Abstract
A Candida glabrata erg1 (Cgerg1) mutant, CgTn201S, was identified by transposon mutagenesis and by increased fluconazole susceptibility. CgERG1 encodes a 489-amino-acid protein which, on the basis of its homology with Saccharomyces cerevisiae ERG1, is a squalene epoxidase essential for ergosterol synthesis. Interruption following codon 475 of CgErg1p decreased the ergosterol content by 50%; caused accumulation of the squalene precursor; increased the levels of susceptibility to fluconazole, itraconazole, and terbinafine; increased the level of resistance to amphotericin B; increased the levels of rhodamine 6G and [3H]-fluconazole uptake; reduced the level of growth; and blocked growth under conditions of low oxygen tension. In addition, CgTn201S efficiently took up exogenous cholesterol from cholesterol-containing serum. Cholesterol constituted 34% of the extractable sterols in CgTn201S when it was grown aerobically on serum-containing medium. Under the same conditions, C. albicans contained only 0.1 to 1.2% cholesterol. Exogenous sterols also restored growth under conditions of low oxygen tension. Finally, complementation of the Cgerg1 mutation restored the levels of [3H]fluconazole uptake and drug susceptibility to wild-type levels.
Transposon-based insertion into Candida glabrata has been used effectively for mutational analysis (3). We adapted a commercial Tn5-based system for insertion of Tn5 into C. glabrata genomic DNA. After rescue cloning of the Tn5 DNA from the C. glabrata transformants with increased fluconazole susceptibilities, we found that one of the fluconazole-susceptible transformants, CgTn201S, had an insertion in the 3′ end of the C. glabrata ERG1 (CgERG1) open reading frame (ORF). We explored the mechanism by which this mutation causes fluconazole susceptibility because of our interest in azole resistance in this species. Prolonged treatment of patients with fluconazole causes substantial increases in the levels of azole resistance in C. glabrata. This species is naturally about 8-fold more resistant to fluconazole than Candida albicans, and following drug exposure, the levels of resistance can increase 16-fold more, leading to clinically significant fluconazole resistance (1, 18). The major mechanisms for clinical resistance described to date have been increased drug efflux and alteration of the azole-binding site, the C-14 sterol demethylase, coded for by ERG11. Recently, an erg1C1228G Saccharomyces cerevisiae mutant was reported to be aerobically viable and to have at least a 10-fold increase in the level of terbinafine resistance but no change in the level of itraconazole resistance (9). Null erg1 mutants of S. cerevisiae are not aerobically viable but are anaerobically viable in the presence of exogenous ergosterol (11). In the absence of data about Cgerg1 mutants, we studied the phenotype of strain CgTn201S, including its azole susceptibility, its ability to grow under conditions of low oxygen tension, and its uptake of exogenous sterols.
MATERIALS AND METHODS
Strains and culture conditions.
Plasmids were maintained in Escherichia coli XL1-Blue (Stratagene, La Jolla, Calif.) and Top 10F′ (Invitrogen, Carlsbad, Calif.) host cells grown in the presence of 50 μg of ampicillin per ml or 12.5 μg of chloramphenicol per ml. EC100D pir+ and EC100D pir-116 (Epicentre, Madison, Wis.) were used as hosts for the EZ::TN transposon (Epicentre), the replication of which is reliant on the R6K origin. C. glabrata strains (Table 1) were cultured on either YPD agar, which contained 1% Bacto Yeast Extract (Difco Laboratories, Detroit, Mich.), 2% Bacto Peptone (Difco), and 2% glucose (Sigma, St. Louis, Mo.), or MIN agar, which contained 0.67% Yeast Nitrogen Base plus 2% glucose without amino acids (Difco). C. glabrata ura3 mutants were selected on MIN agar containing 1 mg of 5-fluoroorotic acid (FOA; Lancaster, Pelham, N.H.) per ml and 50 μg of uracil per ml; and the genotype was confirmed by the restoration of uracil prototrophy with S. cerevisiae URA3 from YEp24 (2). C. albicans was grown on YPADU (1% Bacto Yeast Extract, 2% Bacto Peptone, 2% d-glucose, 0.12% adenine, 0.8% uridine [Sigma]). YPADU containing 20% bovine serum (Sigma) or 20 μg of sterol (ergosterol or cholesterol) per ml was used for sterol uptake analysis. The GasPak Plus system (BBL Microbiology Systems, Cockeysville, Md.) or the BBL GasPak Pouch anaerobic system (BBL Microbiology Systems) was used for cultures grown under conditions of low oxygen tension (hypoxic).
TABLE 1.
Candida strains used in this study
| Strain | Species | Genotype or description | Source or referencea |
|---|---|---|---|
| Cg1660 | C. glabrata | Clinical isolate | FHCRC |
| Cg1660u | C. glabrata | ura3 mutant of Cg1660 | This study |
| CgTn201S | C. glabrata | ura3 cgerg1::Tn5<Cm URA3> | This study |
| CgTn201C | C. glabrata | URA3 CgERG1 | This study |
| Cg4672 | C. glabrata | Clinical isolate | FHCRC |
| Cg13928 | C. glabrata | Clinical isolate | FHCRC |
| NCCLS84 | C. glabrata | Wild type | ATCC 90030 |
| BWP17 | C. albicans | Mutantb | 20 |
| CAI4 | C. albicans | ura3Δ::λimm434/ura3Δ::λimm434 | 4 |
FHCRC, Fred Hutchinson Cancer Research Center, Seattle, Wash. (a kind gift of Kieren Marr); ATCC, American Type Culture Collection, Manassas, Va.
ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG.
Fluconazole susceptibility was determined by Etest (AB Biodisk, Solna, Sweden) on YPD agar. Two milligrams of ergosterol or cholesterol was dissolved in a mixture of 50% ethanol and 50% Tween 80 (Sigma) to give a 2-mg/ml stock solution, which was used to supplement the media with a final sterol concentration of 20 μg/ml. For comparison, the same final concentration of ethanol-Tween 80 without sterol was used. The susceptibilities of the strain to fluconazole (courtesy Pfizer, Sandwich, United Kingdom), itraconazole (Janssen Pharmaceuticals, Titusville, N.J.), amphotericin B (Pharma-Tek, Huntington, N.Y.), and terbinafine (courtesy of Neil Ryder, Novartis, Vienna, Austria) were tested. The MICs for 80% growth reduction (MIC80) were determined by the NCCLS method, modified by addition of glucose 2% to the RPMI medium and incubation at 37°C or with MIN medium at 30°C.
To evaluate the ability of C. glabrata to utilize exogenous sterols, cells were inoculated into 50 ml of liquid medium and incubated at 30°C with shaking at 200 rpm overnight. The cell concentration was counted under a microscope with a hemacytometer and then diluted to give a series cell concentrations of 2 × 107, 2 × 106, 2 × 105, and 2 × 104 cells/ml. Five microliters of each concentration was added onto solid medium to give a series of spots containing 105 to 102 cells.
Construction of a C. glabrata genomic fosmid library.
Large fragments of C. glabrata genomic DNA were often found to be unstable in E. coli when they were maintained on a high-copy-number plasmid. Therefore, a fosmid was used for genomic library construction. To overcome the problem of poor DNA yield due to the low-copy-number fosmid, we used a copy control (CC) fosmid, which carries a second replication origin and which can be induced to replicate at high copy numbers. CC fosmid vector pCC1Fos (Epicentre) was modified by cloning a 1.2-kb HindIII DNA fragment from YEp24, which carries the S. cerevisiae URA3 gene, to give the vector pCC1URA3. Genomic DNA for the fosmid library was obtained from C. glabrata strain Cg13928, a fluconazole-resistant clinical isolate. Size-selected 40- to 60-kb fragments from the sheared genomic DNA of Cg13928 were ligated with Eco72I-digested pCC1URA3 DNA and packaged in vitro. A total of 1,600 colonies were obtained after transfection of E. coli EPI300 (Epicentre). The average insert size of the genomic library was about 40 kb.
Transposon construction, insertion, and rescue.
The custom Tn5 transposon Tn5<Cm URA3> was constructed to carry an E. coli replication origin, a chloramphenicol selection marker in E. coli, and a uracil selection marker in C. glabrata. A 1.2-kb HindIII DNA fragment carrying the S. cerevisiae URA3 gene from YEp24 was cloned into the HindIII site of EZ::TNpMOD-3<R6Kγori/MCS> (Epicentre) to give pMOD-ScURA3. Subsequently, a 1.2-kb XhoI DNA fragment carrying the chloramphenicol resistance gene from pBC KS+ (Stratagene) was blunt ended with T4 DNA polymerase and cloned into the blunt-ended BamHI site of pMOD-ScURA3 to give pMOD-CM1. The 2.9-kb custom transposon Tn5<Cm URA3> was purified from pMOD-CM1 after PshA1 digestion and was used for DNA tagging by transposase-mediated transposition in vitro.
Transposase (Epicentre) was used to mediate the in vitro insertion of Tn5<CmURA3> into the purified sheared 40- to 60-kb genomic DNA from fluconazole-resistant C. glabrata isolate Cg4672 (Fig. 1). DNA containing transposed genomic DNA from a transposon was then electroporated into intermediate fluconazole-resistant clinical isolate Cg1660u, a ura3 mutant of Cg1660. The fluconazole MIC80 for Cg1660 was 32 μg/ml by the standard NCCLS M27-A protocol (16). Tn5<Cm URA> served as both a selection marker and a mutagenic agent during C. glabrata transformation. Transformants exhibiting URA3 prototrophy were selected for altered fluconazole susceptibilities.
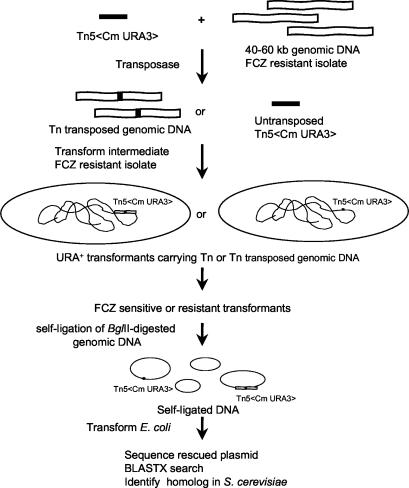
FIG. 1.
In vitro transposon (Tn) insertion and transposon rescue cloning. Sequences were obtained by using the transposon-specific primers SqFP and SqRP (Epicentre), and the sequences were used to search the Génolevures Consortium database with the BLASTX algorithm. FCZ, fluconazole.
