Abstract
Staphylococcal bacteria are a prevalent cause of infections associated with foreign bodies and indwelling medical devices. Bacteria are capable of escaping antibiotic treatment through encapsulation into biofilms. RNA III-inhibiting peptide (RIP) is a heptapeptide that inhibits staphylococcal biofilm formation by obstructing quorum-sensing mechanisms. K4-S4(1-13)a is a 13-residue dermaseptin derivative (DD13) believed to kill bacteria via membrane disruption. We tested each of these peptides as well as a hybrid construct, DD13-RIP, for their ability to inhibit bacterial proliferation and suppress quorum sensing in vitro and for their efficacy in preventing staphylococcal infection in a rat graft infection model with methicillin-resistant Staphylococcus aureus (MRSA) or S. epidermidis (MRSE). In vitro, proliferation assays demonstrated that RIP had no inhibitory effect, while DD13-RIP and DD13 were equally effective, and that the chimeric peptide but not DD13 was slightly more effective than RIP in inhibiting RNA III synthesis, a regulatory RNA molecule important for staphylococcal pathogenesis. In vivo, the three peptides reduced graft-associated bacterial load in a dose-dependent manner, but the hybrid peptide was most potent in totally preventing staphylococcal infections at the lowest dose. In addition, each of the peptides acted synergistically with antibiotics. The data indicate that RIP and DD13 act in synergy by attacking bacteria simultaneously by two different mechanisms. Such a chimeric peptide may be useful for coating medical devices to prevent drug-resistant staphylococcal infections.
Nosocomial infections due to Staphylococcus aureus and Staphylococcus epidermidis have increased over the last decades (36, 41, 42). These common skin bacteria are also known to be a frequent cause of infections related to prosthetic and indwelling medical devices (16, 46), where these infections are related to bacterial biofilm formation on material surfaces, such as venous or urinary catheters, prosthetic heart valves, orthopedic devices, and contact lenses (16, 40, 46, 63). In particular, the late-appearing vascular graft infections are one of the most feared complications that the vascular surgeon treats, frequently resulting in prolonged hospitalization, organ failure, amputation, and death (7, 34).
Effective strategies for the prevention of prosthetic infections vary and include the use of antimicrobials bound in high concentrations to prosthetic grafts (11, 44, 63, 69, 71). Because of the increase in resistance to antibiotics, several studies have focused on developing new prosthetic materials that reduce biofilm formation (28, 71). One of the most interesting studies was based on the fact that the resistance of a biofilm to antimicrobial agents is acquired as a multicellular strategy that relies on exchange of chemical signals between cells in a process known as quorum sensing (48). Thus, interfering with this mechanism of bacterial cell-cell communication could provide a novel approach to prevent biofilm formation. A seven-amino-acid peptide termed the RNA III-inhibiting peptide (RIP) was described to suppress diseases caused by S. aureus and S. epidermidis (2-6, 14, 27, 29, 73).
RIP inhibits pathogenesis via biofilm formation (6) and toxin production (73) by disrupting quorum-sensing mechanisms through inhibition of TRAP phosphorylation (4). TRAP is a protein unique to staphylococci but is highly conserved among strains, and therefore its inhibitor RIP was effective against any staphylococcal strain tested (2, 3, 14, 27, 29, 73). RIP does not directly kill the bacteria but interferes with their signal transduction, thus making them nonpathogenic.
The ubiquitously produced cationic antimicrobial peptides (1, 9, 23, 30, 31, 39, 57, 72) are attracting increasing research and clinical interest for their roles in innate immunity and for their potential uses in various antimicrobial fields (50, 74). The cytoplasmic membrane was proposed to be their ultimate target. Being cationic, the peptides interact electrostatically with the negatively charged phospholipid headgroups and insert into the membrane bilayer in a manner that leads to its disruption (22, 33, 35, 42, 65). Although the steps involved in this mechanism remain to be delineated, there is a large body of experimental data demonstrating that antimicrobial peptides display a direct correlation between antibiotic effect and increased plasma membrane permeability, conductance of ions across lipid bilayers, and dissipation of the transmembrane electric potential (17, 21, 24, 39, 49, 58, 59, 62, 68, 66). Thus, while their precise mechanism of action is not fully understood, their microbicidal effect is believed to result from the peptides' capacity to disrupt the ordered membrane structure of target cells.
