Abstract
Viruses are both inducers and targets of posttranscriptional gene silencing (PTGS), a natural defense mechanism in plants. Here we report molecular evidence of the ability of single-stranded DNA (ssDNA) viruses to induce PTGS in infected plants irrespective of the severity of or recovery from the symptoms. Our results reveal that five distinct species of cassava-infecting geminiviruses were capable of triggering PTGS by producing two classes of virus-specific short interfering RNAs (siRNAs) of 21 to 26 nucleotides in two plant hosts, tobacco (Nicotiana benthamiana) and cassava (Manihot esculenta, Crantz). However, the efficacy of virus-induced PTGS varied depending on the intrinsic features of the virus and its interaction with the plant host. We found that symptom recovery over time in plants infected with the isolates of African cassava mosaic virus (ACMV-[CM]) or Sri Lankan cassava mosaic virus was associated with a much higher level of virus-derived siRNA accumulation compared to plants infected with viruses that do not show symptom recovery. Furthermore, we determined that the C terminus of AC1 that overlaps with the N terminus of AC2 early viral genes involved in virus replication were the primary targets for ACMV-[CM]-induced PTGS, whereas the C terminus of BC1 was targeted for the East African cassava mosaic Cameroon virus. In addition, our results reveal the possibility for double-stranded RNA formation during transcription in ssDNA viruses, which explains in part how these viruses can trigger PTGS in plants.
Posttranscriptional gene silencing (PTGS) operates in sequence-specific recognition and degradation of mRNA in diverse eukaryotes. It was first reported in plants, referred to as PTGS, cosuppression, or RNA-mediated virus resistance (3, 24, 45). Over the years, it has been described for other organisms and is referred to as quelling in fungi (6) and RNA interference (RNAi) in animals (10, 17). Transgenes and viruses can trigger gene silencing in plants, which is recognized as a natural antiviral defense mechanism (41). Although the mechanism by which a virus infection triggers PTGS in plants is not fully understood, double-stranded RNA (dsRNA) is a strong inducer of PTGS (45). Such dsRNA types, or aberrant RNAs that are produced during replication of an RNA virus, become dsRNA by the activity of virus- or host-encoded RNA-dependent RNA polymerase (RdRp) (8, 44). These dsRNAs are cleaved by a dsRNA-specific RNase III enzyme termed Dicer (4) to yield short-interfering RNAs (siRNAs) of 21 to 26 nucleotides (nt), which subsequently serve as complementary guides for cleavage of homologous RNA molecules by forming a multicomponent silencing complex termed RNA-induced silencing complex (RISC) (10, 16, 36, 48). Two classes (21 to 22 nt and 24 to 26 nt) of siRNAs were shown to be involved in RNA silencing (14). Recently, a Dicer-associated protein called R2D2 that is important for siRNA incorporation into RISC linking the initiation and execution phases of RNAi has been identified in Drosophila melanogaster (21). In plants, the signal moves through plasmodesmata and for greater distances through the vascular system (25). The signal would move together with, or in advance of the virus, and mediates silencing of the viral RNA in the newly infected cells. Consequently, the infection would progress slowly or would be arrested. Moreover, the introduction of siRNAs has been shown to downregulate gene expression and viral DNA accumulation in plant cells (40).
As a counterdefense, certain plant viruses encode proteins that can suppress the RNA silencing in order to overcome this defense mechanism (39). Among these, helper-component proteinase (HC-Pro) of potyviruses has been shown to be a strong suppressor of PTGS (2, 20). In geminiviruses, AC2 (transcriptional activator protein [TrAP]) of African cassava mosaic virus isolate from Kenya (ACMV-[KE]) and C2 of Tomato yellow leaf curl China virus were identified as mild suppressors of gene silencing in plants (42, 46).
Most plant RNA viruses replicate through the formation of dsRNA intermediates, the potential inducer of PTGS. Unlike the positive single-stranded RNA (ssRNA) viruses, geminiviruses do not use a dsRNA molecular intermediate in their replication cycle. Geminiviruses have ssDNA genomes which replicate through dsDNA as the replicative intermediates in the infected plant cell nucleus (18). Cassava mosaic disease (CMD) is caused by single and dual infections of whitefly-transmitted begomoviruses (Geminiviridae) belonging to eight distinct species of geminiviruses (12). CMD is a major threat to cassava production, an important food staple for 600 million people in tropical Africa, Asia, and South America (5). Molecular understanding of the cassava mosaic geminiviruses responsible for the complex CMD would pave the way to develop molecular strategies to control these devastating viruses.
Based on symptom severity and viral DNA accumulation cassava-infecting geminiviruses are categorized into recovery type and non-recovery type viruses in Nicotiana benthamiana and cassava plants (J. S. Pita et al., unpublished data). We describe here the ability of isolates of five distinct species of cassava-infecting geminiviruses, African cassava mosaic virus (ACMV-[CM]; isolate from Cameroon), East African cassava mosaic Cameroon virus (EACMCV; isolate from Cameroon), East African cassava mosaic virus (EACMV-[UG]; isolate from Uganda), Sri Lankan cassava mosaic virus (SLCMV; isolate from Sri Lanka), and Indian cassava mosaic virus (ICMV; isolate from India) to induce PTGS in two plant hosts, tobacco (N. benthamiana) and cassava (Manihot esculenta, Crantz) in terms of their ability to produce virus-specific siRNAs. In addition, we demonstrate that there is a strong correlation between the recovery phenotype and the virus-derived siRNA accumulation over a period of 8 weeks. Furthermore, our results clearly show that, in the case of ACMV-[CM], certain early viral genes are preferentially targeted by the virus-induced PTGS (the AC1 that encodes for replication-associated protein [Rep] and the AC2 encodes for transcriptional activator protein [TrAP]). In contrast, EACMCV-induced PTGS targeted the late gene (BC1) encoded by the DNA-B component of the virus. How geminiviruses could trigger PTGS and the PTGS capacity of these viruses in relation to symptom severity and recovery phenotype in infected plants are discussed.
MATERIALS AND METHODS
Multiplication of cassava and N. benthamiana plants.
Cassava plants were multiplied in vitro by micropropagation. Nodal cuttings of the cassava cultivar TMS 60444 from 6-week-old regenerated plants were obtained. They were surface sterilized for 30 min in 15% bleach solution containing two drops of Tween 20 and then washed thoroughly in sterilized water. These sterilized cuttings were cultured in petri dishes containing MS2 medium (4.31 g of MS salt; 20 g of sucrose; 1 ml of Gramborg’s vitamin solution [Sigma]; 0.5 ml of 1-naphthaleneacetic acid from 1 mM stock, pH 5.8 [solidified by Phytagel]) and incubated at 28°C under 16 h light for 3 to 4 weeks. Rooted plantlets were transplanted into pots (7-cm diameter by 6-cm depth) containing growth medium 702 Metro-Mix (Scott's Company, Maryville, Ohio). The plants were maintained in a greenhouse for 3 to 4 weeks before they were inoculated with virus. Seeds of the N. benthamiana were sown in a pot and kept in humidifying chambers. One week later, seedlings were transplanted into individual pots containing a mix of Scott's Co.360 with Coir. The plants were maintained in a greenhouse for 3 to 4 weeks before they were inoculated with virus.
Virus inoculation.
Construction of the infectious clones of DNA-A and DNA-B of ACMV-[CM] and EACMCV (13) and EACMV-[UG] (26) has been previously described. Infectious clones of DNA-A and DNA-B of SLCMV and ICMV were kindly provided by John Stanley, John Innes Centre (United Kingdom). For virus inoculation, gold particles (0.6 μ) were coated separately with a mixture of 10 ng each of DNA-A and DNA-B of infectious clones of ACMV-[CM], SLCMV, EACMCV, EACMV-[UG], and ICMV. Five plants were inoculated for each virus by using a particle delivery system at 1,100 psi (Bio-Rad model PDS-1000/He), and the experiment was repeated three times. To ensure 100% infection, the youngest unfolded leaf was targeted on the adaxial at the point where the leaflets are attached to the petiole. The plants were kept at 25°C in a greenhouse free of whiteflies.
Assessment of symptoms.
