Abstract
A survivin small interfering RNA sequence specific for a human and mouse homogenous sequence was constructed. Survivin small interfering RNA could significantly inhibit glioma cell proliferation and induce apoptosis when it was transfected into either a human glioma cell line U251 or rat glioma C6 cells in vitro. In addition, treatment of rat orthotopic glioma models with survivin small interfering demonstrated the inhibition of glioma growth in vivo. Our experimental findings suggest that the use of RNA interference techniques to target the survivin sequence may be useful in the treatment of glioma.
Keywords: RNA interference, survivin, glioblastoma, apoptosis
Abbreviations:
siRNA, small interfering RNA; HE, hematoxylin-eosin; MTT; methylthiazolyldiphenyl-tetrazolium bromide; AO/EB, acridine orange/ethidium bromide
INTRODUCTION
Glioma is the most aggressive cancer among human malignancies and can develop radiotherapy and chemotherapy resistance. The median survival is less than 1 year[1]. Recently, survivin has been characterized as an important marker in glioma initiation and development. Survivin is of particular interest, as a large comparative analysis of human transcripts in malignant tumors and normal tissues identified it as the fourth-most highly expressed gene in tumors. Survivin is also highly expressed in gliomas[2] and its gene is located on chromosome 17q25[3], which is expressed at high levels in fetal tissue but absent in most normal differentiated cells in adults[4]. The increased expression of survivin in cell nuclei appears to be associated with the aggressive nature and/or poor prognosis of cancer[1]. Survivin mediates its effects by strengthening the survival of tumor cells primarily through its anti-apoptotic function[5].
It is specifically expressed in carcinomas and developing cells, which undergo either inappropriate or programmed cell growth by binding to mitotic spindle fibers. In addition, it causes cells to enter the S phase of the cell cycle and controls apoptosis by inhibiting apoptosis pathways[6]. Survivin is an important oncogene for promoting glioblastoma tumor proliferation, and its inhibition in glioma cells can decrease cell growth and induce cell cycle arrest.
Moreover, survivin is a downstream target of Src kinase and Stat3 activity in different types of cancer. Several reports have shown that survivin mRNA and protein expression levels are markedly increased in gliomas as measured by reverse transcription-PCR, western blot assay, and immunohistochemistry. Moreover, survivin overexpression correlated with the pathologic grade and poor prognosis of this malignancy[7].
Intriguingly, survivin small interfering RNA (siRNA) inhibits tumor growth and induces apoptosis of prostate cancer, lung cancer and bladder cancer cells[8,9]. However, currently there is little evidence addressing the effect of survivin siRNA on glioblastoma growth in vitro and in vivo. A number of studies have documented the increased expression of cytoplasmic and nuclear survivin in human gliomas, and immunohistochemistry and western blot analysis showed that the increased expression in gliomas correlated with an increased pathological grade of tumor[10,11].
In addition, glioma patients with detectable survivin expression measured by western blot analysis have a significantly shorter overall survival rate compared with those without detectable expression[12]. This evidence suggests that survivin might be involved in tumorigenesis and the progression of glioblastoma.
Survivin is a promising target for glioblastoma therapy as it is a convergence point for several signaling pathways that promote glioblastoma growth and maintenance. In addition, it induces the expression of genes that regulate anti-apoptotic pathways and proliferation, such as vascular endothelial growth factor, c-Myc, caspase-3, caspase-7 and cyclin D1[13,14]. The anti-apoptotic molecule Bcl-2 utilizes multiple pathways to inhibit death signals in cancerous cells[15,16]. One of these mechanisms involves the up-regulation of survivin, which acts on caspases directly and mainly suppresses caspase-3 and caspase-7 activity[17].
Disruption of survivin microtubule interactions could result in the loss of its anti-apoptotic function and thus increase caspase-3 activity. Furthermore, survivin also mediates an anti-apoptotic effect through the inhibition of the second mitochondria derived apoptosis inducing factor Smac/DIABLO[18]. Thus, silencing survivin may promote glioma death through different signal pathways[19]. RNA interference is an evolutionarily conserved, post-transcriptional gene-silencing mechanism wherein a double-stranded siRNA directs the sequence specific degradation of its target mRNA[20]. Therefore, the use of siRNA to suppress survivin expression may be a promising treatment for gliomas.
In this study, we observed the expression of survivin and its downstream products, including Bcl-2, cyclin D1, c-Myc and caspase-3 in human U251 and rat C6 cell lines, and explored the biological effect of survivin down-regulation in a rat tumor-transplantation models.