The molecular basis for selective activity between mammalian and bacterial cells is also ill defined but is believed to result from differences in the lipid composition of target versus nontarget cells, such as membrane fluidity and high negative charge density (1, 12, 45, 74). These differences seem to be responsible for the preferred accumulation of antimicrobial peptides on microbial membranes. Numerous studies have demonstrated that the peptides' physicochemical properties, i.e., amphipathy, positive charge content, and hydrophobicity, are the main factors affecting membrane lysis activity (1, 8, 12, 42). Accordingly, isomers composed of all d-amino acids are as active as the l-enantiomers, implying that the mechanism of action is not mediated by interaction with a stereo-specific receptor. These properties enabled several cationic antimicrobial peptides to escape microbial mechanisms involved in multidrug resistance (12, 25, 56).
Dermaseptins are a large family of linear polycationic peptides from frog skin (10, 51, 53, 54) whose cytolytic activity is believed to result from interaction of their N-terminal domain with the plasma membranes of target cells (52, 55). Recent investigations with respect to the relations between physical properties (structure and organization in solution) of dermaseptin S4 and its interaction with target membranes (19, 37) led to the design of short derivatives that maintain the amphipathic alpha-helical structure of the parent peptide, bind avidly to model membranes with association affinity constants (KA) in the range of 105 to 107 M−1 and exert rapid cytolytic activity against a variety of pathogens. Among these, the 13-mer derivative K4-S4(1-13)a is of particular interest as it is the smallest derivative that combines low toxicity and efficient large-spectrum antimicrobial activity both in culture and in animal models of infection (20, 56).
In this paper, we tested the hypothesis that a hybrid construct composed of RIP and dermaseptin would benefit from the properties of both components. We show that RIP and the 13-mer dermaseptin derivative DD13 individually inhibit biofilm formation on a Dacron graft and that their chimeric version eradicates drug-resistant staphylococcal infections in a fashion that is greater than the additive effect of the individual peptides comprising the chimera.
MATERIALS AND METHODS
Peptides
Peptides were synthesized by the solid-phase method as described previously (37), applying the Fmoc active ester chemistry on a fully automated, programmable model 433A peptide synthesizer (Applied Biosystems). 4-Methylbenzhydrylamine-resin (Novabiochem) was used to obtain amidated peptides. The crude peptides were extracted from the resin with 30% acetonitrile in water and purified to chromatographic homogeneity in the range of 98 to >99% by reverse-phase high-pressure liquid chromatography (HPLC) (Alliance-Waters). HPLC runs were performed on a semipreparative C4 column (Vydac) with a linear gradient of acetonitrile in water (1%/min), both solvents containing 0.1% trifluoroacetic acid. The purified peptides were subjected to amino acid analysis and electrospray mass spectrometry in order to confirm their composition. Peptides were stored as a lyophilized powder at −20°C.
CD spectra.
Circular dichroism (CD) spectra were measured in 20% (vol/vol) trifluoroethanol in water with an AVIV model 62A DS spectrometer (Aviv Assoc., Lakewood, N.J.) with a 0.020-cm rectangular QS Hellma cuvette at 25°C. CD data represent average values from three separate recordings.
Bacteria.
Methicillin-resistant S. aureus (MRSA) strain ATCC 43300 was commercially purchased from Oxoid S.p.A., Milan, Italy. Methicillin-resistant S. epidermidis (MRSE) was a clinical strain from the Institute of Infectious Diseases and Public Health, University of Ancona, Ancona, Italy.
Animals.
Adult male Wistar rats, 250 to 300 g (Istituto Nazionale Riposo e Cura Anziani, Istituto di Ricovero e Cura a Carattere Scientifico animal facility, Ancona, Italy) were used, with 15 animals per experimental group.
Susceptibility testing.
The antimicrobial susceptibilities of the strains were determined by the broth microdilution method, according to the procedures outlined by the National Committee for Clinical Laboratory Standards. The MIC was taken as the lowest antibiotic concentration at which observable growth was inhibited. Experiments were performed in triplicate.
Graft infection rat model.
Rats were anesthetized, their back hair was shaved, and the skin was cleansed with 10% povidone-iodine solution. One subcutaneous pocket was made on each side of the median line by a 1.5-cm incision. Aseptically, 1-cm2 sterile albumin-sealed Dacron grafts (Albograft; Sorin Biomedica Cardio, S.p.A., Saluggia VC, Italy) were implanted into the pockets. Before implantation, the grafts were soaked for 20 min in sterile solutions of the various agents. The pockets were closed by means of skin clips, and saline solution (1 ml) containing MRSA or MRSE at a concentration of 2 × 107 CFU/ml was inoculated onto the graft surface with a tuberculin syringe to create a subcutaneous fluid-filled pocket. The animals were returned to individual cages and thoroughly examined daily. All grafts were explanted at 7 days following implantation.