Systemically infected leaves were scored every other day after inoculation for 8 weeks. We decided upon 8 weeks because N. benthamiana is about to set seeds after 8 weeks and cassava becomes too large to handle. A leaf was regarded as systemically infected if there was no scar on the leaf surface as a result of inoculation and symptoms developed first at the joint of the petiole and the leaflets. Symptom severity score was rated on a six-point scale: 0 = no symptoms, 1 = mild chlorosis without leaf deformation, 2 = clear mosaic with or without slight leaf deformation, 3 = strong mosaic all over the leaflets with leaf deformation, 4 = same as for score 3 but with severe leaf deformation, and 5 = severe mosaic and severe reduction of leaf size (11). Inoculated leaves at 2 and 4 dpi (days postinoculation) and symptomatic young leaves from 1 to 8 wpi (weeks postinoculation) were collected on a weekly basis to determine viral DNA, mRNA, and siRNA accumulation by Southern and Northern blot analysis.
Southern and Northern blot analysis.
Southern blotting was performed as described previously (30). Portions (4 μg) of total DNA isolated from a pool of leaf tissue collected from five infected plants as described previously (9) for each of the five viruses were separated by electrophoresis in a 1% agarose gel in 1× Tris-borate-EDTA (TBE) and then transferred to Hybond-N+ membrane (Amersham International, Inc.). For Northern blotting, 2 μg of total RNA was isolated from a pool of leaf tissue collected from five infected plants for each of the five viruses by using the RNA isolation kit (Qiagen). Samples were run on 1% formaldehyde agarose gels after treatment with DNase I (RNase-free) and transferred to Hybond-N+ membrane (Amersham). To make an ACMV-[CM]-specific probe, a 794-bp EcoRI fragment (coordinates nt 1789 to 2583) of ACMV-[UG] DNA-A was used, and to make an EACMCV-specific probe, a 944-bp EcoRI fragment (coordinates nt 1821 to 2765) of EACMV-[UG2] DNA-A (26) was used. These DNA fragments were labeled by using [α-32P]dCTP and a random primer labeling kit (Prime II kit; Stratagene). Hybridization was carried out at 65°C for Southern blots and at 42°C for Northern blots. Posthybridization washes were done sequentially with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M trisodium citrate), 0.5× SSC, and 0.2× SSC, along with 0.1% sodium dodecyl sulfate (SDS), each wash was carried out for 30 min at 65°C (40). Blots were scanned by using a PhosphorImager and quantified by using IQMacV1.2 software (Storm; Amersham, Inc.).
Detection, isolation, and labeling of 21- to 26-nt RNAs.
The low-molecular-weight RNAs, isolated from a pool of leaves (1 gm) collected from five infected plants for each virus, was powdered with a mortar and pestle. To the finely powdered tissue, 5 ml of TRI-Reagent and 1 ml of chloroform (Sigma) were added, followed by thorough mixing. After 15 min at room temperature, the sample was centrifuged; the aqueous phase was extracted with phenol-chloroform and precipitated with ethanol. The pellet was dissolved in 50% formamide and heated to 65°C for 5 min before being loaded on 15% polyacrylamide (19:1) gels containing 7 M urea in 0.5× TBE. The samples were electrophoresed until the bromophenol blue reached the bottom of the gel and then blotted to Hybond-N+ membrane (Amersham International, Inc.) and UV cross-linked. To make probes, full-length viral DNA fragments (DNA-A and DNA-B) of each virus ACMV-[CM], SLCMV, EACMCV, EACMV-[UG], and ICMV were separately labeled with [α-32P]dCTP and a random primer labeling kit (Prime II kit). Hybridization was carried out at 42°C overnight in 5× SSC (0.75 M NaCl, 0.075 M trisodium citrate [pH 7.0]), 1× Denhardt solution (0.1% each Ficoll, polyvinylpyrrolidone, and bovine serum albumin), and 0.5% SDS with competitor DNA (herring sperm DNA; Sigma). Posthybridization washes were done sequentially with 2× SSC, 0.5× SSC, and 0.2× SSC, along with 0.1% SDS; each wash for 30 min. Blots were scanned and quantified by using IQMacV1.2 software and a phosphorimager (Storm; Amersham, Inc.). For molecular size markers, oligomers (21 and 30 nt) were end labeled by using T4-polynucleotide kinase and [γ-32P]ATP.
For preparation of 21- to 26-nt RNA, 40 μg of low-molecular-weight RNA was subjected to electrophoresis through an 15% denaturing polyacrylamide gel in 0.5× TBE buffer, followed by staining in ethidium bromide solution (0.5 μg/ml) for about 20 to 30 min. The 21- to 26-nt RNAs were visualized by UV light and excised from gel. The gel slice was crushed, covered with 3 M NaCl, and incubated overnight at 4°C. The gel residue was removed by centrifugation, and the supernatant was precipitated with ethanol (35). The 21- to 26-nt RNAs (ca. 1 to 2 μg) were dephosphorylated (CIP; NEB) and subsequently labeled in a 30-μl reaction in the presence of [γ-32P]ATP by using T4-polynucleotide kinase (NEB). The labeled ACMV-[CM]- and EACMCV-specific 21- to 26-nt RNAs were used for hybridization of blots containing PCR-amplified (∼400 bp) DNA fragments of DNA-A and DNA-B of ACMV-[CM] and EACMCV, respectively. Blots were scanned and quantified by using IQMacV1.2 software and a Storm phosphorimager.
RESULTS
Recovery from the symptoms induced by ACMV-[CM] positively correlates with the accumulation of ACMV-[CM]-specific siRNAs
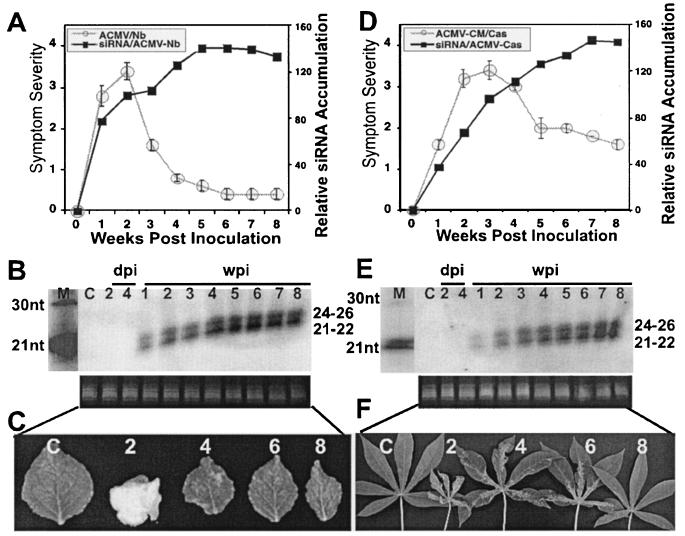
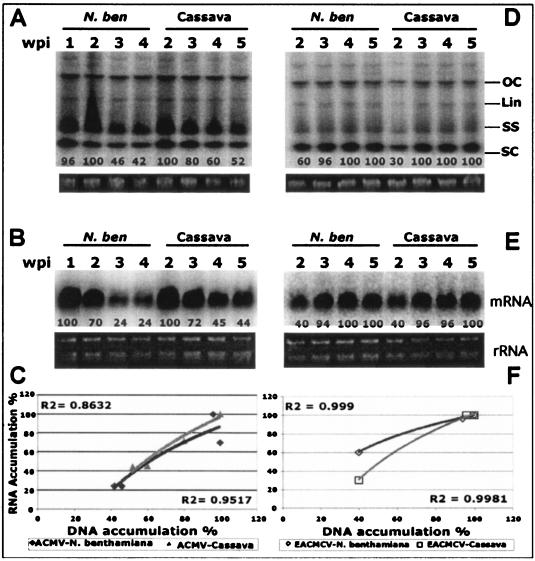
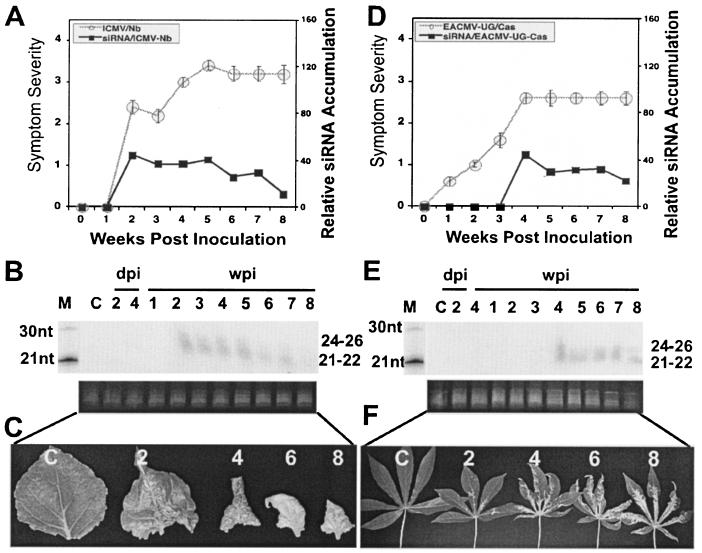
To assess the ability of ACMV-[CM] to induce PTGS, seedlings of N. benthamiana and cassava were inoculated by particle bombardment with a mixture of DNA-A and DNA-B of the virus. Viral symptoms in infected plants were scored as described previously (11). Symptom appearance started within 5 days postinoculation (dpi; 15 infected out of 15 inoculated) with leaf curling and yellow mosaic, reaching maximum severity ca. 10 dpi with leaf distortion. Symptom severity persisted for 2 weeks in N. benthamiana and for 2 to 3 weeks in cassava. However, both infected plants recovered from symptoms at a later stage of the infection cycle. ACMV-[CM]-infected N. benthamiana plants started to show symptom recovery between 2 and 3 weeks postinoculation (wpi), and infected cassava started to show symptom recovery by around 3 to 4 wpi, after which their growth ability was restored and the newly emerging leaves displayed very mild chlorotic spots in N. benthamiana and very mild symptoms in cassava. Nevertheless, the different phases of the ACMV-[CM] infection progression curves are similar in both plant species, with an early increase in symptom severity followed by a symptomless recovery phenotype at the later stage of the infection cycle (Fig. 1A and D). Representative leaves with various levels of symptoms at 2, 4, 6, and 8 wpi in N. benthamiana (Fig. 1C) and in cassava (Fig. 1F) plants are shown. We analyzed the levels of accumulation of both viral DNA and mRNA at 1, 2, 3, and 4 wpi in infected N. benthamiana and at 2, 3, 4, and 5 wpi in infected cassava (since symptom recovery is slower in cassava). Viral DNA accumulation level was at the maximum up to 2 wpi and decreased gradually over time in both infected N. benthamiana and cassava (Fig. 2A). However, the level of mRNA accumulation reached a maximum at 1 and 2 wpi in N. benthamiana and cassava, respectively, and it decreased overtime in both infected plants (Fig. 2B). In addition, there is a positive highly significant correlation between the amount of DNA and mRNA accumulation in the infected plants, with R2 coefficients of 0.95 and 0.86, respectively, for ACMV-[CM] (Fig. 2C).
