Abstract
Failure of anti-Pneumocystis jiroveci prophylaxis with sulfa drugs is associated with mutations within the putative active site of the fungal dihydropteroate synthase (DHPS), an enzyme encoded by the multidomain FAS gene. This enzyme is involved in the essential biosynthesis of folic acid. The most frequent polymorphisms are two mutations leading to two amino acid changes (55Trp-Arg-57Pro to 55Ala-Arg-57Ser), observed as a single or double mutation in the same P. jiroveci isolate. In the absence of a culture method for P. jiroveci, we studied potential resistance to sulfa drugs conferred by these polymorphisms by using Saccharomyces cerevisiae as a model. Single or double mutations identical to those observed in the DHPS domain of the P. jiroveci FAS gene were introduced by in vitro site-directed mutagenesis into alleles of the S. cerevisiae FOL1 gene, which is the orthologue of the P. jiroveci FAS gene. The mutated alleles were integrated at the genomic locus in S. cerevisiae and expressed by functional complementation in a strain with a disrupted FOL1 allele. The single mutation 55Trp to 55Ala conferred resistance to sulfanilamide, whereas the single mutation 57Pro to 57Ser conferred resistance to both sulfanilamide and sulfadoxine. Both single mutations also separately conferred hypersensitivity to sulfamethoxazole and dapsone. The resistance to sulfadoxine is consistent with epidemiological data on P. jiroveci. The double mutation 55Trp-Arg-57Pro to 55Ala-Arg-57Ser conferred on S. cerevisiae a requirement for p-aminobenzoate, suggesting reduced affinity of DHPS for this substrate. This characteristic is commonly observed in mutated DHPS enzymes conferring sulfa drug resistance from other organisms. However, the double mutation conferred hypersensitivity to sulfamethoxazole, which is not in agreement with epidemiological data on P. jiroveci. Taken together, our results suggest that the DHPS polymorphisms observed in P. jiroveci confer sulfa drug resistance on this pathogen.
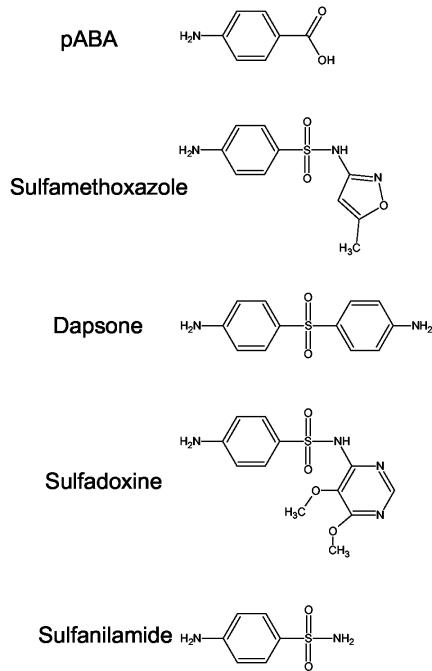
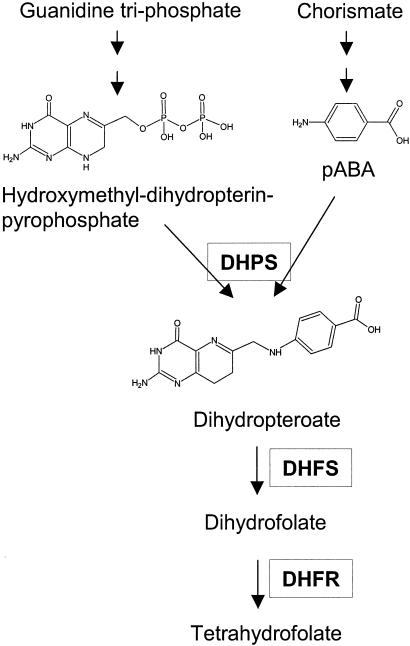
Sulfa drugs are widely used for prophylaxis and treatment of Pneumocystis jiroveci pneumonia, as well as for other microbial pathogens, such as Plasmodium falciparum, Toxoplasma gondii, and Streptococcus pyogenes. Sulfa drugs interfere with the de novo biosynthesis of folic acid by competing with p-aminobenzoate (PABA), the cosubstrate of dihydropteroate synthase (DHPS), to which they are structurally related (Fig. 1). DHPS catalyzes the condensation of PABA and hydroxymethyl-dihydropterin-pyrophosphate to produce dihydropteroate, which is subsequently converted into dihydrofolate by dihydrofolate synthase. Dihydrofolate is then reduced by dihydrofolate reductase into tetrahydrofolate, a cofactor essential for various biochemical pathways (Fig. 2). The sulfa drugs sulfamethoxazole and dapsone are most commonly used against P. jiroveci; sulfadoxine is also sometimes used. Dapsone is used alone, whereas sulfamethoxazole and sulfadoxine are administrated with a drug inhibiting dihydrofolate reductase. P. jiroveci multidomain Fasp has DHPS activity, as well as two other enzymatic activities involved in the folic acid pathway (dihydroneopterin aldolase and hydroxymethyl dihydropterin pyrophosphokinase). During recent years, widespread use of sulfa drugs has been correlated with the appearance of two mutations altering the putative active site of P. jiroveci DHPS. The mutations result in an amino acid change at positions 55 (Trp to Ala; mutation M1) and 57 (Pro to Ser; mutation M2) of the DHPS domain, as either a single or double mutation (M3) in the same isolate. These mutations are