The genomic DNA from the mutants into which the transposon had been inserted was digested with BglII, self-ligated, and used to transform E. coli EC100D pir+ or EC100D pir-116 (Epicentre) (Fig. 1). Plasmid DNA was then isolated from the E. coli transformants and sequenced by using transposon-specific primers SqFP and SqRP (Epicentre). The nucleotide sequences of the DNA were then used to search the GenBank database with the BLASTX algorithm to identify homologs. A plasmid (pTn201S) rescued from fluconazole-susceptible transformant CgTn201S was found to contain sequences homologous to that of S. cerevisiae ERG1. The full nucleotide sequence of the CgERG1 homolog was obtained from the Pasteur Institute genomic database (Génolevures Consortium; http://cbi.labri.fr/Genolevures/), prior to publication of the complete genome sequence of C. glabrata, through the courtesy of Bernard Dujon and Christophe Hennequin.
Cloning of CgERG1.
A 2.2-kb CgERG1 gene was obtained by PCR with PfuUltra DNA polymerase (Stratagene), primer set CgERG1-3S (5′-TAGAGACCACTTCGGTCAAG-3′) and CgERG1-4AS (5′-TTTCTATTGTGTGAGGAAATG-3′), and total DNA of strain NCCLS84 as the template. The PCR parameters were 2 min at 94°C, followed by 30 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 2 min 30 s and then extension at 72°C for 10 min. The PCR product was then cloned into pCR-Script Amp SK(+) (Stratagene) to give plasmid pCgERG1.
CgERG1 complementation.
For functional complementation, a full-length CgERG1 gene was obtained from the C. glabrata fosmid genomic library by screening by PCR with the primer set CgERG1-3S and CgERG1-4AS. A 7-kb HindIII DNA fragment containing CgERG1 from a positive fosmid clone was blunt ended with Klenow DNA polymerase and subcloned into the SmaI site of pCgACU (a gift from K. Kitada) (8) to give pCgERG1-f2. To replace the CgERG1 gene in CgTn201S disrupted by the transposon, pCgERG1-f2 was digested with ApaI and transformed into Tn201S. Transformants were selected on MIN-uracil-100 μg of fluconazole per ml and then replicated on a MIN-uracil-FOA agar plate. FOA-resistant transformants were selected and analyzed by PCR. Southern analysis indicated that in the clone into which Cgerg1 was inserted, Cgerg1 was replaced with the CgERG1 wild-type gene by targeted gene replacement in complemented strain CgTn201C (Fig. 2).
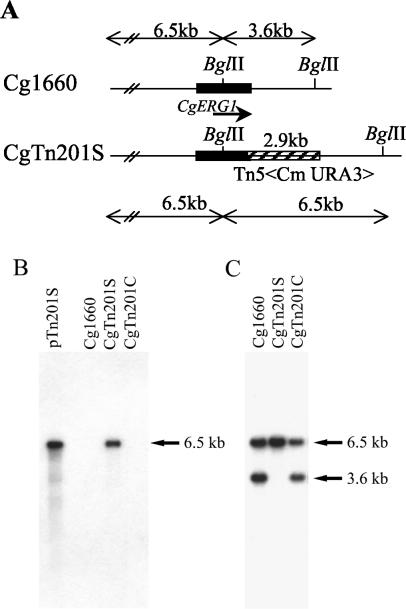
FIG. 2.
Single transposon insertion in CgTn201S. Rescued plasmid pTn210S and the genomic DNAs of Cg1660, CgTn201S, and complemented strain CgTn201C were digested with BglII, electrophoresed, and blotted onto a nylon membrane. The sizes of the DNA fragments detected are indicated in kilobases, and the insertion of a single transposon with its flanking regions of CgERG1 is shown. (A) Restriction enzyme map of CgERG1 locus in the wild-type strain (Cg1660) and the mutant into which the transposon was inserted (CgTn201S); (B) Southern blot analysis with the 2.9-kb transposon Tn5<Cm URA3> (hatched box in panel Biosciences) as the probe; (C) Southern blot analysis with the 2.2-kb CgERG1 sequence (black box in panel A) as the probe.
Sterol analysis.
To measure the sterol contents of the yeast strains both qualitatively and quantitatively, sterols were extracted from yeast cells by saponification, as described previously (13), and were then analyzed by gas chromatography. An HP5890 GC series II instrument equipped with the Hewlett-Packard CHEMSTATION software package was used to analyze the sterol profiles. The capillary column (DB-5) was 15 m by 0.25 mm by 0.25 μm and was programmed to heat from 195 to 280°C (1 min at 195°C, then an increase to 240°C at 20°C/min, and then an increase to 280°C at 2°C/min). The linear velocity was 30 cm/s, nitrogen was used as the carrier gas, and all injections were run in the splitless mode (6). To calculate the percentage of sterol per milligram of cell mass, 10 ml of the original 250-ml culture was harvested by vacuum filtration onto a preweighed 0.45-μm-pore-size Whatman filter. The cells were dried in a 75°C oven overnight, followed by desiccation at room temperature for 4 h. The amount of each individual sterol was calculated on the basis of the area under each peak of the chromatograph relative to the area for a known amount of sterol standard loaded onto the gas chromatograph by using the Hewlett-Packard sterol quantitation program. Each sample was injected twice, and the quantity of each sterol reported is the average from two injections.
Rhodamine 6G accumulation analysis.