FIG. 1.
Relationship between symptom severity, recovery, and siRNA accumulation in ACMV-[CM]-infected N. benthamiana and cassava plants. (A and D) ACMV-[CM] symptom severity and siRNA accumulation trend in N. benthamiana (A) and in cassava (D). A total of 15 N. benthamiana and cassava plants were inoculated as five plants in three experiments. Bars in the symptom severity curve indicate the standard error (SE) values of 15 plants. (B and E) RNA gel blots probed with [∝-32P]dCTP-labeled DNA-A and DNA-B of ACMV-[CM] in N. benthamiana (B) and in cassava (E) at days 2 and 4 in inoculated leaves and at weeks 1 to 8 in systemically infected leaves. The left lane shows [γ-32P]ATP-end-labeled oligonucleotide markers of 21 and 30 nt. Each lane was loaded with 40 μg of low-molecular-weight RNA. Ethidium bromide-stained RNA is shown as a loading control. (C and F) Representative leaves showing different degrees of symptoms and recovery phenotypes at 2, 4, 6, and 8 wpi in N. benthamiana (C) and cassava (F). The C within the blots and leaf panels represents a mock-inoculated, control plant.
FIG. 2.
Levels of viral DNA and mRNA accumulation in ACMV-[CM] and EACMCV-infected N. benthamiana and cassava. (A and D) Southern blot analysis ACMV-[CM] DNA accumulation at 1, 2, 3, and 4 wpi in N. benthamiana and at 2, 3, 4, and 5 wpi in cassava (A) and EACMCV DNA accumulation at 2, 3, 4, and 5 wpi in N. benthamiana and cassava (D). Each lane was loaded with 4 μg of total genomic DNA isolated from virus-infected plants. Different viral DNA forms—supercoiled (SC), single stranded (SS), linear (Lin), and open circular (OC)—are indicated. Ethidium bromide-stained gels at the bottom of each blot serve as loading control. (B and E) Northern blot analysis of ACMV-[CM] mRNA accumulation at 1, 2, 3, and 4 wpi in infected N. benthamiana and 2, 3, 4, and 5 wpi in infected cassava (B) and of EACMCV viral mRNA accumulation at 2, 3, 4, and 5 wpi in infected N. benthamiana and cassava (E). Total RNA (2 μg) isolated from virus-infected plants was loaded into each lane. Blots were hybridized with [∝-32P]dCTP-labeled ACMV-[CM]-specific (A and B) and EACMCV-specific (D and E) probes. Ethidium bromide-stained rRNA was shown as the loading control. (C and F) Correlation of viral DNA and mRNA accumulation in ACMV-[CM]-infected (C) and EACMCV-infected (F) N. benthamiana and cassava plants.
Next, we determined whether the recovery phenotype in ACMV-[CM]-infected N. benthamiana and cassava is associated with virus-induced PTGS in terms of virus-specific siRNA accumulation. Low molecular weight siRNAs were purified from a pool of leaf samples collected from five infected N. benthamiana and cassava plants on a weekly basis over a period of 8 weeks. Northern blots with ACMV-[CM] DNA-A and DNA-B as the probes hybridized with low-molecular-weight RNAs, indicating that ACMV-[CM] was able to induce PTGS with the production of two virus-specific classes of siRNAs (Fig. 1B and E). The siRNA blots were quantified, and it was found that the amount of virus-specific siRNAs gradually increased with time in both plant hosts. However, a slight difference in the rate of siRNA accumulation was observed between the infected N. benthamiana and cassava. In infected N. benthamiana, virus-derived siRNA accumulation was 10% at week 1 and 30% at weeks 2 and 3 (100%) and reached a maximum (141%) at week 5. Later, siRNA accumulation maintained a plateau between weeks 5 and 7, with a slight decrease at week 8 (Fig. 1B). The amount of siRNA at week 2 (30%) of ACMV-[CM]-infected N. benthamiana was considered as 100% for further comparative graphical representation with the virus-derived siRNA accumulation of other viruses. The above-mentioned values in parentheses represent the percentage calculated after normalization with the 2 wpi point as 100% (Fig. 1A). In cassava, even though siRNA accumulation started 1 week after inoculation, it was less abundant compared to the N. benthamiana host until week 4 and then increased to its maximum between weeks 7 and 8 (Fig. 1E). The PTGS capacity of ACMV-[CM] was apparently more efficient in N. benthamiana than in cassava, since we could detect slightly less siRNA accumulation in cassava compared to that of N. benthamiana. Symptom severity and viral DNA and mRNA accumulation were about the maximum level up to the second week and then decreased over time. However, siRNA continued to accumulate over time in both plant species. Virus-specific siRNAs could not be detected in inoculated leaves at 2 and 4 dpi (Fig. 1B and E), even though symptoms were observed on day 4 after inoculation, indicating that either the siRNA accumulation was below the level of detection or infected plants required more than 4 dpi to respond to the ACMV-[CM]-induced PTGS.
These results clearly indicate that virus-derived siRNA accumulation correlates well with the recovery phenotype in infected N. benthamiana (Fig. 1A to C) and in cassava plants (Fig. 1D to F). siRNA molecules were not detected in the RNA samples that were isolated from mock-inoculated control N. benthamiana and in cassava plants, indicating that infected plants responded specifically to the virus.
PTGS induced by SLCMV is regulated differentially in N. benthamiana and cassava.