significantly associated with failure of sulfa prophylaxis (6, 11, 12, 13, 14) and, based on homology to the three-dimensional structure of Escherichia coli DHPS (1), are localized within the highly conserved active site of the protein. Moreover, mutations within the active site of DHPS have been shown to confer resistance to sulfa drugs in other pathogenic microorganisms (18). The mutation M3 is believed to confer resistance to the sulfa drugs sulfamethoxazole and dapsone, because it was the most frequent polymorphism associated with failure of prophylaxis in studies where these drugs were first-choice regimens for prophylaxis (9, 11, 12, 13, 15, 19). Mutation M2 may confer resistance to sulfadoxine, since it has been shown to be associated with failure of prophylaxis using that drug (14). However, in the absence of a long-term culture system for P. jiroveci, the resistance conferred by these mutations cannot be directly assessed.
FIG. 1.
Structures of PABA and sulfa drugs tested in the present work.
FIG. 2.
Schematic representation of de novo folate biosynthesis. DHFS, dihydrofolate synthase; DHFR, dihydrofolate reductase.
To study potential resistance to sulfa drugs conferred by the different P. jiroveci DHPS gene alleles, we used Saccharomyces cerevisiae as a model. This organism is a relative of P. jiroveci, since it also belongs to the phylum Ascomycota, and its DHPS shows high similarity to that of P. jiroveci, with a highly conserved active site (Fig. 3). Alleles of the S. cerevisiae FOL1 gene, the orthologue of the P. jiroveci FAS gene, containing mutations identical to those observed in the DHPS domain of the P. jiroveci FAS gene were created by in vitro site-directed mutagenesis, and potential resistance to sulfa drugs was tested.
FIG. 3.
Alignment of P. jiroveci and S. cerevisiae DHPS domains. The asterisks indicate identical residues (48% of P. jiroveci DHPS residues). Dashes indicate missing residues. Positions mutated in P. jiroveci are indicated in boldface.
MATERIALS AND METHODS
Microbial strains and growth media.
E. coli DH5α (Life Technologies) was used for the construction and amplification of recombinant plasmids. It was grown in Luria-Bertani medium (1% [wt/vol] Difco tryptone, 0.5% Difco yeast extract, 1% NaCl) and transformed using the method of Chung and Miller (4a). S. cerevisiae YH1 (mata leu2-3,112 trp1 tup1 ura3-52) (10) was grown in YEPD medium (1% [wt/vol] Difco yeast extract, 2% [wt/vol] Difco peptone, 2% glucose). S. cerevisiae EHY1 harbors a disruption of the FOL1 gene in the DHPS domain (mata leu2-3,112 trp1 tup1 ura3-52 fol1Δ::LEU2) (2). In the absence of folate synthesis, strain EHY1 requires the pathway end products methionine, adenine, histidine, and dTMP (MAHT). It was grown in YEPD supplemented with dTMP (100 μg/ml) or in yeast nitrogen base (YNB) medium (0.67% [wt/vol] YNB, 2% glucose) supplemented with uracil (20 μg/ml), tryptophan (40 μg/ml), methionine (20 μg/ml), adenine (50 μg/ml), histidine (20 μg/ml), and dTMP (100 μg/ml). Ura+ transformants of EHY1 were selected on solid YNB medium supplemented with tryptophan and MAHT and then streaked on the same medium lacking MAHT to check complementation of the disrupted FOL1. YNB lacking PABA and folic acid (Bio 101) was used in the sulfa resistance assays. Solid media contained 2% agar (Gibco).
Chemicals.
Sulfa drugs and PABA were resuspended at a concentration of 100 mg/ml in dimethyl sulfoxide and stored at −20°C. Sulfamethoxazole was from ICN Biomedicals Inc., sulfadoxine was from Riedel-de Haën, sulfanilamide was from Fluka, and both dapsone and PABA were from Sigma-Aldrich.
Creation of mutated alleles of S. cerevisiae FOL1 and integration into the chromosome.