The accumulation of rhodamine 6G (Sigma) was measured by flow cytometry in a FACS Calibur fluorescence-activated cell cytometer (Becton Dickinson, San Jose, Calif.) (7, 12). Cultures grown overnight at 30°C in MIN medium were diluted with MIN so that the optical density at 600 nm was 1 and were shaken for an additional 2 h at 30°C. Rhodamine 6G was added to give a final concentration of 0.2 μg/ml, and the culture was shaken for an additional 4 h at 30°C. After incubation, 10 μl of the culture was transferred to 0.9 ml of ice-cold phosphate-buffered saline (PBS) at pH 7.0 and incubated on ice for 5 min before fluorescence-activated cell sorter (FACS) analysis. A total of 20,000 cells were scanned with a 488-nm laser and an FL-2 filter. Cultures without rhodamine 6G were also analyzed and served as unstained controls. Data were analyzed with the CellQuest (Becton Dickinson) and FlowJo (Tree Star Inc., San Carols, Calif.) programs. The geometric mean fluorescence was used for calculation.
[3H]fluconazole accumulation assay.
C. glabrata was grown at 30°C overnight in MIN medium with and without 20 μg of ergosterol per ml under aerobic or low-oxygen-tension conditions. Cells were collected by centrifugation and resuspended in the same volume of MIN. The optical densities of the cell suspensions were measured to determine the number of cells used for fluconazole uptake analysis (17). Fluconazole uptake was measured by using drug that had been tritiated by gas exchange to a specific activity of 629 GBq/mmol (Amersham Biosciences, Arlington Heights, Ill.). [3H]fluconazole was added to the cell suspensions at a final concentration of 7.4 kBq/ml (0.2 μCi/ml; 3.6 ng/ml) (1). After 60 min of rotation at 225 rpm and 30°C, triplicate 2.5-ml samples were filtered on a Millipore (Bedford, Mass.) vacuum manifold by using 24-mm-diameter filters (Whatman GF/C, Maidstone, United Kingdom) which had been presoaked in 100 μM unlabeled fluconazole in PBS (pH 7.0). For samples recovered at zero time, the cell suspension was chilled on ice for 30 min and then filtered after the addition of tritiated fluconazole. Cells were washed on the filters twice with 8 ml of PBS containing 100 μM unlabeled fluconazole. The filters were dried at 37°C for 1 h and placed in 10 ml of Hydrofluor scintillation fluid (National Diagnostics, Atlanta, Ga.). After overnight incubation at room temperature, the radioactivity was determined with a Packard (Downer's Grove, Ill.) 2200CA Tricarb liquid scintillation counter. The results are expressed as the counts per minute per 106 cells at 0 and 60 min.
Techniques and reagents.
C. glabrata genomic DNA was isolated from overnight cultures by using glass beads. DNA was sheared by multiple pipetting steps. C. glabrata RNA was isolated from logarithmic-phase cultures with a FastRNA Pro-Red kit (QBIOgene, Carlsbad, Calif.). Purified DNA fragments were recovered by use of a Geneclean II kit (Bio 101, Vista, Calif.). Hybond-N nylon membranes (Amersham, Arlington Heights, Ill.) were used for blot analyses. DNA probes were labeled with [α-32P]dCTP (Amersham Biosciences) by using a Prime-It II kit (Stratagene). DNA cloning and hybridization analyses were done by standard protocols (19). DNA sequencing was done with a DNA sequencing kit with a dRhodamine terminator (Applied Biosystems, Foster City, Calif.) and an automatic DNA sequencing system (Applied Biosystems).
RESULTS
Transposon mutant CgTn201S contains a disrupted CgERG1 gene.
One of the transposon mutants, CgTn201S, was unable to grow on a MIN agar plate containing fluconazole at 100 μg/ml, whereas host strain Cg1660 was able to grow. Genomic DNA from CgTn201S was digested with BglII, self-ligated, and electroporated into E. coli; and the chloramphenicol-resistant clones were analyzed. DNA contiguous to Tn5<CmURA3> was sequenced. A search of the GenBank database with the BLASTX algorithm found sequence homology with S. cerevisiae ERG1, which encodes a squalene epoxidase of 496 amino acids. The full sequence of the CgERG1 ORF was obtained from the Génolevures Consortium. Tn5<Cm URA3> was found to be inserted at the codon for amino acid 476 of CgERG1 (data not shown).
Southern blot analysis with purified 2.9-kb Tn5<Cm URA3> as a probe detected a single 6.5-kb band in CgTn201S as well as in the rescued plasmid, pTn201S, while there was no signal in the parental strain, Cg1660 (Fig. 2B). A 2.2-kb CgERG1 probe detected 3.6- and 6.5-kb fragments in Cg1660 but hybridized only to the 6.5-kb fragments in CgTn201S (Fig. 2C). The 3.6-kb band was absent from CgTn201S. Together, Southern blotting and DNA sequence analyses indicated that CgERG1 is the gene targeted by the transposon in CgTn201S.
Increased expression of CgERG1 and CgERG11 in mutant CgTn201S.