Assessment of the infection.
The explanted grafts were placed in sterile tubes, washed in sterile saline solution, placed in tubes containing 10 ml of phosphate-buffered saline solution, and sonicated for 5 min to remove the adherent bacteria from the grafts (6). After sonication, grafts were microscopically checked to verify that all bacteria were removed. Quantitation of viable bacteria was performed by culturing serial dilutions (0.1 ml) of the bacterial suspension on blood agar plates. All plates were incubated at 37°C for 48 h and evaluated for the presence of the staphylococcal strains. The organisms were quantitated by counting the number of CFU per plate. No significant differences in cell viability were present upon testing the effect of sonication for up to 10 min on both antibiotic-sensitive and -resistant bacteria. The limit of detection for this method was approximately 10 CFU/ml (14).
Peptide binding to Dacron.
To determine how much peptide impregnated the graft, 1-cm2 slices of collagen-sealed Dacron were soaked at room temperature in peptide solutions in quadruplicate for 0.5 and 5 h (1 ml of saline per graft, containing 50 μg/ml of each peptide). Following the incubation period, the Dacron was removed, and residual peptide (unbound fraction) was subjected to reverse-phase HPLC analysis as described above. Peptide identification was based on retention time and spectral analysis. The amount of unbound peptide was calculated by area integration of the UV-absorbing peak (220 nm) and comparison with standard curves of known concentrations for each peptide (19).
RNA III synthesis and bacterial growth.
Early-exponential-phase S. aureus cells containing the rnaiii::blaZ fusion construct (29) were grown in 30 μl (2 × 107 cells) for 2.5 h with 5 μl of peptides (0 to 5 μg) in TSB growth medium at 37°C with shaking. Of the total reaction volume (35 μl), a 5-μl sample was removed, diluted in saline, and streaked on Luria agar plates to determine CFU. The rest of the reaction mixture was used to determine RNA III synthesis (beta-lactamase activity). This was done by adding a substrate of beta-lactamase (nitrocefin), and the optical density (OD) was determined at 490 and 650 nm (29).
Statistical analysis.
MICs are presented as the geometric mean of three separate experiments. Quantitative culture results from all groups are presented as mean ± standard deviation, and the statistical comparisons between groups were made with analysis of variance on the log-transformed data. Significance was accepted when the P value was ≤0.05.
RESULTS
Prevention of graft-associated infections.
Since DD13 is a bactericidal peptide while RIP prevents staphylococcal biofilm formation, a hybrid version, DD13-RIP, was constructed (Table 1) and tested for efficacy in preventing staphylococcal graft-associated infections. The RIP and DD13 concentrations range to be tested was based on their active concentrations from previous studies (2-6, 19, 20, 22, 37, 56). Immediately prior to being implanted in rats, the collagen-coated Dacron grafts were coated with 10, 20, or 50 mg of either RIP or DD13 per liter and compared to DD13-RIP at the same concentrations.
TABLE 1.
Primary structures and designations of the peptides investigated
| Peptide | Amino acid sequence | Designation |
|---|---|---|
| RIP | YSPWTNFCONH2 | RIP |
| K4-S4(1-13)a | ALWKTLLKKVLKACONH2 | DD13 |
| Hybrid | ALWKTLLKKVLKAYSPWTNFCONH2 | DD13-RIP |
Bacteria (MRSA or MRSE) were injected into the implants, the implants were removed after a week, and the bacterial load was determined. The study included a negative control group (untreated graft with no bacterial challenge) and a positive control group (untreated graft with bacterial challenge). The results are summarized in Table 2. None of the animals included in the negative control group had anatomic or microbiological evidence of graft infection (no graft contamination). All rats included in the positive control groups that were implanted with untreated grafts and challenged with MRSA or MRSE (total of 30 rats) demonstrated evidence of bacterial colonization, with quantitative culture results showing 4.4 × 106 ± 1.2 × 106 and 6.9 × 106 ± 1.8 × 106 CFU/ml, respectively.
TABLE 2.