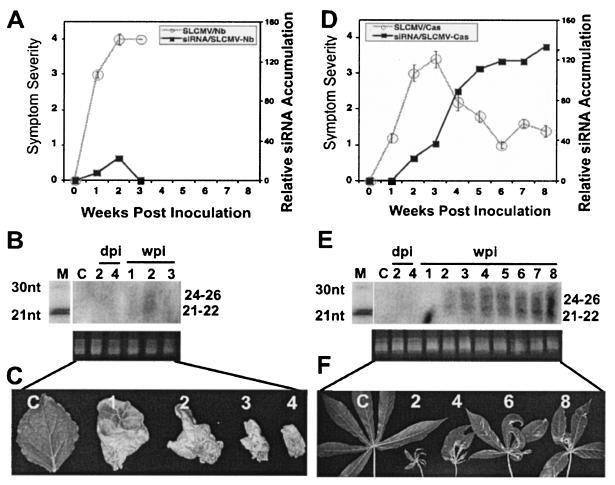
The ability of SLCMV to induce PTGS was tested by inoculating DNA-A and DNA-B of the virus in N. benthamiana and cassava seedlings. Symptom apparition started within 3 dpi in both plant species. In cassava, although the virus causes severe symptoms initially (15 infected out of 15 inoculated), the infected plants showed recovery at a later stage of the infection cycle, as in the case of ACMV-[CM] (Fig. 3D and F). SLCMV-specific siRNA accumulation increased over time from week 2 onward until week 8 (Fig. 3E). A strong correlation was observed between the siRNA accumulation and recovery phenotype in SLCMV-infected cassava plants. However, it behaved differentially in N. benthamiana. SLCMV-infected N. benthamiana (15 infected and 15 inoculated) initially displayed severe chlorotic symptoms, followed by necrotic symptoms and finally plant death by ca. 4 wpi (Fig. 3A and C). Northern blot analysis revealed that SLCMV-specific siRNA accumulation was barely detectable at weeks 1 and 2 and was below the level of detection at week 3 (Fig. 3B). siRNA accumulation was not analyzed after week 3, since the plant was dead. The absence of siRNA after week 2 in these plants suggests that SLCMV might encode a strong suppressor protein, or it might induce early apoptosis of the whole plant. As a result, the infected plants never recovered but died by ca. 4 wpi (Fig. 3C). These results clearly revealed that, although SLCMV is a killer in N. benthamiana, it behaved like ACMV-[CM] in cassava, with severe symptoms in the beginning followed by a recovery phenotype.
FIG. 3.
Relationship between symptom severity, recovery, and siRNA accumulation in SLCMV-infected N. benthamiana and cassava plants. (A and D) SLCMV symptom severity and siRNA accumulation curves in N. benthamiana (A) and cassava (D). A total of 15 N. benthamiana and cassava plants were inoculated as five plants in three experiments. Bars in the symptom severity curve indicate the SE values of 15 plants. (B and E) RNA gel blots probed with [∝-32P]dCTP-labeled DNA-A and DNA-B of SLCMV in N. benthamiana (B) and in cassava (E) days 2 and 4 in inoculated leaves and at weeks 1 to 8 in systemically infected leaves. Each lane was loaded with 40 μg of low-molecular-weight RNA. Ethidium bromide-stained RNA is shown as a loading control. (C and F) Representative leaves showing different degree of symptoms and recovery phenotypes at 1, 2, 3, and 4 wpi in N. benthamiana (C) and at 2, 4, 6, and 8 wpi in cassava (F). The C within the blots and leaf panels represents a mock-inoculated, control plant.
PTGS induced by nonrecovery-type cassava geminiviruses is suppressed over time.
We also investigated the PTGS capacity of nonrecovery phenotype cassava-infecting geminiviruses, namely, EACMCV, EACMV-[UG], and ICMV and its relation to symptom severity in N. benthamiana and in cassava plants.
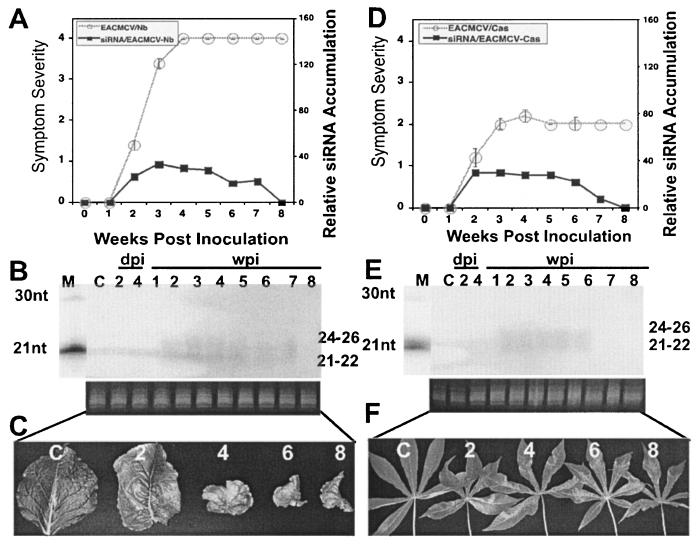
N. benthamiana and cassava plantlets were inoculated by particle bombardment with a mixture of DNA-A and DNA-B of the infectious clones of EACMCV. Infected N. benthamiana and cassava plants showed symptoms between 11 and 14 dpi in all inoculated 15 plants. However, in infected N. benthamiana, symptom severity increased over time with the later stages of infected plant growth exhibiting reduction in internodal length, shorter petioles, and smaller leaves with a rosette-like appearance, which persisted until plant senescence. In contrast, EACMCV-infected cassava displayed a mild mosaic symptom throughout the infection cycle of the virus. Thus, EACMCV behaves differently from ACMV-[CM] in that N. benthamiana (Fig. 4A and C) and cassava plants (Fig. 4D and F) infected with EACMCV never recovered. We analyzed viral DNA and mRNA accumulation levels at 2, 3, 4, and 5 wpi in infected N. benthamiana and cassava. Viral DNA and mRNA were not analyzed at 1 wpi since EACMCV is a slow virus that develops symptoms 1 week after inoculation in both plant hosts. Southern blot analysis revealed that viral DNA accumulation reached the maximum at 3 wpi and maintained the same level until 5 wpi in N. benthamiana and cassava (Fig. 2D). Similarly, a higher level of viral mRNA accumulation was detected at 3 wpi, and this lasted until 5 wpi in both N. benthamiana and cassava (Fig. 2E). There is a positive highly significant correlation between the amount of viral DNA and mRNA accumulation in the infected plants, with R2 coefficients of 0.95 and 0.86 for ACMV-[CM] (Fig. 2C) and of 0.99 and 0.99 for EACMCV, respectively (Fig. 2F), indicating that, irrespective of increasing or decreasing symptoms, the DNA and mRNA accumulations are strictly correlated.
FIG. 4.
Relationship between symptom severity, recovery, and siRNA accumulation in EACMCV-infected N. benthamiana and cassava plants. (A and D) EACMCV symptom severity and siRNA accumulation curves in N. benthamiana (A) and cassava (D). A total of 15 N. benthamiana and cassava plants were inoculated as five plants in three experiments. Bars in symptom severity curve indicate the SE values of 15 plants. (B and E) RNA gel blots probed with [∝-32P]dCTP-labeled DNA-A and DNA-B of EACMCV in N. benthamiana (B) and in cassava (E) at days 2 and 4 in inoculated leaves and at weeks 1 to 8 in systemically infected leaves. Each lane was loaded with 40 μg of low-molecular-weight RNA. Ethidium bromide-stained RNA is shown as a loading control. (C and F) Representative leaves showing different degree of symptoms and recovery phenotype at 2, 4, 6, and 8 wpi in N. benthamiana (C) and in cassava (F). The C within the blots and leaf panels represents a mock-inoculated, control plant.
The PTGS capacity of this virus was evaluated by analyzing low-molecular-weight RNAs isolated from a pool of symptomatic leaves obtained from five infected plants in Northern blots with EACMCV-specific probes. EACMCV-specific siRNA accumulation was detected starting from 2 wpi in both infected N. benthamiana (Fig. 4B) and cassava plants (Fig. 4E), but it was less abundant compared to ACMV-[CM]-derived siRNAs. In addition, it was below the level of detection at 1 wpi, and the level of accumulation did not show an increase as in the case of ACMV-[CM]. The low level of siRNA accumulation correlated with the nonrecovery phenotype associated with this virus. Representative leaves with typical symptoms of this virus at different time points are shown (Fig. 4C and F). Thus, although EACMCV can induce PTGS, it is insufficient to reduce the viral load, and as a result, the infected plants displayed symptoms until senescence in both N. benthamiana and cassava plants.
Infectious clones of ICMV, for unknown reasons, have the ability to infect N. benthamiana (15 infected out of 15 inoculated), but not cassava. Infected plants displayed symptoms around 11 to 14 dpi. Symptom severity associated with green patches in leaves, leaf puckering, curling, reduction in leaf lamina, and leaf distortion increased with time, and the infected plants never recovered (Fig. 5A and C). Low-molecular-weight RNA isolated from a pool of leaves obtained from five infected plants was subjected to Northern blot analysis with ICMV DNA as the probe. siRNA accumulation was detected starting from 2 wpi, maintaining similar levels up to 5 wpi, and then decreased from 6 to 8 wpi (Fig. 5B). The low level of siRNA accumulation correlated with the nonrecovery phenotype associated with this virus.