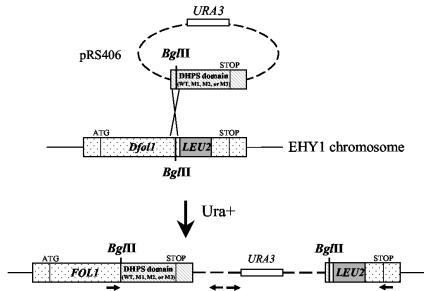
S. cerevisiae multidomain Fol1p encodes DHPS activity and, as P. jiroveci Fasp, the kinase and aldolase activities involved in folic acid biosynthesis. The distal part of FOL1 from position 813 to position 3036 (561 bp downstream of the stop codon), including the DHPS domain, was amplified by PCR from S. cerevisiae YH1 using proofreading Expand Taq polymerase (Boehringer) and the primers CGCGGATCCCTCCATACATGAATCGTCTG and CGGAATTCAAGCAATGTTACAATTG, creating unique BamHI and EcoRI restriction sites (underlined) for the PCR product. The PCR product (2,240 bp) was digested with BamHI and EcoRI and cloned into the integrative vector pRS406 (Stratagene). Mutations in the DHPS active site identical to those observed in P. jiroveci were created by in vitro site-directed mutagenesis using the Stratagene Quickchange XL Site-Directed Mutagenesis kit following the manufacturer's instructions. The mutations A to G and C to T were created at nucleotide positions 1789 and 1795 of the FOL1 gene, respectively, leading to the amino acid changes Trp to Ala and Pro to Ser at positions 597 and 599 of Fol1p. The mutagenic primers were from position 1660 to position 1690. pRS406, with part of the S. cerevisiae FOL1 gene with a mutation in the DHPS domain, was integrated (8) at the genomic locus in strain EHY1 by transformation for uracil prototrophy using the lithium acetate procedure (4). Linearization of the plasmid by restriction with BglII at position 1173 of the FOL1 gene stimulated homologous recombination at this location. The presence of the mutations within the recombinant strains was confirmed by DNA sequencing of PCR products spanning the entire FOL1 gene using an ABI PRISM 310 automated sequencer and a Big Dye Terminator DNA sequencing kit (both from Perkin-Elmer Biosystems). Yeast DNA templates for PCR were prepared as described previously (17). Two clones of each S. cerevisiae recombinant strain were analyzed in each experiment, and similar results were obtained.
Cloning of the wild-type FOL1 gene.
The entire wild-type FOL1 gene from position 725 upstream of the start codon to position 561 downstream of the stop codon was amplified from S. cerevisiae YH1 using proofreading Expand Taq polymerase and the primers AACTGCAGGTTTCTGTAGGGTTCAATTG and CGGAATTCAAGCAATGTTACAATTG, creating unique PstI and EcoRI restriction sites (underlined) for the PCR product. The PCR product (3,771 bp) was digested with PstI and EcoRI, cloned in the centromeric plasmid YCplac22 (5), and introduced into S. cerevisiae by transformation for tryptophan prototrophy.
Sulfa drug resistance assays on solid medium.
Strains with plasmid integrated into the chromosome were grown overnight at 30°C with shaking in liquid YEPD medium. Strains harboring the centromeric plasmid carrying the wild-type FOL1 gene were grown in YNB medium to select for the plasmid. The culture was diluted at an absorbance at 540 nm of 2 (ca. 1.5 × 107 cells/ml), and serial 10-fold dilutions in sterile water were prepared. Five microliters of each dilution was spotted onto solid minimal YNB medium lacking PABA or YNB containing 0.075 μg of PABA/ml. Sulfa drugs were added to the molten medium at the concentrations indicated in Results below. Pictures were taken after 4 to 6 days of incubation at 30°C.
Sulfa drug resistance assays in liquid medium.
Sulfa drug resistance was quantified in liquid minimal YNB medium lacking PABA or YNB containing 0.05 μg of PABA/ml. Cells grown overnight in YEPD medium were diluted to 2.5 × 105/ml in 0.2 ml of medium containing sulfa drugs at various concentrations in a well of a 96-well microplate (Corning Inc.). After 3 or 4 days of growth at 30°C without shaking, the cultures were diluted 10-fold in sterile water, and the absorbance at 540 nm was measured using a microplate reader (3550-UV; Bio-Rad). The amount of growth in the culture without drug was considered to be 100%, and the amount of growth at each sulfa drug concentration was divided by that of the culture without drug to determine the relative growth. Generation times under these conditions were determined by absorbance measurements during incubation.
RESULTS
Construction and phenotype of S. cerevisiae recombinant strains with mutated FOL1.