Effect of peptides on graft-associated infections in rats
| Treatmenta | Soaking solution (mg/liter) | MRSA (mean CFU/ml ± SD) | MRSE (mean CFU/ml ± SD) |
|---|---|---|---|
| None (control) | 0 | 4.4 × 106 ± 1.2 × 106 | 6.9 × 106 ± 1.8 × 106 |
| RIP* | 10 | 4.1 × 104 ± 7.1 × 103 | 6.9 × 103 ± 1.9 × 103 |
| 20 | 5.9 × 103 ± 1.7 × 103 | 8.5 × 102 ± 2.0 × 102 | |
| 50 | 84 ± 36 | 39 ± 16 | |
| DD13* | 10 | 5.8 × 102 ± 2.1 × 102 | 5.2 × 102 ± 1.6 × 102 |
| 20 | 40 ± 17 | 44 ± 13 | |
| 50 | <10 | <10 | |
| DD13-RIP* | 10 | 19 ± 3 | 30 ± 7 |
| 20 | <10 | <10 | |
| 50 | <10 | <10 |
Dacron grafts were presoaked in saline (control), RIP, DD13, or DD13-RIP at the designated concentrations and implanted in rats (15 rats per group). Grafts were then inoculated with MRSA or a clinical isolate of MRSE. Grafts were removed after a week, and CFU were determined. *, P < 0.05 compared to control untreated group. Detection limit, 10 CFU/ml.
Animals implanted with grafts presoaked in 10, 20, or 50 mg of RIP solutions per liter and challenged with MRSA demonstrated reduced evidence of bacterial colonization, with 4.1 × 104 ± 7.1 × 103, 5.9 × 103 ± 1.7 × 103, and 8.4 × 101 ± 3.6 × 101 CFU/ml, respectively, and those challenged with MRSE demonstrated reduced evidence of infection with 6.9 × 103 ± 1.9 × 103, 8.5 × 102 ± 2.0 × 102, and 3.9 × 101 ± 1.6 × 101 CFU/ml, respectively.
Animals implanted with grafts presoaked with DD13 and challenged with MRSA demonstrated reduced or no evidence of bacterial colonization, with 5.8 × 102 ± 1.6 × 102 and 4.0 × 101 ± 1.7 × 101 CFU/ml, respectively, and negative quantitative cultures for the same doses as RIP. Similarly, animals challenged with MRSE demonstrated reduced or no evidence of infection,. with 5.2 × 102 ± 1.6 × 102 and 4.4 × 101 ± 1.3 × 101 CFU/ml, respectively, and negative quantitative cultures.
Animals implanted with in DD13-RIP solutions and challenged with MRSA demonstrated reduced evidence of bacterial colonization, with 1.9 × 101 ± 0.3 × 101 CFU/ml at 10 mg per lite,r and when the grafts were soaked in 20 or 50 mg per liter, animals showed no evidence of staphylococcal infection, with negative quantitative cultures. Similarly, animals challenged with MRSE displayed 3.0 × 101 ± 0.7 × 101 CFU/ml and negative quantitative culturesfor 10 and 20 to 50 mg per liter.
In addition, treatment with these peptides was compared with the conventionally used antibiotic rifampin (63). These experimental groups received grafts presoaked with 5 mg of rifampin per liter alone or rifampin and RIP, DD13, or DD13-RIP at 10 mg per liter. The results are summarized in Table 3. The group of rifampin-treated rats showed quantitative culture results of 6.7 × 103 ± 9.1 × 102 CFU/ml for MRSA and 8.8 × 102 ± 3.0 × 102 CFU/ml for MRSE, but when grafts were soaked in combination with RIP, DD13, or DD13-RIP, the animals showed negative quantitative cultures and no evidence of bacterial colonization. Note that rifampin-resistant bacteria that were exposed to DD13 were previously shown to exhibit a MIC identical to that obtained with rifampin-sensitive bacteria (56).
TABLE 3.
Prevention of graft-associated infections in rats based on combined therapya
| Treatment | MRSA (mean CFU/ml ± SD) | MRSE (mean CFU/ml ± SD) |
|---|---|---|
| None (control) | 4.4 × 106 ± 1.2 × 106 | 6.9 × 106 ± 1.8 × 106 |
| Rifampin* | 6.7 × 103 ± 0.9 × 103 | 0.88 × 103 ± 0.3 × 103 |
| Rifampin + RIP* | <10 | <10 |
| Rifampin + DD13* | <10 | <10 |
| Rifampin + DD13-RIP* | <10 | <10 |
Grafts presoaked in saline (control), rifampin alone (5 mg/liter), or rifampin combined with either RIP, DD13, or DD13-RIP (10 mg/liter) were implanted in rats and inoculated with MRSA or MRSE. Grafts were removed after a week and assessed for viable CFU. Detection limit, 10 CFU/ml. *, P < 0.05 compared to control untreated group.
Of note is that all agents used did not show any signs of toxicity and none of the animals included in any group died or had clinical evidence of drug-related adverse effects, such as local signs of perigraft inflammation, anorexia, vomiting, diarrhea, or behavioral alterations. Reduction in bacterial load was significant (P < 0.05) in all experimental groups compared to positive control groups.