RESULTS
Survivin siRNA inhibited U251 and C6 cell growth and induced apoptosis in vitro
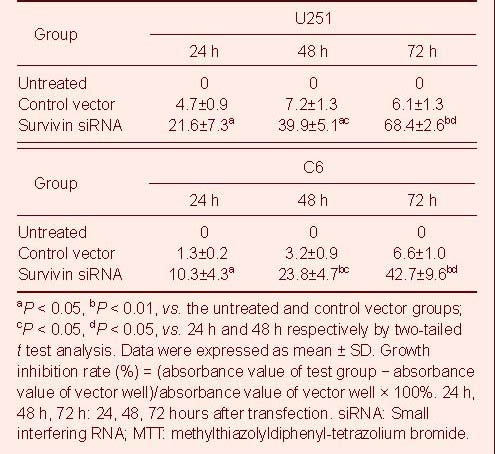
Treatment with survivin siRNA induced a time-dependent inhibition of U251 and C6 cell growth, whereas no inhibitory effect was observed in untreated cells and cells transfected with the vector siRNA plasmid as measured by 3-(4,5-dmethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Table 1).
Table 1.
The growth inhibition (%) effect of survivin siRNA on U251 cells and C6 cells by MTT
Flow cytometry analysis and acridine orange/ethidium bromide (AO/EB) staining (nucleus condensation) were used to determine apoptosis in U251 and C6 cells (Table 2, Figure 1). The apoptosis rate in survivin siRNA-treated cells was significantly higher compared with the untreated and vector siRNA-treated U251 and C6 cells (P < 0.01 or P < 0.05). Although the apoptosis rate in the vector-treated U251 and C6 cells was slightly higher than that in the untreated cells it was not statistically significant (Table 2). This suggested that survivin siRNA induced the apoptosis of glioma cells. Moreover, AO/EB staining revealed that both early apoptotic and late apoptotic cells were seen in the group treated with survivin siRNA (Figure 1).
Table 2.
Flow cytometry analysis of the apoptosis (%) induced by survivin siRNA on U251 cells
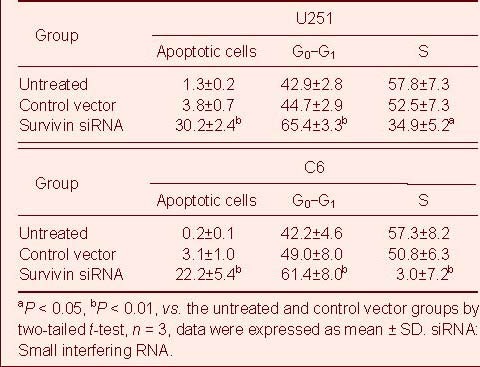
Figure 1.
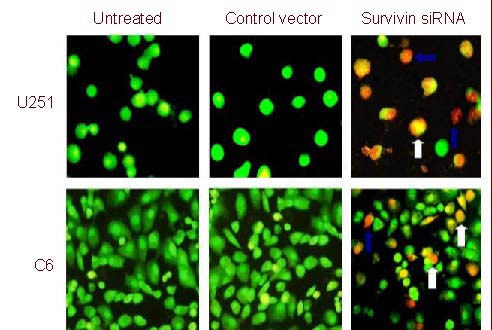
Representative fluoromicrographs of apoptosis detected by acridine orange/ethidium bromide assay in U251 and C6 cells (× 200). White arrows indicate early stage apoptotic cell and blue arrows indicate late stage apoptotic cell. siRNA: Small interfering RNA.
Survivin siRNA effectively reduced Bcl-2, cyclin D1, c-Myc expression and promoted caspase-3 expression in U251 cells
Survivin expression levels have been shown to be significantly higher in glioblastoma cells compared with normal cells[10], so we attempted to determine whether synthetic survivin siRNA could inhibit the expression of survivin protein in U251 cells. We observed a significant decrease of survivin protein levels in U251 cells transfected with siRNA-survivin (Figure 2A) compared with untreated or vector-treated U251 cells. Survivin expression is specifically targeted by survivin siRNA, since survivin siRNA did not change the expression of β-actin. Survivin suppresses the expression of caspase-3, which promotes apoptosis, and induces the expression of Bcl-2, an anti-apoptotic protein, and cyclin D1 and c-Myc, which promote cell division. To determine the expression of these gene products, western blot analysis was performed using protein isolated from U251 cells transfected with survivin siRNA. Western blots and quantitative analysis relative to β-actin levels showed that intracellular Bcl-2, cyclin D1 and c-Myc levels were significantly decreased in survivin siRNA-transfected U251 cells, whereas caspase-3 expression levels significantly increased (Figure 2) compared with control groups. Thus, survivin siRNA down-regulated the expression of Bcl-2, cyclin D1 and c-Myc, but up-regulated caspase-3. Interestingly, these factors are known to be associated with apoptosis or stimulation of cell cycle progression. Thus, the survivin siRNA construct inhibited growth and survival, by inducing apoptosis and G1 arrest of U251 cells in vitro.
Figure 2.
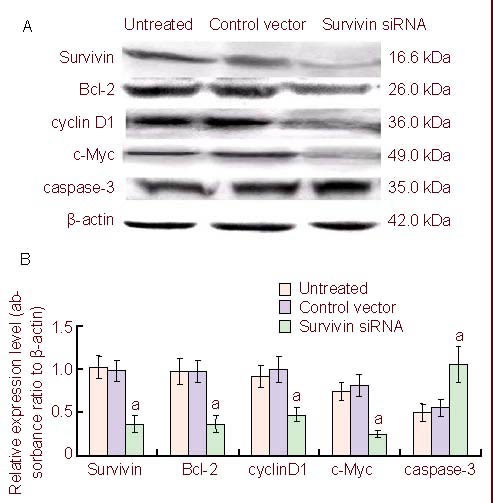
Western blot analysis of proteins expression in U251 cells.