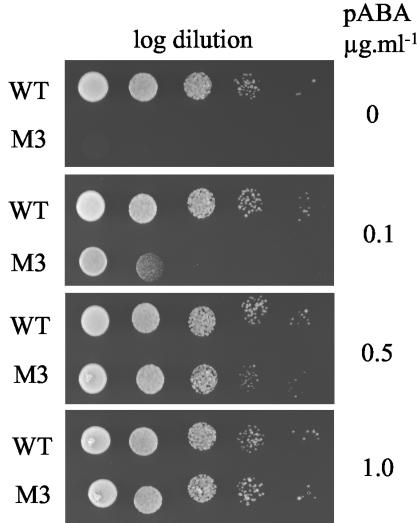
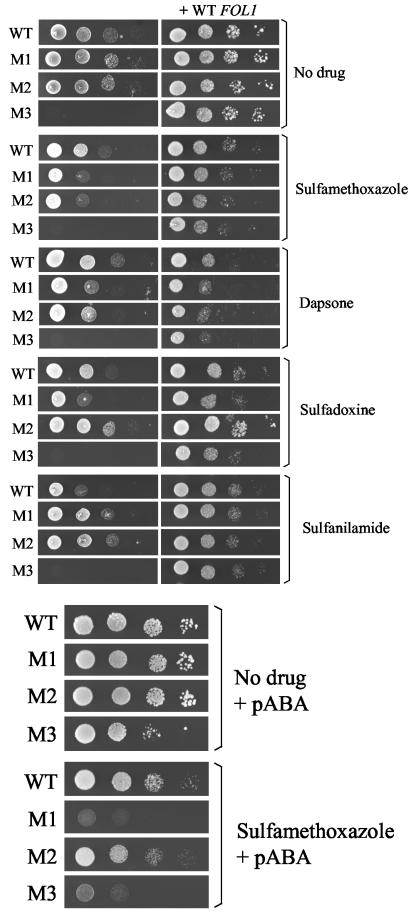
Alleles of the S. cerevisiae FOL1 gene with the mutation M1, M2, or M3 in the DHPS domain were created by site-directed mutagenesis in an integrative plasmid and introduced by chromosomal integration into strain EHY1 harboring a disruption of the FOL1 gene in the DHPS domain (Fig. 4). A strain with a wild-type FOL1 allele was also constructed using the same procedure. The correct integration of the plasmid within the chromosome was checked by specific PCRs using the primers shown in Fig. 4. The recombinant strains are hereafter designated WT, M1, M2, and M3. The requirement for methionine, adenine, histidine, and dTMP due to FOL1 disruption was no longer observed in the four recombinant strains, indicating complementation of the disrupted FOL1 gene by reconstitution of a functional FOL1. Unlike strains WT, M1, and M2, strain M3 required the cosubstrate PABA for growth (Fig. 5), suggesting a reduced affinity for PABA in Fol1p with mutation M3.
FIG. 4.
Construction of S. cerevisiae recombinant strains with FOL1 gene with a mutation in the DHPS domain. Strain EHY1 was transformed with pRS406 linearized with BglII, which carried part of the FOL1 gene with a wild-type or mutated (M1, M2, or M3) DHPS domain. The horizontal arrows represent PCR primers used for controlling the genetic structure of the recombinant strains.
FIG. 5.
pABA requirement of strain M3. Log dilutions of a suspension of cells at 1.5 × 107/ml were spotted on minimal medium with the indicated concentrations of PABA and incubated for 3 days at 30°C. The most concentrated suspension (1.5 × 107 cells/ml) is on the left.
Test of sulfa drug resistance of the recombinant strains by dilution on solid medium.
Because of the competition between sulfa drugs and PABA, sulfa drug resistance is better tested in minimal medium devoid of PABA (3). The requirement for PABA in strain M3 prevented its testing under these conditions. Four different sulfa drugs were tested: (i) sulfamethoxazole, (ii) dapsone, (iii) sulfadoxine, and (iv) sulfanilamide, which is not used against P. jiroveci but is the first sulfa drug isolated from which all other sulfa drugs have been derived artificially. When compared to strain WT, both strains M1 and M2 showed resistance to sulfanilamide but hypersensitivity to sulfamethoxazole and dapsone (Fig. 6, top). Strain M2 showed resistance to sulfadoxine, whereas strain M1 showed hypersensitivity to the drug. The lowest concentration of PABA in solid medium allowing sufficient growth of strain M3 was 0.075 μg/ml. Under these conditions, only sulfamethoxazole inhibited growth of the recombinant strains at the concentrations that could be tested (precipitation of sulfa drugs occurs at higher concentrations); sulfanilamide, dapsone, and sulfadoxine had no effect. In contrast to the situation without PABA in the medium, strain M2 was less hypersensitive to sulfamethoxazole than strain M1 (Fig. 6, bottom). Although its growth was reduced in the absence of drug compared to strain WT, strain M3 appeared as hypersensitive as strain M1 to sulfamethoxazole.