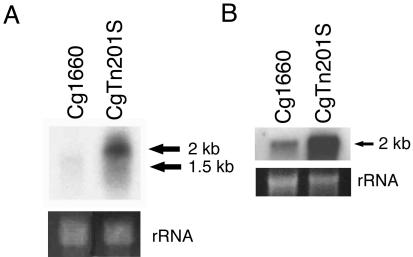
To investigate whether the expression of CgERG1 was affected by the insertion of Tn5<Cm URA3>, the CgERG1 gene was used as a probe for Northern blot analysis. The CgERG1 probe detected a transcript with an expected size of about 1.5 kb in parental strain Cg1660 (Fig. 3A). Although no 1.5-kb transcript was found in mutant CgTn201S, the probe detected abundant transcripts of approximately 2 kb. The alteration in transcript size was likely due to a read-through transcript which included part of Tn5<Cm URA3>, which had been inserted into the 3′ end of the CgERG1 ORF (Fig. 3A). To investigate the possibility of feedback regulation on the ergosterol biosynthetic pathway due to the defect in CgERG1, CgERG11 encoding an alpha-C-14-demethylase was also used as a probe for Northern blot analysis. The results show that the expression of CgERG11 is also up regulated in CgTn201S (Fig. 3B).
FIG. 3.
Expression of CgERG1 and CgERG11. Ten micrograms of total RNA from Cg1660 and CgTn201S was used for Northern blotting analysis. The membranes were hybridized with the 2.2-kb CgERG1 (A) or the 0.9-kb CgERG11 (B) probe. The sizes of the putative transcripts are indicated in kilobases. rRNA stained with ethidium bromide was used as the loading control. The CgERG11 gene was obtained from pDM3 (5).
Accumulation of squalene and reduced ergosterol synthesis in CgTn201S.
Sterol analysis of aerobically grown cultures showed that the level of accumulation of the sterol precursor, squalene, increased dramatically in CgTn201S (Table 2). Squalene is the substrate of squalene epoxidase, encoded by ERG1. While squalene constitutes only 1.6% of the total extractable sterols in Cg1660, squalene comprises 66.6% of the total extractable sterols in CgTn201S. The accumulation of squalene in CgTn201S suggested that squalene epoxidation in ergosterol synthesis was impaired due to the CgERG1 mutation in CgTn201S. The synthesis of ergosterol, which constituted 32.9% of the total extractable sterols in CgTn210S, was reduced but not abolished (63.7%) in Cg1660.
TABLE 2.
Differences in levels of cholesterol uptake and sterol compositions of C. glabrata and C. albicans strains grown aerobically
| Strain | Mediuma | Percent
|
|||
|---|---|---|---|---|---|
| Squalene | Cholesterol | Ergosterol | Sterol intermediates | ||
| BWP17 | YPADU | 0.0 | 0.0 | 79.6 | 20.4 |
| CAI4 | YPADU | 0.0 | 0.0 | 75.9 | 24.3 |
| Cg1660 | YPADU | 1.6 | 0.0 | 63.7 | 34.8 |
| CgTn201S | YPADU | 66.6 | 0.0 | 32.9 | 0.5 |
| NCCLS84 | YPADU | 1.0 | 0.0 | 80.5 | 18.5 |
| BWP17 | YPADU-BSa | 0.1 | 1.2 | 71.5 | 27.3 |
| CAI4 | YPADU-BS | 0.1 | 0.8 | 80.7 | 18.5 |
| Cg1660 | YPADU-BS | 5.0 | 9.5 | 67.5 | 18.1 |
| CgTn201S | YPADU-BS | 38.1 | 34.0 | 26.8 | 1.2 |
| NCCLS84 | YPADU-BS | 5.5 | 21.2 | 59.5 | 13.7 |
YAPDU-BS, YAPDU medium with 20% bovine serum.
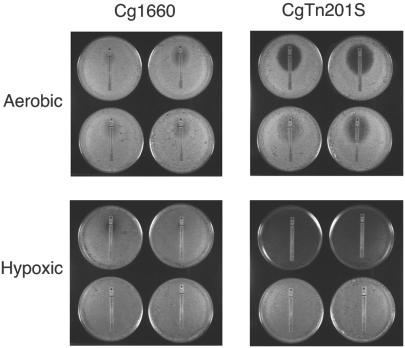
Sterol supplements restored the growth defect of CgTn201S under conditions of low oxygen tension.
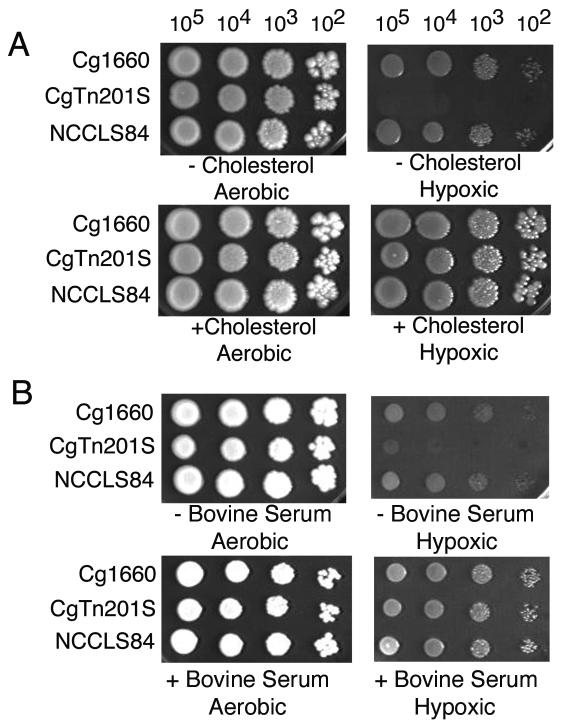
While CgTn201S grew slightly more slowly than Cg1660 under aerobic conditions, growth under conditions of low oxygen tension (hypoxia) required the presence of bovine serum or cholesterol (Fig. 4). In contrast, both parental strain Cg1660 and a standard laboratory strain, NCCLS84, grew slowly but well under conditions of low oxygen tension.