(A) Survivin and its regulatory proteins expression in U251 cells in the untreated, control vector, and survivin siRNA groups at 48 hours after transfection.
(B) Amounts of survivin and its regulatory proteins (Bcl-2, cyclin D1, c-Myc and caspase-3) in U251 cells transfected with survivin siRNAs.
Data are expressed as mean ± SD in three independent experiments. aP < 0.05, vs. untreated and control vector groups (two-tailed t-test). siRNA: Small interfering RNA.
Quantitative analysis of experimental animals
Thirty rats were used for the development of a cerebral glial tumor model, by intracranial injection of C6 cells. Following injection of the C6 cells rats were randomly divided into three groups: untreated group (injected with phosphate buffered saline (PBS) only); control vector group (transfected with blank siRNA plasmid); and survivin siRNA group (transfected with survivin siRNA plasmid). At 21 days post-transplantation, the tumor volume was measured by CT, and the development and growth of C6 cells following different treatments were determined by hematoxylin and eosin (HE) staining, terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) assay and immunohistochemistry.
Survivin siRNA inhibited C6 glioma growth in vivo
To evaluate the effects of survivin siRNA on glioma growth intracranially, we examined its antitumor efficacy using a rat model. Adult rats were intracranially inoculated with 2 × 105 C6 cells. Seven days after C6 glioma transplants, rats treated with PBS had a median survival of 18.4 ± 9.51 days (n = 10) with the longest survival of 36 days, and rats treated with scrambled siRNA had a median survival of 20.1 ± 8.50 days (n = 10) with the longest survival of 32 days. There was no statistical significance between the scrambled control group and PBS group. Intracranial therapy with survivin siRNA significantly improved the survival rate (median survival 31.9 ± 6.04 days, P < 0.01) compared with both controls and with the longest survival of 41 days. The size of C6 gliomas in rats treated with survivin siRNA analyzed by CT scan was 22.8 ± 5.32 mm3 at 21 days post-transplantation, which was significantly lower compared with the PBS group and scrambled siRNA group (42.1 ± 8.08 mm3, 44.2 ± 6.27 mm3, respectively. P < 0.05; Figure 3). Thus, a more pronounced suppression of intracranial C6 glioma growth was observed in survivin siRNA-treated rats.
Figure 3.
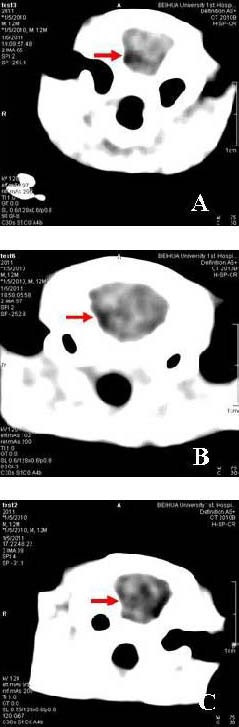
Photomicrographs of rat brains treated with survivin siRNA resulting in significant inhibition of tumor growth through computed tomography scan. Red arrows represent glioblastoma.
The rat C6 glioma treated with phosphate-buffered saline (A), scrambled siRNA (B), and survivin siRNA (C) by day 21.
Compared with untreated and control vector groups, survivin siRNA treatment resulted in obvious growth inhibition of giloma cells. siRNA: Small interfering RNA.
In vitro experiments demonstrated reduced survivin expression levels in U251 and C6 cells treated with survivin siRNA and significant inhibition of cell growth (Table 1; Figure 1). To evaluate the antitumor effect in vivo, C6 glioma cells were inoculated intracranially, thus producing carcinoma in situ, and then treated with survivin siRNA and controls, respectively. Tumors in situ were removed and analyzed for survivin levels. Western blot analysis showed that survivin protein levels were significantly reduced in the survivin siRNA group, confirming survivin siRNA-transfection of the glioma cells. Approximately 2–2.5 folds decrease of survivin protein levels were observed in the survivin siRNA treatment group compared with controls (Figure 4).
Figure 4.
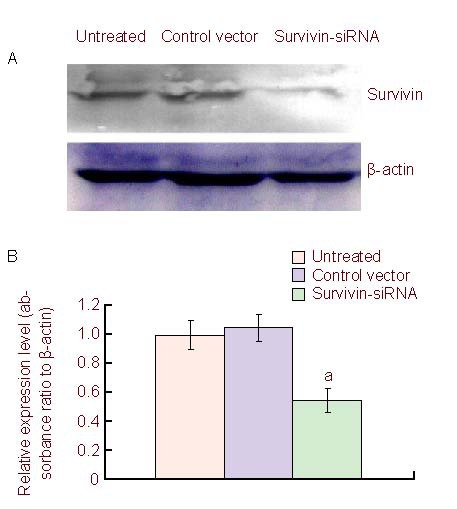
Western blot analysis of survivin protein level from C6 xenograft.