FIG. 5.
Relationship between symptom severity, recovery, and siRNA accumulation in ICMV-infected N. benthamiana and in EACMV-[UG]-infected cassava plants. (A) ICMV symptom severity and siRNA accumulation trend in N. benthamiana. (B) RNA gel blot probed with [∝-32P]dCTP-labeled DNA-A and DNA-B of ICMV in N. benthamiana at days 2 and 4 in inoculated leaves and at weeks 1 to 8 in systemically infected leaves. (C) Representative leaves showing different degree of symptoms and recovery phenotype at 2, 4, 6, and 8 wpi in N. benthamiana. (D) EACMV-[UG] symptom severity and siRNA accumulation curves in cassava plants. (E) RNA gel blot probed with [∝-32P]dCTP-labeled DNA-A and DNA-B of EACMV-[UG] in cassava plants at days 2 and 4 in inoculated leaves and at weeks 1 to 8 in systemically infected leaves. (F) Representative leaves showing different degree of symptoms and recovery phenotype at 2, 4, 6, and 8 wpi in cassava plants. A total of 15 N. benthamiana plants for ICMV and 15 cassava plants for EACMV-[UG] were inoculated as five plants in three experiments. Ethidium bromide-stained RNA is shown as a loading control. Bars in symptom severity curve indicate the SE values of 15 plants. The C within the blots and leaf panels represents a mock-inoculated, control plant.
Conversely, EACMV-[UG] can infect cassava but not N. benthamiana (Pita et al., unpublished). Cassava plantlets inoculated with EACMV-[UG] needed a minimum of 7 to 14 dpi to establish systemic infection (15 infected out of 15 inoculated). However, symptom severity increased over time and the infected plants never recovered (Fig. 5D). The leaves displayed chlorotic symptoms throughout the duration of the infection cycle (Fig. 5F). Low-molecular-weight RNAs were isolated from a pool of five EACMV-[UG]-infected plant leaves at different time points to test the PTGS capacity of EACMV-[UG]. Northern blot analysis detected very low levels of virus-specific siRNA accumulation starting from week 4 and maintained the same level until week 8 (Fig. 5E). The nonrecovery phenotype of this virus correlated with low level of siRNA accumulation in infected plants.
Identification of target viral sequences for ACMV-[CM] and EACMCV-induced gene silencing.
Next, we determined whether geminivirus-induced PTGS spreads homogeneously along the viral genome or has a sequence preference in targeting specific viral gene(s). Toward that goal, we compared the composition of siRNAs of plants infected with ACMV-[CM] and EACMCV representing recovery and nonrecovery-type viruses, respectively. The peak of infection based on symptom scoring, at 2 wpi for ACMV-[CM] and at 4 wpi for EACMCV, was chosen as a test point for each virus. We followed two approaches. (i) Viral genome was PCR amplified into uniform-sized fragments, and gel blots were probed with purified virus-derived siRNAs; and (ii) gel blots carrying virus-derived siRNAs were probed with PCR-amplified DNA fragments representing different viral genes as the probe.
ACMV-[CM] and EACMCV have a divided genome defined as DNA-A and DNA-B (Fig. 6A and E). Both components are required to establish infection in plants (32). DNA-A contains six open reading frames (ORFs): AC1, AC2, AC3, AC4, AV1, and AV2. AC1 encodes a replication-associated protein (Rep), the only virus-encoded protein indispensable for viral replication; AC2 codes for a transcriptional activator protein (TrAP); AC3 codes for the replication-enhancer protein (REn); and AV1 codes for the coat protein (CP) (Fig. 6A). DNA-B has only two ORFs, BV1 (nuclear shuttle protein) and BC1 (movement protein), which are involved, respectively, in cell-to-cell and systemic infections (Fig. 6A). The nucleotide sequence of DNA-A and DNA-B are different except for a region of ∼200 nt, which shares >90% nucleotide sequence identity defined as the common region (CR). The CR carries regulatory sequences essential for viral replication and transcription (37). The two components of both viruses differ, DNA-A by 60% and DNA-B by 33%, and are consequently recognized as different species (12).
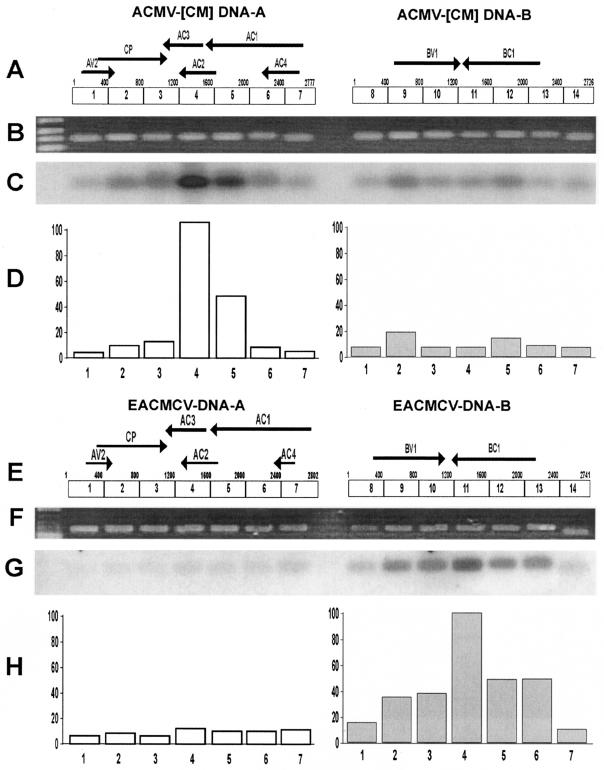
FIG. 6.
Identification of the origin of small 21- to 26-nt guide RNAs derived from ACMV-[CM] and EACMCV-infected N. benthamiana. (A) ACMV-[CM] genome consists of DNA-A (2,777 nt) and DNA-B (2,726 nt). PCR-amplified ∼400-bp DNA fragments 1 through 7 for DNA-A and PCR-amplified ∼400-bp DNA fragments 8 through 14 for DNA-B of ACMV-[CM] separated in an ethidium bromide-stained 1% agarose gel (B) were blotted and hybridized (C) with 5′-end-labeled small 21- to 26-nt guide RNAs purified from ACMV-[CM]-infected N. benthamiana. (D) Intensity of the obtained signals expressed as values of 0 to 100 (y axis), where the highest signal was scored as 100% by using ImageQuant (IqMacV1.2) software. Numbers in the x axis indicate the PCR-amplified DNA fragments. EACMCV genome comprises DNA-A (2,802 nt) and DNA-B (2,741 nt) (E). PCR-amplified ∼400-bp DNA fragments 1 through 7 for DNA-A and PCR-amplified ∼400-bp DNA fragments 8 through 14 for DNA-B, separated in an ethidium bromide-stained 1% agarose gel (F), were blotted and hybridized (G) with 5′-end-labeled small 21- to 26-nt guide RNAs isolated from EACMCV-infected N. benthamiana. (H) Intensity of the obtained signals expressed as values of 0 to 100, where the highest was scored as 100% by using ImageQuant software.
The DNA-A and DNA-B of ACMV-[CM] and EACMCV genomes were PCR amplified into ∼400-bp DNA fragments by using specific primers (Fig. 6B and F). Gel blots of these fragments were hybridized with labeled ACMV-[CM]-specific (Fig. 6C) or EACMCV-specific (Fig. 6G) siRNAs purified from the respective virus infected N. benthamiana plants. In the case of ACMV-[CM] blot, the results revealed that the majority of siRNAs were derived from the DNA-A component, particularly regions (fragments 4 [100%] and 5 [46%]) that correspond to AC1, AC2, and AC3 genes of the DNA-A component of ACMV-[CM] (Fig. 6D). DNA-B is almost uniformly targeted, with the production of siRNAs being <20% compared to that of DNA-A.