FIG. 4.
Effect of sterol on growth of CgTn201S under conditions of low oxygen tension. Two C. glabrata wild-type strains, Cg1660 and NCCLS84, as well as Cgerg1 mutant CgTn201S, were grown aerobically or under conditions of low oxygen tension (hypoxic). (A) YPD with or without 20 μg of cholesterol per ml; (B) YPD with or without 20% bovine serum.
CgTn201 had more efficient cholesterol uptake than wild-type C. glabrata and C. albicans strains.
Cholesterol is not synthesized by fungi, but both wild-type C. glabrata strains tested, Cg1660 and NCCLS84, were able to take up cholesterol from bovine serum-supplemented medium under aerobic conditions, with the levels of uptake constituting 9.5 and 21.2% of total extractable sterols, respectively (Table 2). By comparison, two wild-type strains of C. albicans, BWP17 and CAI4, showed negligible cholesterol uptake under the same conditions. Strain CgTn201S was the most efficient at taking up cholesterol from serum-containing medium, with the levels of uptake constituting 34% of the total extractable sterols.
Altered drug susceptibility in CgTn201.
The susceptibilities to fluconazole, terbinafine, itraconazole, and amphotericin B were assessed in Cg1660, CgTn201S, and the CgERG1-complemented strain (CgTn201C) by using either MIN or RPMI medium (Table 3). CgTn201S was remarkably more susceptible to the sterol synthesis inhibitors fluconazole, itraconazole, and terbinafine; but there was a slight decrease in susceptibility to the sterol-binding antifungal amphotericin B. Complementation of CgTn201S with the CgERG1 gene restored the levels of susceptibility of the CgTn201C strain to the wild-type levels. As ergosterol constituted only 32.9% of the total extractable sterols in CgTn201S, whereas it constituted 63.7% in Cg1660 (Table 2), it is possible that fewer membrane ergosterol targets for amphotericin B led to the slight reduction in susceptibility to amphotericin B.
TABLE 3.
Drug susceptibilities of C. glabrata strains
| Strain | MIC80 (μg/ml)
|
|||
|---|---|---|---|---|
| Fluconazolea | Terbinafinea | Itraconazolea | Amphotericin Bb | |
| Cg1660 | 256 | 256 | >16 | 0.125 |
| CgTn201S | 16 | 64 | 2 | 0.25 |
| CgTn201C | 256 | 256 | >16 | 0.125 |
With MIN agar at 30°C.
With RPMI at 37°C.
Sterol supplements reduced the fluconazole susceptibility of CgTn201S.
As determined by Etest, the fluconazole MIC for Cg1660 without sterol was approximately 48 μg/ml under aerobic conditions, while the fluconazole MIC for CgTn201S was dramatically reduced to approximately 4 μg/ml and a distinct inhibition zone was detected, in contrast to the fuzzy inhibition zone detected for strain Cg1660 (Fig. 5). Addition of ethanol-Tween 80 without sterol slightly increased the fluconazole susceptibilities of both Cg1660 and CgTn201S. Sterol supplementation reduced the fluconazole susceptibility of CgTn201S. No inhibition zone around fluconazole strips was observed with CgTn201S or Cg1660 under conditions of low oxygen tension and with exogenous ergosterol or cholesterol.
FIG. 5.
Ergosterol or cholesterol reduced the fluconazole susceptibility of CgTn201S. Plates were incubated aerobically or under conditions of low oxygen tension at 30°C for 2 days. The fluconazole susceptibilities of Cg1660 and CgTn201S were analyzed by Etest. The intercept of the zone of growth inhibition with the paper strip indicates the MIC. The addition of ergosterol or cholesterol is indicated by the position within each of the four groups, as follows: upper left corner, YPD; upper right corner; YPD with ethanol-Tween 80 solvent alone; lower left corner, YPD with ergosterol; and lower right corner, YPD with cholesterol. Hypoxic, conditions of low oxygen tension.
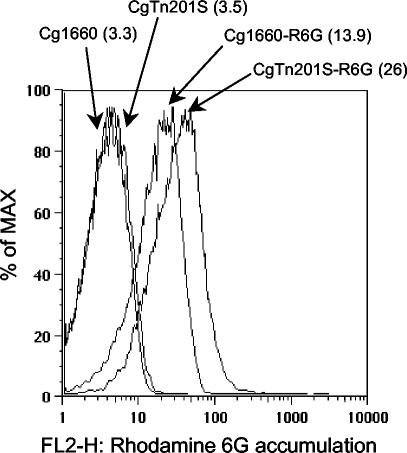
CgTn201S had increased levels of uptake of rhodamine 6G and [3H]fluconazole.
Reduced ergosterol synthesis in CgTn201S could lead to an altered membrane composition and altered permeability. Therefore, the levels of both rhodamine 6G and [3H]fluconazole accumulation were analyzed to determine uptake efficiency. The accumulation of rhodamine 6G, as determined by fluorescence cytometry, was higher in strain CgTn201S than in strain Cg1660, with geometric mean values of 26 and 13.9 arbitrary units, respectively (Fig. 6). In addition, four separate experiments demonstrated that CgTn201S had much higher levels of [3H]fluconazole uptake than Cg1660, with the increases ranging from 5.4-fold (Fig. 7A) to 7.5-fold (data not shown). The increase in the level of [3H]fluconazole uptake in CgTn201S was consistent with the finding that CgTn201S had higher levels of rhodamine 6G uptake.