FIG. 6.
Sulfa drug resistance assay on solid medium of the recombinant S. cerevisiae strains with mutated or wild-type FOL1. Log dilutions of a suspension of cells at 1.5 × 107/ml were spotted on minimal medium with and without a sulfa drug and incubated for 4 to 6 days of growth at 30°C. (Top) strains harboring (+WT FOL1) or not harboring wild-type FOL1 cloned in a centromeric plasmid spotted on minimal YNB medium devoid of PABA. Sulfamethoxazole, dapsone, sulfadoxine, and sulfanilamide were at 20, 100, 100, and 200 μg/ml, respectively. (Bottom) Strains without plasmid spotted on minimal YNB medium containing 0.075 μg of PABA/ml with or without 400 μg of sulfamethoxazole/ml.
To confirm that the phenotypes of the recombinant strains were due to the mutations in the DHPS domain of FOL1, we attempted their complementation by introducing a centromeric plasmid expressing the entire wild-type FOL1 gene under the control of its own promoter and terminator. When compared to strain WT harboring the plasmid with wild-type FOL1, the phenotypes of all strains were complemented, although the resistance of M2 to sulfadoxine was not fully complemented (Fig. 6, top).
Test of sulfa drug resistance of recombinant strains in liquid medium and determination of IC50.
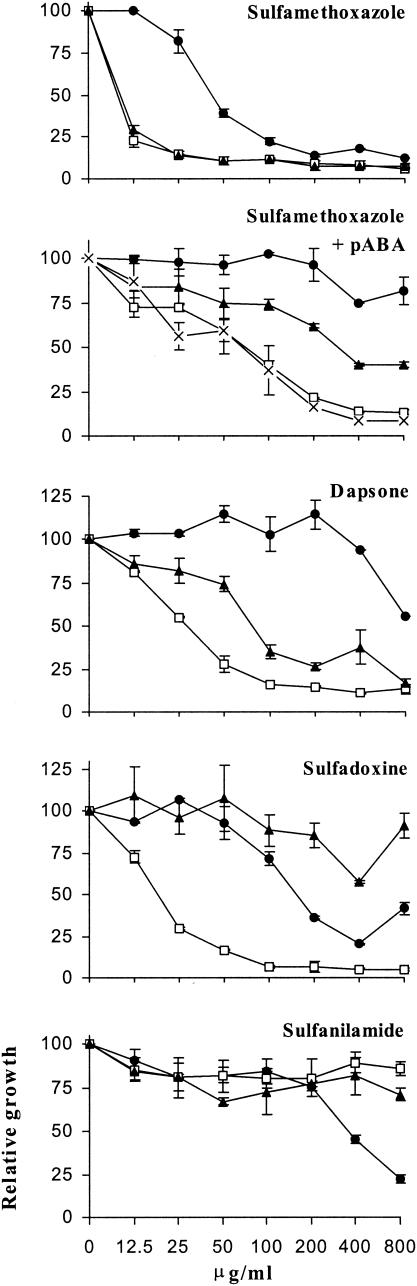
To quantify the resistance and hypersensitivity to sulfa drugs conferred by the mutations M1, M2, and M3, susceptibility assays were performed in liquid YNB medium devoid of PABA or in YNB medium supplemented with 0.05 μg of PABA/ml to allow the growth of strain M3 (Fig. 7). Cells were cultured in the presence of sulfa drugs within wells of microplates for 3 to 4 days, and growth was measured by absorbance. The increase of absorbance at the highest concentrations of sulfadoxine is due to precipitation of the drug. These assays confirmed the resistance and hypersensitivity phenotypes of each recombinant strain observed in the tests on solid medium. Resistance of strains M1 and M2 to sulfanilamide occurred only at the highest concentrations of the drug. In the presence of PABA, only sulfamethoxazole inhibited the growth of the recombinant strains, as on solid medium. In the presence of PABA but with no drug, the doubling time of strain M3 (8 h) was greater than those of strains WT, M1, and M2 (5 h), so that strain M3 reached saturation only after 4 days of incubation, whereas only 3 days was necessary for the other strains. The concentrations inhibiting growth by 50% (IC50s) observed in these experiments are given in Table 1. Several values were >800 μg of sulfa drug/ml, the highest drug concentration that could be tested. The important standard deviations of these values reflect variations from experiment to experiment, presumably due to uncontrolled physiological variations of the cells.
FIG. 7.