In vitro studies. (i) Peptide binding to Dacron cuffs.
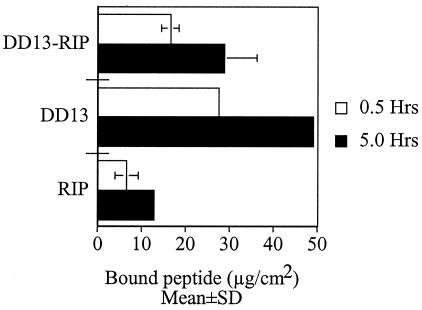
To verify the relationships between the observed in vivo activity and the amount of peptide present on the Dacron grafts, the grafts were soaked in peptide solutions as described above prior to being grafted in rats. After removal of the grafts, the solutions were subjected to HPLC analysis, from which the amount of Dacron-bound peptide was deduced from the calculated amount of residual unbound fraction. The results are summarized in Fig. 1 for the peptide solution of 50 mg per liter. Among the three peptides, RIP bound the least, with a mean bound amount of 6.5 ± 2.5 μg/cm2. DD13 was found to bind the most and significantly more than RIP, with a mean bound amount of 27 ± 0.5 μg/cm2, whereas DD13-RIP bound more than RIP but less than DD13 with a mean bound amount of 16 ± 2 μg/cm2. The data indicate that longer soaking periods enabled uptake of larger amounts of each peptide. For instance, nearly 100% (i.e., 50 μg) of DD13 was found to bind after 5 h of soaking (Fig. 1). While these experiments indicate that longer soaking times result in higher peptide concentrations on the grafts, they also demonstrate that the high protective efficacy of the chimeric peptide compared to the peptides alone is not due to its relative concentration but rather to its specific activity (see below).
FIG. 1.
Peptides binding to Dacron. Collagen-sealed Dacron grafts were soaked in peptide solutions (50 mg per liter) for either 0.5 or 5 h. Bound peptide was estimated from the unbound fraction, which was subjected to analytical reverse-phase HPLC as described previously (19). Peptide identification was based on retention time and spectral analysis. Unbound peptide was determined after area integration of the UV-absorbing peak (220 nm) and comparison with standard curves of known concentrations. Error bars indicate standard deviations of the mean determined from four independent experiments.
(ii) Bacterial growth and RNA III synthesis.
To better understand the molecular mechanisms involved in the synergistic activity, the three peptides were investigated for their effect(s) in vitro on RNA III synthesis (known to be inhibited by RIP) and for bacterial proliferation (known to be inhibited by DD13). These experiments were performed by growing S. aureus cells containing an rnaiii::blaZ fusion construct in the presence of either one of the peptides. Cultures were monitored by determination of RNA III synthesis by a colorimetric method with nitrocefin as a substrate, while the peptides' effect on bacterial viability was assessed by performing CFU counts with conventional microbiological methods. The results are summarized in Fig. 2.
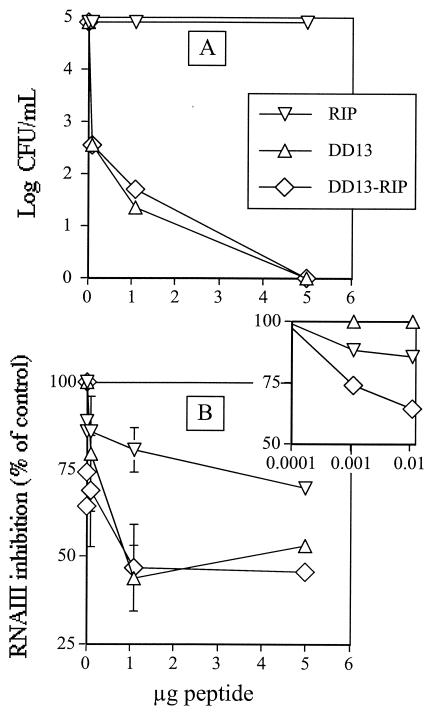
FIG. 2.
In vitro effects of peptides on bacterial growth and RNA III synthesis. Early-exponential-phase S. aureus cells containing the rnaiii::blaZ fusion construct (≈2 × 107 cells) were grown with peptides (0 to 5 μg) in a total volume of 35 μl in TSB and then divided into two portions. One portion (5 μl) was diluted in saline and streaked on LB agar plates to determine CFU (A). The other portion (5 μl) was used to determine RNA III synthesis (B) by adding the beta-lactamase substrate nitrocefin and measuring the optical density (OD) at 490 and 650 nm (29). Error bars indicate standard deviations from the mean as determined from two independent experiments performed in triplicate. If no error bar is shown, the standard deviation was smaller than the diameter of the symbol. The inset is the same as panel B but focuses on low peptide concentrations. Symbols in panel B are the same as in panel A.