(A) Survivin protein expression level in the untreated, control vector, and survivin siRNA groups from xenograft.
(B) The relative amounts of survivin after treatment, data are expressed as mean ± SD in three independent experiments. aP < 0.05, vs. untreated and control vector groups (two-tailed t test). siRNA: Small interfering RNA.
To determine the mechanism underlying tumor growth inhibition in vivo, C6 tumors treated with different vectors were excised for HE staining and analyzed using TUNEL assay and Ki67 immunohistochemistry. HE staining showed that the cells proliferated actively in the mock and empty vector treatment group, while glioma growth was significantly inhibited in the survivin siRNA-treated group (Figures 5A–C). TUNEL staining indicated that a large number of apoptotic cells were present in the survivin siRNA group compared with controls (Figures 5D–F). The immunohistochemistry results showed that numbers of Ki67+ cells were markedly reduced in the survivin siRNA-treated group compared with controls (Figures 5G–I).
Figure 5.
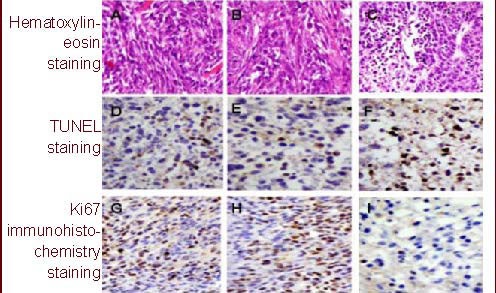
Hematoxylin-eosin staining, terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling and Ki67 immunohistochemistry analysis for rat glioma (× 100).
(A, D, G) In the untreated groups, the tumors grow very strong and apoptosis cells are little.
(B, E, H) In control vector group, apoptotic cells are also little, Ki67 positive cells are everywhere.
(C, F, I) In survivin small interfering RNA group, apoptotic cells increase significantly while Ki67 immuno-positive cells are reduced greatly. Positive cells: brown.
DISCUSSION
To determine the exact role of survivin in gliomas, we used RNA interference to specifically inhibit the expression of survivin in U251 cells. We found that survivin was highly expressed in U251 cells, and that its expression could be significantly decreased when cells were transfected with a survivin siRNA plasmid. To determine whether synthetic survivin siRNA had an inhibitory effect on human U251 and rat C6 cell growth, we determined cell proliferation by MTT assay. The survival of both cell types was significantly reduced compared with controls following survivin siRNA transfection. Moreover, quantification of apoptotic cells by flow cytometry assay determined that survivin siRNA significantly increased the number of apoptotic cells.
Additionally, visualization of nucleus condensation using AO/EB dyes suggested that the synthetic survivin siRNA has effects on both early and late apoptosis. Western blot analysis showed that survivin siRNA could inhibit the protein levels of survivin, Bcl-2, cyclin D1 and c-Myc and promote caspase-3 levels in U251 cells.
This study also demonstrated that the synthetic survivin siRNA effectively suppressed U251 cell growth as evidenced by the reduced growth rate of U251 cells. Thus, reduced survivin expression could inhibit the downstream gene expression of cyclin D1 and c-Myc, which would induce cell cycle arrest and slow cell proliferation, overall reducing glioma cell survival[21,22,23,24].
This study showed that survivin siRNA treatment could cause long-term target gene inhibition, leading to growth suppression and induction of apoptosis in U251 and C6 cells. Treatment of U251 cells with survivin siRNA resulted in morphologic and biochemical changes indicating apoptosis. However, previous studies have reported that U251-MG glioma cells may also undergo caspase-dependent cell death upon survivin RNAi-treatment[25,26]. In this study, when U251 cells were transfected with survivin siRNA, a caspase dependent apoptosis pathway was observed. To evaluate the effects of survivin siRNA on glioblastoma growth in vivo, we examined the antitumor efficacy of survivin siRNA in a rat tumor model. CT scan analysis demonstrated that survivin siRNA-treatment delayed glioma growth and significantly prolonged the life span of rats inoculated with glioma cells. Tumors isolated from survivin siRNA-treated rats demonstrated reduced survivin expression, confirming that survivin siRNA had been successfully transfected into glioma cells. Additionally, massive apoptosis of the tumor cells was detected by TUNEL, Ki67 immunohistochemistry and HE staining assays in survivin siRNA-treated rats. These data suggested that survivin siRNA injection into the tumor could exert significant antitumor effects in vivo similar with those in vitro. Therefore, survivin is an ideal target for cancer therapy.
In conclusion, data presented in the current study show that the blockade of survivin signaling using RNA interference significantly suppressed survivin expression in vitro and in vivo, suggesting that survivin signaling is a potential molecular target for glioblastoma therapy. Plasmid based siRNA therapy for tumor suppression may offer an effective and inexpensive approach for the treatment of glioblastoma.
MATERIALS AND METHODS
Design
A randomized, controlled study in vivo and parallel experiments in vitro.