Sulfa drug resistance assay in liquid medium of the recombinant S. cerevisiae strains with mutated or wild-type FOL1. Cells at 2.5 × 106/ml were incubated in minimal medium lacking PABA or containing 0.05 μg of PABA/ml (+ pABA) with increasing concentrations of sulfa drugs for 3 to 4 days at 30°C, and the absorbance at 540 nm was measured. Circles, strain WT; squares, strain M1; triangles, strain M2; ×, strain M3. One representative experiment out of three is shown. The error bars indicate standard deviations of duplicate measurements.
TABLE 1.
IC50s of sulfa drugs in liquid media for recombinant S. cerevisiae strains with wild-type or mutated FOL1 genes
| Sulfa drug | IC50 (μg/ml)a for strain:
|
|||
|---|---|---|---|---|
| WT | M1 | M2 | M3 | |
| Sulfamethoxazole | 61.5 ± 24.8 | 9.1 ± 1.4 | 11.0 ± 2.9 | −b |
| Sulfamethoxazole + 0.05 μg of PABA/ml | >800c | 63.7 ± 31.8 | 268.1 ± 28.7 | 60.5 ± 23.7 |
| Dapsone | >800 | 54.8 ± 42.7 | 102.9 ± 26.8 | − |
| Sulfadoxine | 123.8 ± 53.1 | 17.2 ± 7.3 | >800 | − |
| Sulfanilamide | 286.8 ± 115.6 | >800 | >800 | − |
Sulfa drug resistance was tested in liquid minimal medium lacking PABA or supplemented with 0.05 μg of PABA/ml as described in the legend to Fig. 7. Each value is the average of at least two independent experiments ± standard deviation.
−, not measurable because there was no growth in the absence of PABA.
Highest concentration without precipitation of sulfa drug.
DISCUSSION
To study potential resistance conferred by P. jiroveci DHPS mutations associated with failure of prophylaxis with sulfa drugs, we used S. cerevisiae as a model. We constructed S. cerevisiae strains carrying identical mutations and tested their resistance to sulfa drugs. Introduction of the DHPS mutations into the chromosomal FOL1 location led to a single mutated copy of the FOL1 gene, ensuring an expression level similar to that of the wild type. Recombinant S. cerevisiae strains displayed resistance and hypersensitive phenotypes toward different sulfa drugs. Recently, site-directed mutagenesis of S. cerevisiae has also been used to show that the mutations observed in P. jiroveci mitochondrial cytochrome b confer resistance to atovaquone, a second-choice drug against P. jiroveci (7).
When introduced in S. cerevisiae, mutation M2 conferred resistance to both sulfadoxine and sulfanilamide but caused hypersensitivity to sulfamethoxazole and dapsone. Mutation M1 exhibited the same phenotypes as M2 except that it conferred hypersensitivity to sulfadoxine. The resistance to sulfadoxine conferred by mutation M2 is consistent with previously reported epidemiological data on P. jiroveci. Indeed, all 14 patients investigated who experienced failure of prophylaxis with sulfadoxine (as part of sulfadoxine and pyrimethamine [Fansidar]) were infected with P. jiroveci carrying mutation M2 (14). The interpretation of the results obtained with mutation M3 (mutation M1 plus mutation M2) is more complicated. This polymorphism resulted in a requirement for PABA, strongly suggesting a reduced affinity of the mutated DHPS for this substrate. Such a phenotype has been reported for sulfa drug-resistant DHPS with point mutations from others organisms (E. coli, Streptococcus pneumoniae, and Campylobacter jejuni) (18). It is generally accompanied by a reduction of affinity for sulfa drugs, but to a higher extent than for PABA, explaining the resistance phenotype. Therefore, this phenotype suggests that mutation M3 may confer sulfa drug resistance. Moreover, epidemiological data suggest that mutation M3 in P. jiroveci may confer resistance to sulfamethoxazole and dapsone, because it was the most frequent polymorphism associated with failure of prophylaxis in studies where these drugs were first-choice regimens for prophylaxis (9, 11, 12, 13, 15, 19). However, in our assays, mutation M3 conferred hypersensitivity to sulfamethoxazole. This contradiction suggests the following hypothesis. The alteration of DHPS due to mutation M3 may be less pronounced in P. jiroveci than in S. cerevisiae, so mutation M3 may confer resistance to sulfa drugs in P. jiroveci but not in S. cerevisiae because the enzyme is too altered. However, other hypotheses cannot be excluded. For example, mutation M3 may also be very deleterious in P. jiroveci. This mutation may inactivate the folate biosynthesis pathway and select for cells able to use folate from the human host, thereby circumventing the effects of sulfa drugs. Alternatively, mutation M3 may be linked in P. jiroveci to a mechanism conferring sulfa drug resistance other than target modification. Site-directed mutagenesis of DHPS enzymes of other fungi may be useful to further investigate this issue.
