteinase K, 0.5 M ethylenediamine tetraacetic acid, and 1 M Tris-HCl, the mixtures were incubated at 45°C for 1 hour. The DNA associated with acetylation histones was purified and collected in an elution buffer. Most ChIP experiments were performed twice on two independent tissue samples for confirmation.
Quantification of DNA by qRT-PCR: Levels of specific histone acetylation at the htr3a and gapdh promoter regions were determined by measuring the amount of acetylated histone-associated DNA using qRT-PCR. Specific primers were designed to amplify proximal regions. For htr3a, the primers were 5’-TGT TTT CTG TGA AAG GAC TTG AGG-3’ and 5’-TCT CTG CAC TTT GAA TCT GTA GCC-3’. For gapdh, which was examined as a control, the primers were 5-CGT AGC TCA GGC CTC TGC GCC CTT-3’and 5’-CTG GCA CTG CAC AAG AAG ATG CGG CTG-3’. The PCR cycling conditions were 95°C for 5 minutes, 35 cycles of 95°C for 30 seconds, 55°C for 45 seconds, 72°C for 45 seconds, and 72°C for 5 minutes. The products were stored at 4°C. The input and immunoprecipitated DNA amplification reactions were repeated three times independently. Relative quantification of samples was performed using the delta-delta method as described by Godmann et al[40] with some modifications.
The fold change over control was calculated as 2-ΔΔCT, wherein ΔCT = CTanti-H3K9 acetylation–CTinput, ΔΔCT = ΔCTtreatment–ΔCTcontrol.
Several controls were performed to confirm the specificity and validity of these assays. Chromatin samples were immunoprecipitated with anti-RNA polymerase II as a positive control and normal mouse IgG as a negative control. To ensure that histone acetylation is specific for the htr3a promoter region, the levels of histone acetylation in the gapdh promoter region, which was examined to establish the specificity of the effects of the treatments were measured.
Statistical analysis
All data are shown as mean ± SEM, and were analyzed utilizing SPSS 15.0 statistical software (SPSS, Chicago, IL, USA). Significant differences among multiple groups were analyzed by one-way analysis of variance followed by least significant difference or Dunnett's post hoc test. Pearson's correlation test was used to conduct the correlation analyses between the CPP scores and htr3a mRNA expression levels and between the htr3a mRNA expression levels and the levels of H3K9 acetylation in the htr3a promoter region. Statistical significance level was accepted at P < 0.05.
Acknowledgments:
The authors wish to thank professor Yilang Tang from Department of Human Genetics at Emory University School of Medicine, Atlanta, GA 30322, USA, for his critical comments and suggestions.
Footnotes
Funding: This study was supported by the National Key Basic Research and Development Program (NKBRDP) of China (No. 2009CB522000), the National Natural Science Foundation of China (No. 30971050), the State Key Program of the National Natural Science of China (No. 81130020) and the Key Program on Basic Science of Henan Science and Technology Department (No. 094200510005).
Conflicts of interest: None declared.
Ethical approval: This study was approved by the Animal Ethics Committee, Central South University, China, and Animal Ethics Committee, Xinxiang Medical University, China.