Time and setting
All experimental procedures were performed in the Research Center of Molecular Medicine, Basic Medical College of Beihua University, China between October 2009 and June 2011.
Materials
Animals
Male Wistar cleaning rats, weighing 160–180 g were supplied by the Animal Experimental Center of Jilin University Bethune Medical College (license No. SCXK (Ji) 2003-0001). Rats were housed under standardized pathogen-free conditions allowing access to food and water ad libitum. All the experimental protocols were performed in accordance with the Guidance Suggestions for the Care and Use of Laboratory Animals, formulated by the Ministry of Science and Technology of China[26].
Cells
Human U251 cells and rat C6 cells[27,28] were supplied by the American Type Culture Collection (Manassas, Virginia, USA).
Methods
Construction of pSilence™ neo3.1-H1-Survivin siRNA
DNA templates for the synthesis of siRNAs were constructed under control of the H1 promoter in the plasmid pSilence™ neo3.1. First, the double stranded DNA template encoding survivin siRNA oligonucleotides (Gene Bank: access number for human survivin: NM-001168) that contained a sense strand of a 19 nucleotide sequence followed by a short space sequence (TTC AAG AGA), the reverse complement of the sense strand, and five thymidines as a RNA polymerase III transcriptional stop signal were synthesized. The forward sequence was 5’-GAT CCC GGA CCA CCG CAT CTC TAC ATT CAA GAG ATG TAG AGA TGC GGT GGT CCT TTT TGG AT-3’ and reverse sequence was 5’-AGC TATC CAA AAA GGA CCA CCG CAT CTC TAC ATC TCT TGA ATG TAG AGA TGC GGT GGT CCG G-3’. The survivin siRNA was specific for both human and rat survivin sequences. The oligonucleotides were annealed in a buffer (potassium acetate 100 mM, 30 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid-KOH pH 7.4, and magnesium acetate 2 mM) and the mixture was incubated at 90°C for 3 minutes and then at 37°C for 1 hour. The double stranded oligonucleotides were cloned into the pSilence™ neo3.1-H1 vector with BamHI and HindIII sites, where siRNA were expressed under the control of the H1 promoter.
Cell culture and transfection
U251 and C6 cells were cultured in Dulbecco's Modified Eagle's Medium (Invitrogen Inc, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (MO BIO Inc, Carlsbad, CA, USA), penicillin (100 kU/L) and streptomycin (100 mg/L) at 37°C in a humidified incubator with 5% CO2. Lipofectamine 2000 (Invitrogen Branch Company, Shanghai, China) was used for cell transfection as previously described[28]. The cells were cultured for 5-20 hours and then transferred to fresh medium with 10% fetal bovine serum and lysed for 24–72 hours after transfection.
In vitro proliferation and apoptosis assays
U251 and C6 cells were incubated in 96-well plates. Cell proliferation was determined by MTT assay (Sigma, Santa Clara, USA), whereby cell numbers were counted by hemocytometer (All-Time Commercial Co, Shanghai, China) at 24, 48 and 72 hours after transfection. The absorbance values at 570 nm were determined on a multi-well plate reader (Sunostik Medical Technology Co, Changchun, China). Inhibition of cell growth rate was calculated according to the following formula: growth inhibition rate (%) = (absorbance value of test group - absorbance value of vector well)/absorbance value of vector well × 100%.
For flow cytometry analysis of apoptosis, U251 and C6 cells were transfected with siRNA-survivin or siRNA-scramble plasmids. After 24 hours, cells were collected and washed with cold PBS containing 4 mM ethylene diamine tetraacetic acid (EDTA). Cells were fixed in 70% cold ethanol, collected by centrifugation, and washed once again with PBS/EDTA. Cells were resuspended in PBS/EDTA with 0.2% Triton X-100, 20 mL/L of propidium iodide (Sigma) and 40 mg/L RNase A (Biocolor BioScience & Technology Company, Shanghai, China), then the cells were incubated for 30 minutes at 4°C and analyzed by flow cytometry (FACScan, Becton Dickinson, Franklin Lakes, NJ, USA), using Cell Quest software (Becton, Dickinson and Company, Franklin, USA).
For fluoromicroscopic determination of positive apoptosis cells, 95 μL of suspension cells (approximately 1 × 105/μL) were mixed with 0.1% acridine orange/ethidium bromide (Sigma) and observed under a fluorescence microscope (Olympus Corporation, Tokyo, Japan).