The DHPS enzymes of S. cerevisiae and P. jiroveci show high similarity (Fig. 3), and based on homology to the three-dimensional structure of E. coli DHPS (1), their Trp-Arg-Pro motifs are located within the active site of the enzyme. Our observations suggest that mutations M1, M2, and M3 in the Trp-Arg-Pro motif alter the affinity of S. cerevisiae DHPS for its cosubstrate PABA and for sulfa drugs. The replacement of Trp by Ala and/or of Pro by Ser in the Trp-Arg-Pro motif probably modifies the position of the Arg which is involved in binding to PABA and sulfa drugs and thus may alter the affinity for these molecules. Mutations M1 and M2 conferred resistance to some sulfa drugs but hypersensitivity to others. This suggests that the decrease in affinity for a given sulfa drug is accompanied by an increase in affinity for another sulfa drug. This contrasts with P. falciparum, in which the different mutant DHPS enzymes confer cross-resistance to various sulfa drugs (20). Thus, a P. jiroveci strain carrying mutation M2 might be more sensitive to sulfamethoxazole than a wild-type strain, suggesting that DHPS genotyping may help to improve treatment of P. jiroveci pneumonia.
The increase in P. falciparum resistance to sulfa drugs seems to be correlated with an accumulation of DHPS mutations over time, some of them not localized in the active site (16). P. jiroveci Fasp comprises four domains, fasA and -B, encoding the aldolase; fasC, encoding the pyrophosphokinase; and fasD, encoding DHPS. The fact that the fasA domain seems to be necessary for maximum activity of the fasD domain suggests that mutations in the fasA domain may influence the properties of DHPS (22). Thus, additional mutations in the P. jiroveci FAS gene may further increase sulfa drug resistance in the future. In the majority of the studies, only a small portion spanning the DHPS active site has been analyzed. It may be interesting to investigate the entire P. jiroveci FAS gene from patients with clinical resistance to treatment with sulfa drugs. The description of isolates of Pneumocystis carinii infecting mice that are resistant to sulfamethoxazole and that do not show alteration of the DHPS domain of the FAS gene suggests that other mechanisms than target alteration may confer resistance to sulfa drugs on P. jiroveci (B. Lane, P. Hossler, M. Bartlett, S. Queener, T. Oreilly, J. Smith, and S. Meshnick, Abstr. 4th International Workshops Opportunistic Protists, 1996). Alteration of drug uptake or detoxification may be involved. A major facilitator membrane translocase has been shown to be involved in sulfa drug resistance of E. coli (21).
In conclusion, our study demonstrates that the DHPS mutations associated with failure of prophylaxis in P. jiroveci confer resistance to the sulfa drugs sulfadoxine and sulfanilamide on S. cerevisiae. This suggests that these mutations confer sulfa drug resistance on P. jiroveci and further supports the emergence of this resistance.
Acknowledgments
We thank Sophie Chevalley for excellent technical assistance. We are grateful to I. Macreadie who kindly provided strains YH1 and EHY1.
This study was supported by the Swiss National Program on AIDS Research grant 3345-65407 and the Swiss Federal Office for Education and Science for participation in the EUROCARINII Project, Framework V Program, European Commission, contract QLK2-CT-2000-01369.
REFERENCES
- 1.Achari, A., D. O. Somers, J. N. Champness, P. K. Bryant, J. Rosemond, and D. K. Stammers. 1997. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat. Struct. Biol. 4:490-497. [DOI] [PubMed] [Google Scholar]
- 2.Bayly, A. M., J. M. Berglez, O. Patel, L. A. Castelli, E. G. Hankins, P. Coloe, C. Hopkins Sibley, and I. G. Macreadie. 2001. Folic acid utilisation related to sulfa drug resistance in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 204:387-390. [DOI] [PubMed] [Google Scholar]
- 3.Castelli, L. A., N. P. Nguyen, and I. G. Macreadie. 2001. Sulfa drug screening in yeast: fifteen sulfa drugs compete with p-aminobenzoate in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 199:181-184. [DOI] [PubMed] [Google Scholar]
- 4.Chen, D. C., B. C. Yang, and T. T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83-84. [DOI] [PubMed] [Google Scholar]
- 4a.Chung, C. T., and R. H. Miller. 1988. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 16:3580. [DOI] [PMC free article] [PubMed]