(Edited by Jia SH, Duan HB/Song LP)
REFERENCES
- 1.Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–598. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- 2.Kelz MB, Chen J, Carlezon WA, Jr, et al. Expression of the transcription factor deltaFosB in the brain controls sensitivity to cocaine. Nature. 1999;401(6750):272–276. doi: 10.1038/45790. [DOI] [PubMed] [Google Scholar]
- 3.Lu L, Dempsey J, Liu SY, et al. A single infusion of brain-derived neurotrophic factor into the ventral tegmental area induces long-lasting potentiation of cocaine seeking after withdrawal. J Neurosci. 2004;24(7):1604–1611. doi: 10.1523/JNEUROSCI.5124-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graham DL, Edwards S, Bachtell RK, et al. Dynamic BDNF activity in nucleus accumbens with cocaine use increases self-administration and relapse. Nat Neurosci. 2007;10(8):1029–1037. doi: 10.1038/nn1929. [DOI] [PubMed] [Google Scholar]
- 5.Roth TL, Sweatt JD. Regulation of chromatin structure in memory formation. Curr Opin Neurobiol. 2009;19(3):336–342. doi: 10.1016/j.conb.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renthal W, Maze I, Krishnan V, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56(3):517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 7.Cheung P, Lau P. Epigenetic regulation by histone methylation and histone variants. Mol Endocrinol. 2005;19(3):563–573. doi: 10.1210/me.2004-0496. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh J, Gage FH. Chromatin remodeling in neural development and plasticity. Curr Opin Cell Biol. 2005;17(6):664–671. doi: 10.1016/j.ceb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Pokholok DK, Harbison CT, Levine S, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122(4):517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 10.Groblewskia PA, Frankena FH, Cunningham CL. Inhibition of extracellular signal-regulated kinase (ERK) activity with SL327 does not prevent acquisition, expression, and extinction of ethanol-seeking behavior in mice. Behav Brain Res. 2011;217:399–407. doi: 10.1016/j.bbr.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar A, Choi KH, Renthal W, et al. Chromatin remodeling is a key mechanism underlying cocaine- induced plasticity in striatum. Neuron. 2005;48(2):303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 12.Levine AA, Guan Z, Barco A, et al. CREB-binding protein controls response to cocaine by acetylating histones at the fosB promoter in the mouse striatum. Proc Natl Acad Sci U S A. 2005;102(52):19186–19191. doi: 10.1073/pnas.0509735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalda A, Heidmets LT, Shen HY, et al. Histone deacetylase inhibitors modulates the induction and expression of amphetamine-induced behavioral sensitization partially through an associated learning of the environment in mice. Behav Brain Res. 2007;181(1):76–84. doi: 10.1016/j.bbr.2007.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchis-Segura C, Lopez-Atalaya JP, Barco A. Selective boosting of transcriptional and behavioral responses to drugs of abuse by histone deacetylase inhibition. Neuropsychopharmacology. 2009;34(13):2642–2654. doi: 10.1038/npp.2009.125. [DOI] [PubMed] [Google Scholar]
- 15.Chu LF, Liang DY, Li X, et al. From mouse to man: the 5-HT3 receptor modulates physical dependence on opioid narcotics. Pharmacogenet Genomics. 2009;19(3):193–205. doi: 10.1097/FPC.0b013e328322e73d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enoch MA, Gorodetsky E, Hodgkinson C, et al. Functional genetic variants that increase synaptic serotonin and 5-HT3 receptor sensitivity predict alcohol and drug dependence. Mol Psychiatry. 2011;16(11):1139–1146. doi: 10.1038/mp.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le AD, Funk D, Harding S, et al. Effects of dexfenfluramine and 5-HT3 receptor antagonists on stress-induced reinstatement of alcohol seeking in rats. Psychopharmacology (Berl) 2006;186(1):82–92. doi: 10.1007/s00213-006-0346-y. [DOI] [PubMed] [Google Scholar]
- 18.Dyr W, Kostowski W. Evidence that the amygdala is involved in the inhibitory effects of 5-HT3 receptor antagonists on alcohol drinking in rats. Alcohol. 1995;12(4):387–391. doi: 10.1016/0741-8329(95)00023-k. [DOI] [PubMed] [Google Scholar]