DD13 and DD13-RIP displayed quasipotent bactericidal activity, whereas RIP was virtually inactive (Fig. 2A). According to the broth microdilution method, DD13 and DD13-RIP exhibited MICs of 2 mg per liter for both staphylococcal strains (compared to susceptibility to rifampin, with MICs of 0.5 mg per liter for both of the organisms). RIP did not demonstrate any in vitro bactericidal activity against either of the two strains when tested at up to 128 mg per liter (data not shown).
As also shown in Fig. 2, RIP efficiently inhibited RNA III synthesis, while both DD13 and DD13-RIP appeared to be more efficient than RIP alone. However, closer inspection of the data revealed that at high peptide concentrations, most of the inhibitory activity observed was attributable to cell death. Moreover, at low peptide concentrations, where no cell death occurred, DD13 was unable to affect RNA III synthesis, unlike DD13-RIP, which displayed the highest efficacy (Fig. 2B, inset).
CD measurements.
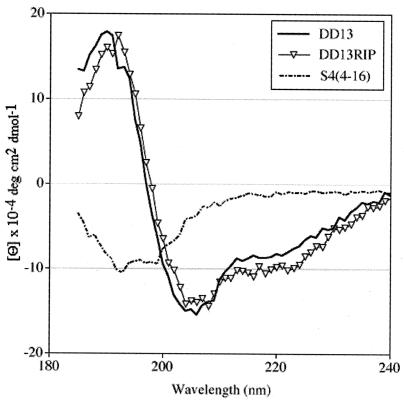
Indication of the molecular organization of the peptides in solution was obtained from CD measurements in trifluoroethanol (Fig. 3). Since the helical organization of the 13-mer dermaseptin derivative was established by nuclear magnetic resonance (37), the present study aimed at comparing detectable differences in the CD spectrum of its elongated analog (DD13-RIP) which would point to alteration(s) of the structure. For a control peptide, another 13-mer dermaseptin S4 derivative of was used, S4 (4-16), KTLLKKVLKAAAK, corresponding to three residues truncated from the DD13 N terminus and elongation by three residues at the C terminus. As shown in Fig. 3, whereas the control peptide displayed a typical spectrum for a random structure, both DD13 and DD13-RIP displayed typical ellipticity spectra of an α-helix as characterized by double minima at 208 and 222 nm. DD13-RIP displayed a slight increase in ellipticity signals around 222 nm, suggesting an eventual slight stabilization of the α-helix but essentially a molecular organization similar to that of DD13.
FIG. 3.
Far-UV CD spectra of DD13 and DD13-RIP and a control peptide. CD spectra were taken for peptide samples of 300 μM (determined by UV by using standard curves of known concentrations for each peptide) in 20% trifluoroethanol-water. CD data represent average values from three separate recordings.
DISCUSSION
In order to prove efficacy in preventing bacterial adhesion and biofilm formation in vivo, a well-characterized experimental Dacron graft rat model was used. For comparison purposes, the antibiotic rifampin was chosen for its current utilization in clinical practice against staphylococci (63). Rifampin was very effective in our experimental model, as expected from previous literature (3, 26). However, as with RIP alone, rifampin did not eradicate bacterial infection; 100% inhibition was reached only when rifampin was combined with a peptide. The reason RIP did not cause total inhibition when used alone could be due to the actual amount of RIP bound to the graft. It is estimated that soaking the graft in 50 mg of RIP per liter resulted in 6.5 ± 2.5 μg of RIP bound to the graft before implantation, which may not be enough to prevent adhesion of the 10 million cells that were injected. Perhaps if the grafts were coated with a higher concentration of RIP, such as after a longer soaking time, better prevention could have been reached with RIP and the other peptides alone. Note, however, that such a high level of a single dose of bacterial contamination is unlikely to occur in clinical settings, suggesting that the amounts of bound peptides utilized in the present in vivo experiments could be sufficient to coat grafts in a clinical practice.