To further confirm these results, we performed a reverse blotting experiment, wherein gel blots of purified siRNAs from N. benthamiana plants infected with ACMV-[CM] were hybridized with PCR-amplified DNA fragments representing different genes as the probes. Among all genes of DNA-A and DNA-B and the CR that were tested, AC1 and AC2 showed the highest levels of siRNA accumulation (data not shown). In fact, the AC4 is embedded within the N-terminal region of AC1, and AC3 overlaps with the N terminus of AC2 (Fig. 6A). Further dissection with a nonoverlapping probe (AC1-2 that are devoid of AC4 and AC2 sequences) revealed that the C-terminal region of the AC1 coding sequence, which overlaps with the N-terminal region of AC2 coding sequence, was preferentially targeted by the ACMV-[CM]-induced PTGS. However, the siRNA accumulation corresponding to the AC3 and AC4 genes was very weak compared to the C terminus of AC1 and the N terminus of AC2 genes. These results were consistent with the previous experiment, indicating that the C terminus of AC1 and AC2 are the primary targets of ACMV-[CM]-induced PTGS.
In contrast, gel blots of PCR fragments of EACMCV genome (Fig. 6E and F) hybridized with labeled EACMCV-derived siRNAs revealed that higher amounts of siRNAs were derived from the DNA-B component of the virus, especially corresponding to the C terminus of the BC1 gene (Fig. 6G and H). BC1 protein is involved in cell-to-cell movement of the virus in plants (43). It appears that the DNA-A component of EACMCV is uniformly targeted, unlike in ACMV-[CM] (Fig. 6G and H), with <10% compared to DNA-B. Overall, the quantity of ACMV-[CM]-derived siRNA accumulation is significantly higher than that of EACMCV-derived siRNAs.
How do ssDNA viruses trigger PTGS in plants?
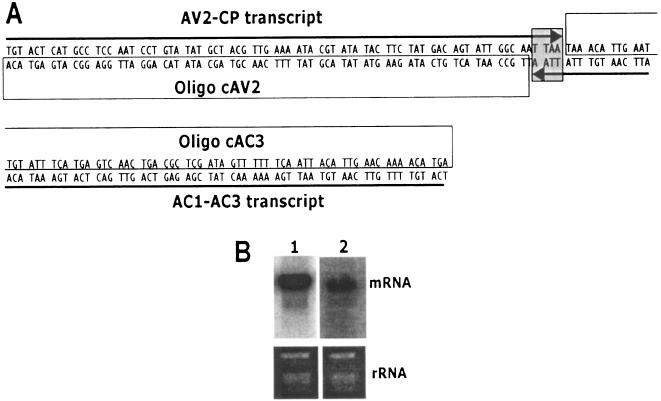
Plant RNA viruses replicate through dsRNA, a strong inducer of PTGS. Geminiviruses contain an ssDNA genome that replicates in the nucleus of the infected plant cells through a rolling circle mechanism via dsDNA intermediates called replicative forms and do not encounter a dsRNA phase in its replication cycle. Replicative forms serve as the template for both replication and transcription (37). Transcription is bidirectional with two major polycistronic transcripts in opposite orientations occurring from the CR that contains the bidirectional promoter sequences. The virion-sense AV1 (CP) ORF and the complementary-sense AC3 ORF overlap by 4 bp at their 3′ ends (Fig. 7A). However, the exact localization of each geminivirus transcript termination is not known. It was suggested that the overlapping transcripts in opposite polarity at the 3′ ends might generate dsRNA due to complementary base pairing, which could induce PTGS (41). In order to demonstrate this hypothesis, we used strand-specific 70-nt 5′-end-labeled oligomers as the probe to detect the extension of mRNA on either side, which could generate dsRNA. Total RNA isolated from ACMV-[CM]-infected N. benthamiana plants was blotted and hybridized separately with two strand-specific probes covering the AC3 and AV1 (CP) possible extensions without including the overlapping 4-bp sequence (Fig. 7A). The AC3 strand-specific probe (oligonucleotide cAV2) sequence was derived from the CP-sense strand which was expected to hybridize with the polycistronic mRNA (AC1-AC3 transcript) encompassing the AC1, AC2, and AC3 genes. Likewise, the CP-strand-specific probe (oligonucleotide cAC3) sequence was derived from the AC3 complementary strand, which would hybridize with the sense strand polycistronic transcript (AV2-CP transcript) encompassing the AV2 and AV1 (CP) genes. Interestingly, we detected that both the AC1-AC3 transcript (Fig. 7B lane 1) and the AV2-CP transcript (Fig. 7B lane 2) hybridized, respectively, with the AC3- and the CP strand-specific probes, indicating that these two opposite polarity transcripts overlap at their 3′ regions. This result reveals the possibility that these two overlapping opposite polarity transcripts could generate dsRNAs, the potential inducer of the plant's PTGS system. More detailed analysis on the overlap transcripts occurring in opposite orientation, which could make dsRNA, is under investigation for both DNA-A and DNA-B.
FIG. 7.
Transcript overlap at the 3′ ends of CP and AC3 genes in ACMV-[CM]. (A) Schematic representation of virion-sense CP gene and complementary-sense AC3 gene overlapped by 4 nt in ACMV-[CM]; (B) Northern blot analysis of total RNA (5 μg) isolated from ACMV-[CM]-infected N. benthamiana. An RNA blot (left) hybridized with CP strand-specific [γ-32P]ATP-labeled 70-mer (oligonucleotide cAV2), and the RNA blot (right) hybridized with AC3 strand-specific [γ-32P]ATP-labeled 70-mer (oligonucleotide cAC3) as the probe. Ethidium bromide-stained rRNA at the bottom of the blot serve as a loading control.
DISCUSSION
We describe here the ability of five distinct species of cassava-infecting geminiviruses, namely, ACMV-[CM], EACMCV, EACMV-[UG], SLCMV, and ICMV, to trigger PTGS in two plant hosts: N. benthamiana and cassava. In each case, we detected the presence of symptoms and the corresponding virus-specific siRNAs at between 7 and 14 days after inoculation. Among the five viruses that we investigated, ACMV-[CM] and SLCMV are recovery-type viruses, whereas EACMCV, EACMV-[UG], and ICMV are nonrecovery-type viruses (Pita et al., unpublished). We noticed that the efficiency of virus-induced PTGS based on the amount of virus-derived siRNAs detected varied but was strictly correlated with the recovery- or nonrecovery-type virus.
For some viruses, the host response to virus infection is characterized by the initial development of systemic symptoms, followed by recovery from symptoms (1, 7). ACMV-[CM]-infected N. benthamiana and cassava plants displayed symptoms within 5 dpi. The infected plants showed severe symptoms of yellow mosaic and leaf distortion in the systemically infected leaves until 2 weeks after inoculation; later, the plants started to show recovery. This symptom recovery phenomenon is quite unusual for geminiviruses, which prompted us to look into the possibility of the involvement of virus-induced PTGS in ACMV-[CM]-infected plants. Virus-derived siRNAs implicating virus-induced PTGS has been shown for RNA and DNA viruses, such as Potato virus X (15) Cymbidium ringspot virus (35) and for Tomato yellow leaf curl Sardinia virus (22). A similarity between viral defense and gene silencing has been predicted for a nepovirus, since recovered plants showed cross-protection to the same virus or to closely related viruses but not to unrelated viruses (28, 29). In the case of ACMV-[CM], which induces a natural recovery phenotype in infected plants, virus-derived siRNA accumulation started at 1 wpi, gradually increased over time, and was abundant in the newly developed symptom recovered leaves, both in N. benthamiana and cassava (Fig. 1), accompanied by a reduction in the levels of viral DNA and mRNA accumulation (Fig. 2A and B). We predicted that in ACMV-[CM]-infected plants the presence of virus-specific siRNAs promotes the degradation of the corresponding mRNAs in a sequence-specific manner, which in turn affects viral replication and transcription. As a result, there was a reduction in virus titer and fewer symptoms in newly developed leaves. The role of viral suppressor is important in determining recovery phenotype (35). In ACMV-[KE], a related strain to ACMV-[CM], the AC2 has been shown to be a mild suppressor of PTGS (14, 42). In Tomato yellow leaf curl China virus, a monopartite geminivirus, C2 a positional homologue of AC2, the zinc finger and DNA-binding properties were shown to be essential for mediating PTGS suppression (46, 47).