- 5.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia-coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking 6-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 6.Helweg-Larsen, J., T. L. Benfield, J. Eugen-Olsen, J. D. Lundgren, and B. Lundgren. 1999. Effects of mutations in Pneumocystis carinii dihydropteroate synthase gene on outcome of AIDS-associated P. carinii pneumonia. Lancet 354:1347-1351. [DOI] [PubMed] [Google Scholar]
- 7.Hill, P., J. Kessl, N. Fisher, S. Meshnick, B. L. Trumpower, and B. Meunier. 2003. Recapitulation in Saccharomyces cerevisiae of cytochrome b mutations conferring resistance to atovaquone in Pneumocystis jiroveci. Antimicrob. Agents Chemother. 47:2725-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinnen, A., J. B. Hicks, and G. R. Fink. 1978. Transformation of yeast. Proc. Natl. Acad. Sci. USA 75:1929-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, L., C. B. Beard, J. Creasman, D. Levy, J. S. Duchin, S. Lee, N. Pieniazek, J. L. Carter, C. del Rio, D. Rimland, and T. R. Navin. 2000. Sulfa or sulfone prophylaxis and geographic region predict mutations in the Pneumocystis carinii dihydropteroate synthase gene. J. Infect. Dis. 182:1192-1198. [DOI] [PubMed] [Google Scholar]
- 10.Huang, T., B. J. Barclay, T. I. Kalman, R. C. Vonborstel, and P. J. Hastings. 1992. The phenotype of a dihydrofolate reductase mutant of Saccharomyces cerevisiae. Gene 121:167-171. [DOI] [PubMed] [Google Scholar]
- 11.Kazanjian, P., W. Armstrong, P. A. Hossler, W. Burman, J. Richardson, C. H. Lee, L. Crane, J. Katz, and S. R. Meshnick. 2000. Pneumocystis carinii mutations are associated with duration of sulfa or sulfone prophylaxis exposure in AIDS patients. J. Infect. Dis. 182:551-557. [DOI] [PubMed] [Google Scholar]
- 12.Kazanjian, P., A. B. Locke, P. A. Hossler, B. R. Lane, M. S. Bartlett, J. W. Smith, M. Cannon, and S. R. Meshnick. 1998. Pneumocystis carinii mutations associated with sulfa and sulfone prophylaxis failures in AIDS patients. AIDS 12:873-878. [DOI] [PubMed] [Google Scholar]
- 13.Ma, L., L. Borio, H. Masur, and J. A. Kovacs. 1999. Pneumocystis carinii dihydropteroate synthase but not dihydrofolate reductase gene mutations correlate with prior trimethoprim-sulfamethoxazole or dapsone use. J. Infect. Dis. 180:1969-1978. [DOI] [PubMed] [Google Scholar]
- 14.Nahimana, A., M. Rabodonirina, G. Zanetti, I. Meneau, P. Francioli, J. Bille, and P. M. Hauser. 2003. Association between a specific Pneumocystis jiroveci dihydropteroate synthase mutation and failure of pyrimethamine/sulfadoxine prophylaxis in human immunodeficiency virus-positive and -negative patients. J. Infect. Dis. 188:1017-1023. [DOI] [PubMed] [Google Scholar]
- 15.Navin, T. R., C. B. Beard, L. Huang, C. del Rio, S. Lee, N. J. Pieniazek, J. L. Carter, T. Le, A. Hightower, and D. Rimland. 2001. Effect of mutations in Pneumocystis carinii dihydropteroate synthase gene on outcome of P. carinii pneumonia in patients with HIV-1: a prospective study. Lancet 358:545-549. [DOI] [PubMed] [Google Scholar]
- 16.Olliaro, P. 2001. Mode of action and mechanisms of resistance for antimalarial drugs. Pharmacol. Ther. 89:207-219. [DOI] [PubMed] [Google Scholar]
- 17.Sanglard, D., and A. Fiechter. 1992. DNA transformations of Candida tropicalis with replicating and integrative vectors. Yeast 8:1065-1075. [DOI] [PubMed] [Google Scholar]
- 18.Skold, O. 2000. Sulfonamide resistance: mechanisms and trends. Drug Resist. Updat. 3:155-160. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi, T., N. Hosoya, T. Endo, T. Nakamura, H. Sakashita, K. Kimura, K. Ohnishi, Y. Nakamura, and A. Iwamoto. 2000. Relationship between mutations in dihydropteroate synthase of Pneumocystis carinii f. sp. hominis isolates in Japan and resistance to sulfonamide therapy. J. Clin. Microbiol. 38:3161-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Triglia, T., J. G. T. Menting, C. Wilson, and A. F. Cowman. 1997. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 94:13944-13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vedantam, G., G. G. Guay, N. E. Austria, S. Z. Doktor, and B. P. Nichols. 1998. Characterization of mutations contributing to sulfathiazole resistance in Escherichia coli. Antimicrob. Agents Chemother. 42:88-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volpe, F., S. P. Ballantine, and C. J. Delves. 1993. The multifunctional folic acid synthesis fas gene of Pneumocystis carinii encodes dihydroneopterin aldolase, hydroxymethyldihydropterin pyrophosphokinase and dihydropteroate synthase. Eur. J. Biochem. 216:449-458. [DOI] [PubMed] [Google Scholar]