Chimeric peptides with enhanced activity compared to the individual peptides were used in previous studies (13, 18, 47, 60, 61, 67). Here, we constructed a chimeric molecule composed of two small peptides that individually produce their antibacterial effects by distinct mechanisms of action. To choose the chimeric sequence, we hypothesized that the N and C termini of the chimera must be occupied by DD13 and RIP, respectively, to preserve their individual properties. Indeed, previous studies suggested that the antimicrobial activity of the dermaseptin derivative (16, 19, 26, 52, 54) and its alpha-helical structure (37) would best be maintained at the N terminus of the chimeric molecule. On the other hand, studies with RIP evoked the importance of the C terminus of RIP for RNA III-inhibiting activity (29). The sequence DD13-RIP was therefore selected in this study, although chimeric peptides with other arrangements will be investigated in the future.
The present study also demonstrates that, like RIP, DD13 is highly efficient in inhibiting staphylococcal colonization whether used alone or in combination with the conventional antibiotic. In fact, DD13 was significantly more efficient than RIP. The chimeric construct, however, displayed further efficiency, endowing the Dacron graft with near sterility at the lowest doses. At present, the molecular basis for this synergistic activity is not well understood, as it could result from a number of reasons. The simplest reason that comes to mind is that DD13-RIP is able to express the typical properties of both building units, i.e., TRAP antagonist activity of RIP and cytolytic activity based on membrane disruption by DD13. This is hypothesized because the viable CFU assays showed that RIP had no cytolytic activity and did not enhance or reduce the bactericidal activity of DD13 since DD13-RIP and DD13 displayed equal activity.
The CD data suggested that DD13 and DD13-RIP have rather similar molecular organizations. On the other hand, at nontoxic concentrations, DD13 was unable to affect RNA III synthesis, unlike DD13-RIP, which displayed the highest inhibitory efficacy. These results suggest that the profound in vivo inhibitory activity of DD13-RIP is a result of synergistic activities based on the ability of DD13 to enhance RIP's RNA III-inhibiting activity. According to our estimate, the amount of peptide present on the graft excludes the possibility that efficacy was proportional to a much higher quantity of bound peptide. For instance, while RIP and DD13-RIP bound to the graft to practically the same extents (about a twofold difference), the chimeric peptide had a much higher efficacy in vivo (more than an order of magnitude). Similarly, DD13 displayed the highest binding but not the highest efficacy in vivo. This indicates that the high efficacy of the chimeric peptide in vivo is not due to its relative concentration on the graft but rather to its specific activity. Taken together, these data indicate that the extreme efficacy of DD13-RIP in vivo could result from its ability to launch an overwhelming attack on the target cells, with either one or both of two different mechanisms of action simultaneously.
In conclusion, we show in this paper an alternative approach to preventing device-associated infection by S. aureus and S. epidermidis, including those caused by antibiotic-resistant strains. It is probably through modulating the bacterial mechanism of communication by RIP combined with the cytotoxic effect of DD13 that their pathogenesis is inhibited. We therefore postulate that RIP and DD13 act in synergy by attacking bacteria simultaneously by different mechanisms. Since DD13-RIP is extremely effective in preventing staphylococcal infections, our data suggest that DD13-RIP could be used to coat medical devices known to be associated with staphylococcal infections.
Acknowledgments
This research was supported in part by the Israel Science Foundation (grant number 387/03) and in part by Technion's President Research Fund.
REFERENCES
- 1.Andreu, A., and L. Rivas. 1998. Animal antimicrobial peptides: an overview. Biopolymers 47:415-433. [DOI] [PubMed] [Google Scholar]
- 2.Balaban, N., L. V. Collins, J. S. Cullor, E. B. Hume, E. Medina-Acosta, O. Vieira da Motta, R. O'Callaghan, P. V. Rossitto, M. E. Shirtliff, L. Silveira, A. Tarkowski, and J. V Torres. 2000. Prevention of diseases caused by Staphylococcus aureus using the peptide RIP. Peptides 21:1301-1311. [DOI] [PubMed] [Google Scholar]