Western blot analysis of U251 cells
Total protein was extracted from 1 × 106 harvested sample cells with protein lysis buffer (5 M EDTA, 300 mM NaCl, 0.5 mM NaF, 0.1% Igepal, 0.5 mM Na3VO4, 0.5 mM phenylmethanesulfonyl fluoride and antiprotease mixture) using the freeze-thaw method[29]. The lysates were centrifuged at 15 000 × g for 30 minutes. Determination of protein concentrations of the supernatants was performed by the Bradford procedure[30] (Bio-Rad Laboratory, Hercules, CA, USA). The rabbit anti-survivin, Bcl-2, cyclin D1, c-Myc and caspase-3 polyclonal antibodies were obtained from Santa Cruz Biotech Inc (Santa Cruz, CA, USA). For survivin, Bcl-2, cyclin D1, c-Myc and caspase-3 analysis, the supernatant containing 50 μg total protein was separated by electrophoresis on 10% SDS-polyacrylamide gels, transferred onto polyvinylidine fluoride membranes (Milipore, Bedford, MA, USA) and blocked with 5% nonfat dry milk in PBS with 0.1% Tween-20 (Sigma). Blots were incubated with specific antibodies against the above described antigens and rabbit anti-β-actin antibody (Santa Cruz) at 4°C overnight, washed with PBS containing 0.1% Tween-20 and subjected to corresponding horseradish peroxidase-conjugated mouse anti-rabbit secondary antibodies (1:2 000; Health Research (Shanghai) Biotechnology Co., Ltd., Shanghai, China) at room temperature for 2 hours. Blots were then washed with PBS and 0.1% Tween-20, and the absorbance was visualized using the enhanced chemiluminescence detection system (Amersham International, Buckinghamshire, United Kingdom).
Intracranial glioblastoma inoculation of rats
Thirty adult Wistar rats were anesthetized with 25% urethane solution (4 mL/kg) and their heads were shaved. C6 glioma cells were trypsinized and washed in complete medium followed by a PBS wash. 2 mm of the galea was dissected laterally, and a 3 mm burr hole was made over the left parietal bone with its center 2 mm posterior to the coronal suture and 2 mm lateral to the sagittal suture. Next, 2 × 105 cells (resuspended in 40 μL PBS) were implanted stereotactically into the rat brain through the coronal suture left of the midline, to a depth of 3 mm[24]. At 7 days after implantation, rats were treated with either mock (treated with PBS only), scrambled siRNA control (20 μg plasmid mixed with lipofectamine and without serum fresh medium, total volume 40 μL) or survivin siRNA (20 μg plasmid mixed with lipofectamine and fresh medium, total volume 40 μL). A volume of 40 μL solution was stereotactically implanted into the coronal suture left of the midline, to a depth of 3 mm. The skin was closed using surgical staples, and animals were returned to their cage and closely monitored daily. At 21 days after transplantation, the tumor volumes were monitored by CT and magnetic resonance imaging (supplementary Figure 1 online) and median survival and long-term survivors were considered as end points. The tumor volume was calculated as length × width × depth (mm3), as had been done for the subcutaneous tumors.
HE staining and TUNEL assays
Serial sections of tumor tissue excised from rats were fixed in formalin, stained with HE, and processed for routine histological examination. The TUNEL assay was performed using the in situ Cell Death Detection Kit (Roche, Switzerland), which relies on the TUNEL assay. Five-micrometer sections from paraffin-embedded tissues (tumor tissue) were dewaxed and hydrated according to the standard protocol. After incubation with proteinase K (200 μg/mL) at 21°C for 30 minutes, the TUNEL reaction mix containing DIG-dUTP, terminal deoxynucleotidyl transferase, and reaction buffer was added to the slides, and they were incubated in a humidified chamber for 2 hours at 37°C, followed by washing and incubation. Next, the slices were incubated with biotin anti-digoxin-specific antibody (1:100; Abcam Inc, Cambridge, UK). Diaminobenzidine staining was used to visualize the TUNEL-positive cells (brown color) by microscopy (Olympus, Tokyo, Japan).
Ki67 immunohistochemistry staining
Tumor cell proliferation was determined by digital image analysis using an Automated Cellular Imaging System (ACIS III®, DakoCytomation, San Juan Capistrano, CA, USA) using 5 μm-thick formalin-fixed, paraffin embedded sections. Detection of Ki-67 was determined by immunohistochemistry on formalin-fixed, paraffin embedded samples, which were deparaffinized with three changes of xylene and rehydrated in a series of alcohols (100%, 95%, 70%) and rinsed well in running distilled water. 5 μm slides were then placed in a preheated 0.1 mM EDTA, pH 8.0 retrieval buffer for 30 minutes and then cooled in the buffer for 5 minutes followed by a 5-minute rinse in running distilled water. After the heat-inactivated epitope retrieval step, sections were incubated with 3% H2O2 in ethanol for 5 minutes to inactivate endogenous peroxides. Sections were incubated in mouse anti-rat Ki67 (1:100; Yaji Biocompany, Shijiazhuang, Hebei Province, China) for 30 minutes at room temperature. Sections were rinsed with wash buffer. Secondary incubation with goat-anti mouse DUAL+/HRP-labeled polymer (1:1 000; K4061, DAKO, Carpenteria, CA, USA) for 15 minutes at room temperature was performed. The slides were rinsed with wash buffer. Sections were then incubated in 3,3’-diaminobenzidine (K3467, DAKO) for 5 minutes, counterstained with modified Schmidt's hematoxylin for 5 minutes, followed by a 3-minute tap water rinse to blue sections, dehydrated through graded alcohols, cleared in three changes of xylene and mounted using a permanent mounting media. The relative absorbance of the target protein was determined with an automatic imaging system (Beijing TOP Biotek Co., Ltd., Beijing, China).