- 19.Wolstenholme JT, Warner JA, Capparuccini MI, et al. Genomic analysis of individual differences in ethanol drinking: evidence for non-genetic factors in C57BL/6 mice. PLoS One. 2011;6(6):e21100. doi: 10.1371/journal.pone.0021100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandey SC, Ugale R, Zhang H, et al. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008;28(14):3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renthal W, Nestler EJ. Epigenetic mechanisms in drug addiction. Trends Mol Med. 2008;14(8):341–350. doi: 10.1016/j.molmed.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JS, Shukla SD. Acute in vivo effect of ethanol (binge drinking) on histone H3 modifications in rat tissues. Alcohol Alcohol. 2006;41(2):126–132. doi: 10.1093/alcalc/agh248. [DOI] [PubMed] [Google Scholar]
- 23.Pascual M, Boix J, Felipo V, et al. Repeated alcohol administration during adolescence causes changes in the mesolimbic dopaminergic and glutamatergic systems and promotes alcohol intake in the adult rat. J Neurochem. 2009;108(4):920–931. doi: 10.1111/j.1471-4159.2008.05835.x. [DOI] [PubMed] [Google Scholar]
- 24.Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28(8):436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Pandey SC. The gene transcription factor cyclic AMP-responsive element binding protein: role in positive and negative affective states of alcohol addiction. Pharmacol Ther. 2004;104(1):47–58. doi: 10.1016/j.pharmthera.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89(1):121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koob GF, Sanna PP, Bloom FE. Neuroscience of addiction. Neuron. 1998;21(3):467–476. doi: 10.1016/s0896-6273(00)80557-7. [DOI] [PubMed] [Google Scholar]
- 28.Moonat S, Starkman BG, Sakharkar A, et al. Neuroscience of alcoholism: molecular and cellular mechanisms. Cell Mol Life Sci. 2010;67(1):73–88. doi: 10.1007/s00018-009-0135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheng M, McFadden G, Greenberg ME. Membrane depolarization and calcium induce c-fos transcription via phosphorylation of transcription factor CREB. Neuron. 1990;4(4):571–582. doi: 10.1016/0896-6273(90)90115-v. [DOI] [PubMed] [Google Scholar]
- 30.Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42(6):961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkins A, Franks NP, Lieb WR. Actions of general anaesthetics on 5-HT3 receptors in N1E-115 neuroblastoma cells. Br J Pharmacol. 1996;117(7):1507–1515. doi: 10.1111/j.1476-5381.1996.tb15314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishii T, Hashimoto E, Ukai W, et al. Epigenetic regulation in alcohol-related brain damage. Nihon Arukoru Yakubutsu Igakkai Zasshi. 2008;43(5):705–713. [PubMed] [Google Scholar]
- 33.Agudelo M, Gandhi N, Saiyed Z, et al. Effects of alcohol on histone deacetylase 2 (HDAC2) and the neuroprotective role of trichostatin A (TSA) Alcohol Clin Exp Res. 2011;35(8):1550–1556. doi: 10.1111/j.1530-0277.2011.01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malvaez M, Sanchis-Segura C, Vo D, et al. Modulation of chromatin modification facilitates extinction of cocaine-induced conditioned place preference. Biol Psychiatry. 2010;67(1):36–43. doi: 10.1016/j.biopsych.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang R, Zhang Y, Qing H, et al. The extinction of morphine-induced conditioned place preference by histone deacetylase inhibition. Neurosci Lett. 2010;483(2):137–142. doi: 10.1016/j.neulet.2010.07.080. [DOI] [PubMed] [Google Scholar]
- 36.Bie B, Zhu W, Pan ZZ. Ethanol-induced delta-opioid receptor modulation of glutamate synaptic transmission and conditioned place preference in central amygdala. Neuroscience. 2009;160(2):348–358. doi: 10.1016/j.neuroscience.2009.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Febo M, Akbarian S, Schroeder FA, et al. Cocaine- induced metabolic activation in cortico-limbic circuitry is increased after exposure to the histone deacetylase inhibitor, sodium butyrate. Neurosci Lett. 2009;465(3):267–271. doi: 10.1016/j.neulet.2009.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rezayof A, Zarrindast MR, Sahraei H, et al. Involvement of dopamine receptors of the dorsal hippocampus on the acquisition and expression of morphine-induced place preference in rats. J Psychopharmacol. 2003;17(4):415–423. doi: 10.1177/0269881103174005. [DOI] [PubMed] [Google Scholar]
- 39.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Godmann M, May E, Kimmins S. Epigenetic mechanisms regulate stem cell expressed genes Pou5f1 and Gfra1 in a male germ cell line. PLoS One. 2010;5(9):e12727. doi: 10.1371/journal.pone.0012727. [DOI] [PMC free article] [PubMed] [Google Scholar]