FIG. 6.
Rhodamine 6G accumulation by Cg1660 and CgTn201S as determined by FACS analysis. The geometric mean fluorescence (in parentheses) is given in arbitrary units. Curves on the left are for unstained strains; curves on the right are for the strains after dye uptake.
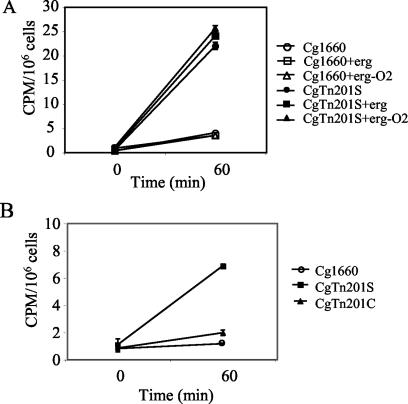
FIG. 7.
[3H]fluconazole uptake with and without ergosterol (erg) or growth under conditions of low oxygen tension (−O2). Bars indicate standard errors of the mean. (A) The level of uptake by wild-type strain Cg1660 was low under all conditions. Note that two of the three lines for Cg1660 overlap. For all conditions the level of uptake was higher in Cgerg1 mutant CgTn201S and was not restored to wild-type levels by ergosterol under aerobic conditions or conditions of low oxygen tension. (B) The level of [3H]fluconazole uptake by CgTn201S was lowered nearly to wild-type levels by CgERG1 complementation (CgTn201C).
CgERG1 complementation but not ergosterol supplementation reduced the levels of [3H]fluconazole uptake in CgTn201S.
Ergosterol supplementation was used to investigate the possibility that membrane integrity could be restored to reduce the levels of [3H]fluconazole uptake. Neither Cg1660 nor CgTn201S showed significant differences in the levels of [3H]fluconazole accumulation with exogenous ergosterol under aerobic conditions or under low oxygen tension (Fig. 7A). In contrast, the CgERG1-complemented strain, CgTn201C, had a 3.5-fold reduction in the level of [3H]fluconazole accumulation (Fig. 7B). These results may indicate a quantitative difference between the amount of ergosterol needed to restore growth under conditions of low oxygen tension and the amount needed to restore drug transport.
DISCUSSION
Transposon insertion into the coding region for the CgErg1p C terminus decreased the level of but did not ablate ergosterol synthesis. It is likely that an abnormal protein was formed, judging from the increased size and abundance of the CgERG1 transcript. As expected from a defective squalene epoxidase, squalene accumulated in the mutant. Several phenotypic consequences were unanticipated on the basis of previous studies of the ERG1 mutations in S. cerevisiae, a closely related species. The S. cerevisiae ERG1 null mutant was not viable under aerobic conditions and grew anaerobically only with addition of ergosterol to the medium (10). Aerobically viable S. cerevisiae mutants have also been reported. By mapping, the sequences of five of six different erg1 mutants selected for terbinafine resistance under aerobic conditions have been found to have single base changes in codons 402 to 433 of the 496 amino acids in S. cerevisiae Erg1p (9, 11). Squalene accumulated in mutant W/H10, and the level of ergosterol synthesis was diminished in but not absent from W/H10 (10). In CgTn201S, the C. glabrata erg1 mutant reported here, the CgERG1 ORF was interrupted at codon 476, only 13 amino acids from the carboxy terminus. The abundance of CgERG1 and CgERG11 mRNA in CgTn201S was increased significantly, which is likely due to ergosterol feedback regulation. Unlike the S. cerevisiae mutants, the susceptibilities of CgTn201S to terbinafine, itraconazole, and fluconazole were increased. Additional findings for CgTn201S were increased uptake of tritiated fluconazole and extremely poor growth under conditions of low oxygen tension. Perhaps mutant CgErg1p had a decreased efficiency of oxygen utilization. The Cgerg1 mutation in CgTn201S is unlikely to be a null mutation because CgTn201S still synthesizes some ergosterol aerobically, allowing aerobic growth of CgTn201S. In contrast, the poor growth of CgTn201S under conditions of low oxygen tension is likely due to the altered C terminus on CgErg1p, which requires high levels of oxygen tension to function properly. Exogenous sterol restored the growth of CgTn201S under conditions of low oxygen tension but did not return the levels of [3H]fluconazole uptake to the levels for the wild type. Complementation of CgERG1, however, did decrease the levels of [3H]fluconazole uptake nearly to the levels for the wild type. The altered sterol composition of CgTn201S may have interfered with drug transport, a defect not sufficiently ameliorated by exogenous ergosterol, even under conditions of low oxygen tension. Even though transport was not restored by exogenous sterol in CgTn201S, fluconazole susceptibility was reduced by exogenously added ergosterol or cholesterol under aerobic conditions and was more dramatically reduced under conditions of low oxygen tension (Fig. 5).
Decreased squalene epoxidation in CgTn210S can be inferred from the accumulation of squalene and the 50% reduction in the ergosterol content of CgTn201S. Decreased growth under hypoxic conditions and slower growth can be attributed to a decreased ergosterol content, on the basis of the ability of cholesterol-containing serum to restore both capacities. Complementation with a functioning copy of CgERG1 offered reassurance that the phenotypic changes in CgTn201S were due solely to changes in this one gene.