Survivin expression in tumor-implanted brain
To analyze whether survivin played any role in tumor growth in vivo, western blot analysis for survivin protein levels was performed from brain tissues harvested from the injection site of C6 tumor-implanted rats. The expression of survivin was normalized to β-actin levels. Rabbit anti-mouse survivin antibody was obtained from Cell Signaling Company (USA) and goat anti rabbit IgG from Boaosen Company (Beijing, China). Blots were washed with TBS-1% Tween and visualized using a enhanced chemiluminescence detection system (Amersham International, Buckinghamshire, United Kingdom). The western blot products were analyzed by relative absorbance of target bands compared with β-actin and were quantified using Image Quant 5.0 software (Molecular Dynamics, Sunnyvale, CA, USA).
Statistical analysis
Data are expressed as mean ± SD. All statistical analysis were performed using SPSS 14.0 software (SPSS, Chicago, IL, USA). The two-tailed t-test was used to determine the mean differences between treatment and control. The level of significance was set at P < 0.05.
Acknowledgments:
We would like to thank Dr. Yan Dong from Tulane University for help with the manuscripts final modification.
Footnotes
Funding: The study was supported by the Natural Science Foundation of Jilin Province, No. 2011146, 2012150 and the Natural Science Foundation of Jilin Technology Division, No. 201133139 and 201032235.
Conflicts of interest: None declared.
Ethical approval: All the experimental protocols were approved by the Animals Ethics Committee for Animal Research, Beihua University, China.
Supplementary information: Supplementary data associated with this article can be found, in the online version, by visiting www.nrronline.org, and entering Vol. 7, No. 12, 2012 after selecting the “NRR Current Issue” button on the page.
(Edited by Song JN, Qiu YM/Yang Y/Song LP)
REFERENCES
- 1.Shirai K, Suzuki Y, Oka K, et al. Nuclear survivin expression predicts poorer prognosis in glioblastoma. J Neurooncol. 2009;91(3):353–358. doi: 10.1007/s11060-008-9720-4. [DOI] [PubMed] [Google Scholar]
- 2.Das A, Tan WL, Teo J, et al. Expression of survivin in primary glioblastomas. J Cancer Res Clin Oncol. 2002;128(6):302–306. doi: 10.1007/s00432-002-0343-4. [DOI] [PubMed] [Google Scholar]
- 3.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3(8):917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 4.Adida C, Berrebi D, Peuchmaur M, et al. Anti-apoptosis gene, survivin, and prognosis of neuroblastoma. Lancet. 1998;351(9106):882–883. doi: 10.1016/S0140-6736(05)70294-4. [DOI] [PubMed] [Google Scholar]
- 5.Zhou R, Zhang LZ, Wang RZ. Effect of celecoxib on proliferation, apoptosis, and survivin expression in human glioma cell line U251. Chin J Cancer. 2010;29(3):294–299. doi: 10.5732/cjc.009.10290. [DOI] [PubMed] [Google Scholar]
- 6.Hussain SF, Kong LY, Jordan J, et al. A novel small molecule inhibitor of signal transducers and activators of transcription 3 reverses immune tolerance in malignant glioma patients. Cancer Res. 2007;67(20):9630–9636. doi: 10.1158/0008-5472.CAN-07-1243. [DOI] [PubMed] [Google Scholar]
- 7.Grzelinski M, Urban-Klein B, Martens T, et al. RNA interference-mediated gene silencing of pleiotrophin through polyethylenimine-complexed small interfering RNAs in vivo exerts antitumoral effects in glioblastoma xenografts. Hum Gene Ther. 2006;17(7):751–766. doi: 10.1089/hum.2006.17.751. [DOI] [PubMed] [Google Scholar]
- 8.Koike H, Morikawa Y, Sekine Y, et al. Survivin is associated with cell proliferation and has a role in 1a,25-dihydroxyvitamin D3 induced cell growth inhibition in prostate cancer. J Urol. 2011;185(4):1497–1503. doi: 10.1016/j.juro.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Kim YS, Jin HO, Seo SK, et al. Sorafenib induces apoptotic cell death in human non-small cell lung cancer cells by down-regulating mammalian target of rapamycin (mTOR)-dependent survivin expression. Biochem Pharmacol. 2011;82(3):216–226. doi: 10.1016/j.bcp.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 10.Sasaki T, Lopes MB, Hankins GR, et al. Expression of survivin, an inhibitor of apoptosis protein, in tumors of the nervous system. Acta Neuropathol. 2002;104(1):105–109. doi: 10.1007/s00401-002-0532-x. [DOI] [PubMed] [Google Scholar]