SLCMV another recovery-type virus, behaved differentially in the two hosts, N. benthamiana and cassava. The infected cassava plants recovered at a later stage of the infection cycle with siRNA accumulation to levels comparable to ACMV-[CM]. However, SLCMV-infected N. benthamiana plants died at about 3 wpi due to complete necrosis of the plant. We could detect a very low level of virus-specific siRNAs in these plants at 2 wpi; however, the accumulation fell below the level of detection at 3 wpi, suggesting that either SLCMV encodes a very strong viral suppressor of gene silencing or that the plant apoptosis pathway was induced early and did not allow PTGS to function normally. As in the case of the potyvirus HC-Pro, which differentially regulates the siRNAs involved in PTGS and the micro-RNAs involved in developmental regulation in plants (19, 23), we propose a differential interaction of the viral suppressor(s) of SLCMV with the host plants that is now under investigation.
Nonrecovery-type cassava-infecting viruses (EACMCV, EACMV-[UG], and ICMV) were also able to trigger the host's PTGS system with the production of virus-derived siRNAs; however, it was apparently not sufficient to induce recovery phenotype in infected plants. The possible explanation is that either these viruses might encode a strong suppressor of PTGS (under investigation) such as HC-Pro of TEV (2, 20) or these viruses may accumulate to such a high level that the plants still show symptoms despite the action of PTGS (27). However, the level of EACMCV DNA accumulation was less compared to that of ACMV-[CM] DNA accumulation in infected plants, suggesting an apparent difference in the strength and differential mode of action of virus-encoded suppressors on the silencing pathway.
A good deal of evidence suggests that plant viruses are capable of inducing gene silencing with the production of virus-derived siRNAs; however, at least in the case of Cymbidium ringspot virus, the siRNAs are not homogeneously distributed throughout the viral genome (35). We investigated the target gene(s) or sequences of the geminivirus-induced PTGS phenomenon in ACMV-[CM] and EACMCV, representing recovery- and nonrecovery-type viruses, respectively. Our results clearly demonstrate that siRNAs are not evenly distributed along the viral genome but are either mostly concentrated on specific sequences of viral genes or located in a specific spot of the genome. In ACMV-[CM], the DNA-A component of the genome has been preferentially targeted, particularly the region corresponding to the C termini of the AC1, AC2, and AC3 genes. In DNA-A, among the four genes (AC1, AC2, AC3, and AC4) present on the complementary-sense strand, AC1, AC2, and AC3 are highly overlapping (Fig. 6A) and are encoded by a single polycistronic transcript (38). Based on the reverse blotting experiment (siRNA containing gel blot probed with different DNA fragments as the probe), the targeted region was delineated to the C terminus of AC1 (Rep), which is indispensable for viral DNA replication and that overlaps with the N terminus of AC2 (TrAP), essential for transactivation of late genes such as CP (AV1) and BV1 (33). The fact that this target zone is located on the AC1 transcript is therefore not surprising. Although no precise quantification has been made, it is a transcript made early in the infection process and is probably more abundant than that of the AV1 transcript in the beginning of the infection. Alternatively, the identified target region of the transcript may simply be more accessible to a Dicer (a host RNase III family enzyme) (4, 36). If so, the short dsRNA segments produced may serve as a template for the RISC complex to cleave the RNA and produce more siRNAs in the same region. In addition, these siRNAs might act as primers to synthesize complementary strands of the transcript to further amplify the PTGS signal (31, 40). Although we could not detect unique high-energy secondary hairpin structures in this region by using the Zucker program, there were several stem-loops consistently associated with all of the folding predictions made in this region (data not shown). Experiments in conjunction with RNA folding prediction will be necessary to determine whether these putative secondary structures play any role in the preferred localization of siRNAs.
The results of these experiments indicate that 3′-AC1 and 5′-AC2 genes are mainly targeted by ACMV-[CM] induced PTGS. Therefore, the possible explanation for a reduced virus load at 3 wpi could be that since AC1 (an early gene indispensable for viral replication) is targeted, there would be a decrease in production of the DNA-B component and consequently poor movement of both the DNA-A and DNA-B of the virus. In addition, targeting AC2 would affect the transcriptional activation of the CP required for ssDNA accumulation and of BV1 essential protein for nuclear shuttling and long-distance movement of the viral DNA (33), which in turn affect the functions of DNA-B, thereby reducing the overall viral DNA accumulation. Therefore, it is obvious that downregulation of the AC1 and AC2 genes would result in less viral DNA accumulation, and this initial effect might get further amplified, establishing recovery phenotype in ACMV-[CM]-infected leaves. We have previously shown that the introduction of siRNAs targeting the AC1 of ACMV-[CM] downregulated the viral DNA accumulation by 70% in cultured plant cells (40).
In contrast, in the case of EACMCV, DNA-B is mainly targeted but the overall level of accumulation of virus-derived siRNAs was ∼10-fold less compared to ACMV-[CM]-derived siRNAs. The nature of EACMCV not to target the DNA-A component of the viral genome coupled with less virus-derived siRNA accumulation could possibly allow the virus to replicate all of the time at a sufficient level, resulting in nonrecovery-type in infected plants. It is noteworthy that most of the siRNAs for EACMCV were located, similarly to the ACMV-[CM], at the middle of the genome but on a different viral component. However, the mode of action of suppressors might definitely play a major role in differentiating the recovery- and nonrecovery-type viruses.
How geminiviruses trigger PTGS has been a mystery since they are nuclear replicating single-stranded, circular, DNA viruses with no known dsRNA form present in their replication cycle. However, transcription in geminiviruses is bidirectional with the production of polycistronic mRNAs occurring from the CR, which contains the promoter sequences. These polycistronic mRNAs of opposite polarity could overlap at their 3′ ends. In DNA-A of bipartite geminiviruses, two major polycistronic mRNA transcripts have been identified for ACMV-[KE] (38) and Tomato golden mosaic virus (34): one on the virion-sense strand encompassing the AV1 and AV2 genes (AV2-CP transcript) and a second on the complementary-sense strand encompassing AC1, AC2, AC3, and AC4 genes (AC1-AC3 transcript). In ACMV-[CM], the virion-sense AV1 (CP) gene and the complementary-sense AC3 gene overlap by 4 bp at their 3′ ends. Using strand-specific probes, we elucidated that both AV2-CP and AC1-AC3 transcripts extended at their 3′ ends by more than the 4-bp overlapping region by a number of nucleotides, since the probes used were devoid of the overlapping nucleotides. The resulting overlap region can form a dsRNA, which would be more than enough to potentially induce the plant's PTGS system. Our results provide insight into the ability of ssDNA viruses to trigger PTGS in plants.
In addition, the fact that each virus, although on a different viral component, seems to target the middle of the genome also supports the idea that an overlap between the two messengers, possibly extended by the host RdRp, triggers the production of specific primary siRNAs from that region, as exemplified by our results. In the case of wheat, an RdRp can even synthesize dsRNA by using exogenous ssRNA as a template without an exogenous primer (36). However, the reason why the DNA-A component for ACMV-[CM] and the DNA-B component for EACMCV are targeted remains unclear and will necessitate further investigation.
In summary, all of the geminiviruses tested here were able to induce PTGS within 1 to 2 weeks after inoculation in two different hosts, including SLCMV in N. benthamiana, which died eventually. A strong correlation between siRNA accumulation and recovery phenotype was observed for ACMV-[CM] over a period of 8 weeks. In contrast, the nonrecovery-type viruses (EACMCV, EACMV-[UG], and ICMV) do not accumulate higher levels of virus-derived siRNAs, as does ACMV-[CM]. The ability of geminiviruses to trigger PTGS, although they have an ssDNA genome and a nuclear-replication cycle, is possibly explained by demonstrating that the virion-sense (AV2-CP) and complementary-sense (AC1-AC3) transcripts overlap at their 3′ ends, which could form a short dsRNA stretch that could also serve as a primer for the extension of long dsRNAs, the potential inducer of PTGS (Fig. 7). Certainly, the capacity of each geminivirus to differentially suppress PTGS is an important component of the final viral pathogenesis and is currently under investigation. Furthermore, we determined that the C-terminal region of the AC1 gene that overlaps with the N-terminal region of AC2 of the viral genome was preferentially targeted by the ACMV-[CM]-induced (recovery-type virus) PTGS system. In the case of EACMCV (a nonrecovery-type virus) virus-induced PTGS targeted the DNA-B; however, the level of siRNA accumulation is much lower than that of ACMV-[CM]-derived siRNAs. The ability of geminiviruses to trigger gene silencing indicates that this natural phenomenon could be used to control these devastating viruses.
Acknowledgments
We thank John Stanley, John Innes Institute (United Kingdom), for providing SLCMV and ICMV and David Baulcombe and Louise Chappell, also of the John Innes Institute, for sharing details of the siRNA isolation protocol. We thank Nigel Taylor, Kathy Kahn, and Patricia Cosgrove for careful reading of the manuscript. We acknowledge our greenhouse staff, Edward Fischer and his team, for excellent care of the plants.