The ability of wild-type C. glabrata to incorporate cholesterol from serum under aerobic conditions has been noted previously (14). We confirmed the ability of C. glabrata to utilize exogenous cholesterol. Uptake of exogenous cholesterol was greatly enhanced by interruption of CgERG1 in our experiments and by the decreased expression of either CgERG9 or CgERG11 in the work of Nakayama et al. (14, 15). The antifungal mechanism of action of azoles is to block ergosterol synthesis. If wild-type C. glabrata were to take up sufficient cholesterol from host tissue, the chemotherapeutic effects of azoles could be impaired.
Acknowledgments
M.B. is a recipient of a Burroughs Wellcome Fund Mycology Scholar Award.
REFERENCES
- 1.Bennett, J. E., K. Izumikawa, and K. A. Marr. 2004. Mechanism of increased fluconazole resistance in Candida glabrata during prophylaxis. Antimicrob. Agents Chemother. 48:1773-1777. [DOI] [PMC free article] [PubMed]
- 2.Botstein, D., S. C. Falco, S. E. Stewart, M. Brennan, S. Scherer, D. T. Stinchcomb, K. Struhl, and R. W. Davis. 1979. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene 8:17-24. [DOI] [PubMed] [Google Scholar]
- 3.Castano, I., R. Kaur, S. Pan, R. Cregg, L. Penas Ade, N. Guo, M. C. Biery, N. L. Craig, and B. P. Cormack. 2003. Tn7-based genome-wide random insertional mutagenesis of Candida glabrata. Genome Res. 13:905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geber, A., C. A. Hitchcock, J. E. Swartz, F. S. Pullen, K. E. Marsden, K. J. Kwon-Chung, and J. E. Bennett. 1995. Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob. Agents Chemother. 39:2708-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hongay, C., N. Jia, M. Bard, and F. Winston. 2002. Mot3 is a transcriptional repressor of ergosterol biosynthetic genes and is required for normal vacuolar function in Saccharomyces cerevisiae. EMBO J. 21:4114-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izumikawa, K., H. Kakeya, H. F. Tsai, B. Grimberg, and J. E. Bennett. 2003. Function of Candida glabrata ABC transporter gene, PDH1. Yeast 20:249-261. [DOI] [PubMed] [Google Scholar]
- 8.Kitada, K., E. Yamaguchi, and M. Arisawa. 1996. Isolation of a Candida glabrata centromere and its use in construction of plasmid vectors. Gene 175:105-108. [DOI] [PubMed] [Google Scholar]
- 9.Klobucnikova, V., P. Kohut, R. Leber, S. Fuchsbichler, N. Schweighofer, F. Turnowsky, and I. Hapala. 2003. Terbinafine resistance in a pleiotropic yeast mutant is caused by a single point mutation in the ERG1 gene. Biochem. Biophys. Res. Commun. 309:666-671. [DOI] [PubMed] [Google Scholar]
- 10.Landl, K. M., B. Klosch, and F. Turnowsky. 1996. ERG1, encoding squalene epoxidase, is located on the right arm of chromosome VII of Saccharomyces cerevisiae. Yeast 12:609-613. [DOI] [PubMed] [Google Scholar]
- 11.Leber, R., S. Fuchsbichler, V. V. Klobucnikova, N. Schweighofer, E. Pitters, K. Wohlfarter, M. Lederer, K. Landl, C. Ruckenstuhl, I. Hapala, and F. Turnowsky. 2003. Molecular mechanism of terbinafine resistance in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 47:3890-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maesaki, S., P. Marichal, H. Vanden Bossche, D. Sanglard, and S. Kohno. 1999. Rhodamine 6G efflux for the detection of CDR1-overexpressing azole-resistant Candida albicans strains. J. Antimicrob. Chemother. 44:27-31. [DOI] [PubMed] [Google Scholar]
- 13.Molzahn, S. W., and R. A. Woods. 1972. Polyene resistance and the isolation of sterol mutants in Saccharomyces cerevisiae. J. Gen. Microbiol. 72:339-348. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama, H., M. Izuta, N. Nakayama, M. Arisawa, and Y. Aoki. 2000. Depletion of the squalene synthase (ERG9) gene does not impair growth of Candida glabrata in mice. Antimicrob. Agents Chemother. 44:2411-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakayama, H., N. Nakayama, M. Arisawa, and Y. Aoki. 2001. In vitro and in vivo effects of 14-α-demethylase (ERG11) depletion in Candida glabrata. Antimicrob. Agents Chemother. 45:3037-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. 1995. Reference method for broth dilution antifungal susceptibility testing of yeast. Approved standard M27-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 17.Parkinson, T., D. J. Falconer, and C. A. Hitchcock. 1995. Fluconazole resistance due to energy-dependent drug efflux in Candida glabrata. Antimicrob. Agents Chemother. 39:1696-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfaller, M. A., S. A. Messer, R. J. Hollis, R. N. Jones, and D. J. Diekema. 2002. In vitro activities of ravuconazole and voriconazole compared with those of four approved systemic antifungal agents against 6,970 clinical isolates of Candida spp. Antimicrob. Agents Chemother. 46:1723-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook, J., and W. Russell (ed.). 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]