- 11.Xie D, Zeng YX, Wang HJ, et al. Expression of cytoplasmic and nuclear Survivin in primary and secondary human glioblastoma. Br J Cancer. 2006;94(1):108–114. doi: 10.1038/sj.bjc.6602904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakravarti A, Noll E, Black PM, et al. Quantitatively determined survivin expression levels are of prognostic value in human gliomas. J Clin Oncol. 2002;20(4):1063–1068. doi: 10.1200/JCO.2002.20.4.1063. [DOI] [PubMed] [Google Scholar]
- 13.Kelly RJ, Lopez-Chavez A, Citrin D, et al. Impacting tumor cell-fate by targeting the inhibitor of apoptosis protein survivin. Mol Cancer. 2011;10:35. doi: 10.1186/1476-4598-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papanikolaou V, Iliopoulos D, Dimou I, et al. Survivin regulation by HER2 through NF-κB and c-myc in irradiated breast cancer cells. J Cell Mol Med. 2011;15(7):1542–1550. doi: 10.1111/j.1582-4934.2010.01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Llambi F, Green DR. Apoptosis and oncogenesis: give and take in the BCL-2 family. Curr Opin Genet Dev. 2011;21(1):12–20. doi: 10.1016/j.gde.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plati J, Bucur O, Khosravi-Far R. Apoptotic cell signaling in cancer progression and therapy. Integr Biol (Camb) 2011;3(4):279–296. doi: 10.1039/c0ib00144a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin S, Sung BJ, Cho YS, et al. An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and -7. Biochemistry. 2001;40(4):1117–1123. doi: 10.1021/bi001603q. [DOI] [PubMed] [Google Scholar]
- 18.Song Z, Yao X, Wu M. Direct interaction between survivin and Smac/DIABLO is essential for the anti-apoptotic activity of survivin during taxol-induced apoptosis. J Biol Chem. 2003;278(25):23130–23140. doi: 10.1074/jbc.M300957200. [DOI] [PubMed] [Google Scholar]
- 19.Uren AG, Wong L, Pakusch M, et al. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol. 2000;10(21):1319–1328. doi: 10.1016/s0960-9822(00)00769-7. [DOI] [PubMed] [Google Scholar]
- 20.Temme A, Rieger M, Reber F, et al. Localization, dynamics, and function of survivin revealed by expression of functional survivinDsRed fusion proteins in the living cell. Mol Biol Cell. 2003;14(1):78–92. doi: 10.1091/mbc.E02-04-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia Q, Li Y, Xu D, et al. Radiosensitivity of glioma to Gamma Knife treatment enhanced in vitro and in vivo by RNA interfering Ku70 that is mediated by a recombinant adenovirus. J Neurosurg. 2010;113(Suppl):228–235. doi: 10.3171/2010.7.GKS10972. [DOI] [PubMed] [Google Scholar]
- 22.Wang MT, Chen G, An SJ, et al. Prognostic significance of cyclinD1 amplification and the co-alteration of cyclinD1/pRb/ppRb in patients with esophageal squamous cell carcinoma. Dis Esophagus. doi: 10.1111/j.1442-2050.2011.01291.x. in press. [DOI] [PubMed] [Google Scholar]
- 23.Zheng D, Gu S, Li Y, et al. A global genomic view on LNX siRNA-mediated cell cycle arrest. Mol Biol Rep. 2011;38(4):2771–2783. doi: 10.1007/s11033-010-0422-6. [DOI] [PubMed] [Google Scholar]
- 24.He SQ, Rehman H, Gong MG, et al. Inhibiting survivin expression enhances TRAIL-induced tumoricidal activity in human hepatocellular carcinoma via cell cycle arrest. Cancer Biol Ther. 2007;6(8):1247–1257. doi: 10.4161/cbt.6.8.4444. [DOI] [PubMed] [Google Scholar]
- 25.Uchida H, Tanaka T, Sasaki K, et al. Adenovirus-mediated transfer of siRNA against survivin induced apoptosis and attenuated tumor cell growth in vitro and in vivo. Mol Ther. 2004;10(1):162–171. doi: 10.1016/j.ymthe.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 26.The Ministry of Science and Technology of the People's Republic of China. Guidance Suggestions for the Care and Use of Laboratory Animals. 2006-09-30 [Google Scholar]
- 27.Zhen HN, Li LW, Zhang W, et al. Short hairpin RNA targeting survivin inhibits growth and angiogenesis of glioma U251 cells. Int J Oncol. 2007;31(5):1111–1117. [PubMed] [Google Scholar]
- 28.Marucci G, Lammi C, Buccioni M, et al. Comparison and optimization of transient transfection methods at human astrocytoma cell line 1321N1. Anal Biochem. 2011;414(2):300–302. doi: 10.1016/j.ab.2011.02.028. [DOI] [PubMed] [Google Scholar]
- 29.Long KH, Gomez FJ, Morris RE, et al. Identification of heat shock protein 60 as the ligand on Histoplasma capsulatum that mediates binding to CD18 receptors on human macrophages. J Immunol. 2003;170(1):487–494. doi: 10.4049/jimmunol.170.1.487. [DOI] [PubMed] [Google Scholar]
- 30.Guerlava P, Izac V, Tholozan JL. Comparison of different methods of cell lysis and protein measurements in Clostridium perfringens: application to the cell volume determination. Curr Microbiol. 1998;36(3):131–135. doi: 10.1007/pl00006756. [DOI] [PubMed] [Google Scholar]