This study was funded by the Donald Danforth Plant Science Center.
REFERENCES
- 1.Al-Kaff, N. S., S. N. Covey, M. M. Kreike, A. M. Page, R. Pinder, and P. J. Dale. 1998. Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279:2113-2115. [DOI] [PubMed] [Google Scholar]
- 2.Anandalakshmi, R., G. J. Pruss, X. Ge, R. Marathe, A. C. Mallory, T. H. Smith, and V. B. Vance. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95:13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baulcombe, D. C. 1996. RNA as a target and an initiator of posttranscriptional gene silencing in transgenic plants. Plant Mol. Biol. 32:79-88. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 5.Cock, J. H. 1985. Cassava: new potential for a neglected crop. Westview Press, Boulder, Colo.
- 6.Cogoni, C., and G. Macino. 1997. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl. Acad. Sci. USA 94:10233-10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covey, S. N., N. S. Al-Kaff, A. Langara, and D. S. Turner. 1997. Plants combat infection by gene silencing. Nature 385:781-782. [Google Scholar]
- 8.Dalmay, T., A. Hamilton, S. Rudd, S. Angell, and D. C. Baulcombe. 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101:543-553. [DOI] [PubMed] [Google Scholar]
- 9.Dellaporta, S. L., J. Wood, and J. B. Hicks. 1983. A plant DNA minipreparation: version II. Plant Mol. Biol. Reporter 1:19-21. [Google Scholar]
- 10.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 11.Fauquet, C. M., and D. Fargette (ed.). 1988. Proceedings of the International Seminar: African cassava mosaic disease and its control. CTA/ORSTOM, Ede, Holland.
- 12.Fauquet, C. M., and J. Stanley. 2003. Geminivirus classification and nomenclature: progress and problems. Ann. Appl. Biol. 142:165-189. [Google Scholar]
- 13.Fondong, V. N., J. S. Pita, M. E. Rey, A. de Kochko, R. N. Beachy, and C. M. Fauquet. 2000. Evidence of synergism between African cassava mosaic virus and a new double-recombinant geminivirus infecting cassava in Cameroon. J. Gen. Virol. 81:287-297. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton, A., O. Voinnet, L. Chappell, and D. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamilton, A. J., and D. C. Baulcombe. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950-952. [DOI] [PubMed] [Google Scholar]
- 16.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates posttranscriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 17.Hammond, S. M., A. A. Caudy, and G. J. Hannon. 2001. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2:110-119. [DOI] [PubMed] [Google Scholar]
- 18.Hanley-Bowdoin, L., S. B. Settlage, B. M. Orozco, S. Nagar, and D. Robertson. 1999. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 18:71-106. [PubMed] [Google Scholar]
- 19.Kasschau, K., D., Z. Xie, E. Allen, C. Llave, E. J. Chapman, K. A. Krizan, and J. C. Carrington. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4:205-217. [DOI] [PubMed] [Google Scholar]
- 20.Kasschau, K. D., and J. C. Carrington. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461-470. [DOI] [PubMed] [Google Scholar]
- 21.Liu, Q., T. A. Rand, S. Kalidas, F. Du, H. Kim, E., D. P. Smith, and X. Wang. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301:1921-1925. [DOI] [PubMed] [Google Scholar]
- 22.Lucioli, A., E. Noris, A. Brunetti, R. Tavazza, V. Ruzza, A. G. Castillo, E. R. Bejarano, G. P. Accotto, and M. Tavazza. 2003. Tomato yellow leaf curl Sardinia virus rep-derived resistance to homologous and heterologous geminiviruses occurs by different mechanisms and is overcome if virus-mediated transgene silencing is activated. J. Virol. 77:6785-6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mallory, A. C., B. J. Reinhart, D. Bartel, V. B. Vance, and L. H. Bowman. 2002. A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc. Natl. Acad. Sci. USA 99:15228-15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Napoli, C., C. Lemieux, and R. Jorgensen. 1990. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2:279-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palauqui, J. C., T. Elmayan, J. M. Pollien, and H. Vaucheret. 1997. Systemic acquired silencing: transgene-specific posttranscriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16:4738-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pita, J. S., V. N. Fondong, A. Sangare, G. W. Otim-Nape, S. Ogwal, and C. M. Fauquet. 2001. Recombination, pseudorecombination, and synergism of geminiviruses are determinant keys to the epidemic of severe cassava mosaic disease in Uganda. J. Gen. Virol. 82:655-665. [DOI] [PubMed] [Google Scholar]
- 27.Pruss, G., X. Ge, X. M. Shi, J. C. Carrington, and V. B. Vance. 1997. Plant viral synergism: the potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 9:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratcliff, F., B. D. Harrison, and D. C. Baulcombe. 1997. A similarity between viral defense and gene silencing in plants. Science 276:1558-1560. [DOI] [PubMed] [Google Scholar]
- 29.Ratcliff, F. G., S. A. MacFarlane, and D. C. Baulcombe. 1999. Gene silencing without DNA:RNA-mediated cross-protection between viruses. Plant Cell 11:1207-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 31.Sijen, T., J. Fleenor, F. Simmer, K. L. Thijssen, S. Parrish, L. Timmons, R. H. A. Plasterk, and A. Fire. 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107:465-476. [DOI] [PubMed] [Google Scholar]
- 32.Stanley, J., and M. R. Gay. 1983. Nucleotide sequence of cassava latent virus DNA. Nature 301:260-262. [Google Scholar]
- 33.Sunter, G., and M. Bisaro. 1992. Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4:1321-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sunter, G., W. E. Gardiner, and D. M. Bisaro. 1989. Identification of tomato golden mosaic virus-specific RNAs in infected plants. Virology 170:243-250. [DOI] [PubMed] [Google Scholar]
- 35.Szittya, G., A. Molnar, D. Silhavy, C. Hornyik, and J. Burgyan. 2002. Short defective interfering RNAs of tombusviruses are not targeted but trigger posttranscriptional gene silencing against their helper virus. Plant Cell 14:359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang, G., B. J. Reinhart, D. P. Bartel, and P. D. Zamore. 2003. A biochemical framework for RNA silencing in plants. Genes Dev. 17:49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timmermans, M. C. P., O. P. Das, and J. Messing. 1994. Geminiviruses and their uses as extrachromosomal replicons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45:79-112. [Google Scholar]
- 38.Townsend, R., J. Stanley, S. J. Curson, and M. N. Short. 1985. Major polyadenylated transcripts of cassava latent virus and location of the gene encoding coat protein. EMBO J. 4:33-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vance, V., and H. Vaucheret. 2001. RNA silencing in plants: defense and counterdefense. Science 292:2277-2280. [DOI] [PubMed] [Google Scholar]
- 40.Vanitharani, R., P. Chellappan, and C. M. Fauquet. 2003. Short interfering RNA-mediated interference of gene expression and viral DNA accumulation in cultured plant cells. Proc. Natl. Acad. Sci. USA 100:9632-9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 42.Voinnet, O., Y. M. Pinto, and D. C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward, B. M., R. Medville, S. G. Lazarowitz, and R. Turgeon. 1997. The geminivirus BL1 movement protein is associated with endoplasmic reticulum-derived tubules in developing phloem cells. J. Virol. 71:3726-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waterhouse, P. M., M. W. Graham, and M. Wang. 1998. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95:13959-13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waterhouse, P. M., M. Wang, and T. Lough. 2001. Gene silencing as an adaptive defense against viruses. Nature 411:834-842. [DOI] [PubMed] [Google Scholar]
- 46.Wezel, R. v., X. Dong, H. Liu, P. Tien, J. Stanley, and Y. Hong. 2002. Mutation of three cysteine residues in tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and posttranscriptional gene-silencing suppression. Mol. Plant-Microbe Interact. 15:203-208. [DOI] [PubMed] [Google Scholar]
- 47.Wezel, R. v., H. Liu, Z. Wu, J. Stanley, and Y. Hong. 2003. Contribution of the zinc finger to zinc and DNA binding by a suppressor of posttranscriptional gene silencing. J. Virol. 77:696-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zamore, P. D., T. Tuschl, P. A. Sharp, and D. P. Bartel. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21- to 23-nucleotide intervals. Cell 101:25-33. [DOI] [PubMed] [Google Scholar]