- 3.Balaban, N., A. Giacometti, O. Cirioni, Y. Gov, et al.. 2003. Use of the quorum-sensing inhibitor RNA III-inhibiting peptide to prevent biofilm formation in vivo by drug-resistant Staphylococcus epidermidis. J. Infect. Dis. 187:625-630. [DOI] [PubMed] [Google Scholar]
- 4.Balaban, N., T. Goldkorn, Y. Gov, M. Hirshberg, N. Koyfman, H. R. Matthews, R. T. Nhan, B. Singh, and O. Uziel. 2001. Regulation of Staphylococcus aureus pathogenesis via target of RNA III-activating protein (TRAP). J. Biol. Chem. 276:2658-2667. [DOI] [PubMed] [Google Scholar]
- 5.Balaban, N., T. Goldkorn, R. T. Nhan, et al. 1998. Autoinducer of virulence as a target for vaccine and therapy against Staphylococcus aureus. Science 280:438-440. [DOI] [PubMed] [Google Scholar]
- 6.Balaban, N., Y. Gov, A. Bitler, and J. R. Boelaert. 2003. Prevention of Staphylococcus aureus biofilm on dialysis catheters and adherence to human cells. Kidney Int. 63:340-345. [DOI] [PubMed] [Google Scholar]
- 7.Barie, P. S. 1998. Antibiotic-resistant gram-positive cocci: implications for surgical practice. World J. Surg. 22:118-126. [DOI] [PubMed] [Google Scholar]
- 8.Blondelle, E. S., and K. Lohner. 2000. Combinatorial libraries: a tool to design antimicrobial and antifungal peptide analogues having lytic specificities for structure-activity relationship studies. Biopolymers 55:74-87. [DOI] [PubMed] [Google Scholar]
- 9.Boman, H. G. 1995. Peptide antibiotics and their role in innate immunity. Annu. Rev. Immunol. 13:61-92. [DOI] [PubMed] [Google Scholar]
- 10.Brand, G. D., J. R. Leite, L. P. Silva, et al. 2002. Dermaseptins from Phyllomedusa oreades and Phyllomedusa distincta. Anti-Trypanosoma cruzi activity without cytotoxicity to mammalian cells. J. Biol. Chem. 277:49332-49340. [DOI] [PubMed] [Google Scholar]
- 11.Carratala, J. 2002. The antibiotic-lock technique for therapy of ‘highly needed’ infected catheters. Clin. Microbiol. Infect. 8:282-289. [DOI] [PubMed] [Google Scholar]
- 12.Chen, J., T. J. Falla., H. Liu., M. A. Hurst., et al. 2000. Development of protegrins for the treatment and prevention of oral mucositis: structure-activity relationships of synthetic protegrin analogues. Biopolymers 55:88-98. [DOI] [PubMed] [Google Scholar]
- 13.Chicharro, C., C. Granata, R. Lozano, D. Andreu, and L. Rivas. 2001. N-terminal fatty acid substitution increases the leishmanicidal activity of CA(1-7)M(2-9), a cecropin-melittin hybrid peptide. Antimicrob. Agents Chemother. 45:2441-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cirioni, O., A. Giacometti, R. Ghiselli, et al. 2003. Prophylactic efficacy of topical temporin A and RNA III-inhibiting peptide in a subcutaneous rat pouch model of graft infection attributable to staphylococci with intermediate resistance to glycopeptides. Circulation 108:767-771. [DOI] [PubMed] [Google Scholar]
- 15.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 16.Dagan, A., L. Efron, L. Gaidukov, A. Mor, and H. Ginsburg. 2002. In vitro antiplasmodium effects of dermaseptin S4 derivatives. Antimicrob. Agents Chemother. 46:1059-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duclohier, H., and H. Wroblewski. 2001. Voltage-dependent pore formation and antimicrobial activity by alamethicin and analogues. J. Membr. Biol. 184:1-12. [DOI] [PubMed] [Google Scholar]
- 18.Edlund, C., M. Hedberg, A. Engstrom, J. I. Flock, and D. Wade. 1998. Antianaerobic activity of a cecropin-melittin peptide. Clin. Microbiol. Infect. 4:181-185. [DOI] [PubMed] [Google Scholar]
- 19.Feder, R., A. Dagan, and A. Mor. 2000. Structure-activity relationship study of antimicrobial dermaseptin S4 showing the consequences of peptide oligomerization on selective cytotoxicity. J. Biol. Chem. 275:4230-4238. [DOI] [PubMed] [Google Scholar]
- 20.Feder, R., R. Nehushtai, and A. Mor. 2001. Affinity driven molecular transfer from erythrocyte membrane to target cells. Peptides 22:1683-1690. [DOI] [PubMed] [Google Scholar]
- 21.Friedrich, C. L., D. Moyles, T. J. Beveridge, and R. E. Hancock. 2000. Antibacterial action of structurally diverse cationic peptides on gram-positive bacteria. Antimicrob. Agents Chemother. 44:2086-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaidukov, L., A. Fish, and A. Mor. 2003. Analysis of membrane-binding properties of dermaseptin analogues: relationships between binding and cytotoxicity. Biochemistry 42:12866-12874. [DOI] [PubMed] [Google Scholar]
- 23.Ganz, T., and R. Lehrer. 1998. Antimicrobial peptides of vertebrates. Curr. Opin. Immunol. 10:41-44. [DOI] [PubMed] [Google Scholar]
- 24.Gazit, E., A. Boman, H. G. Boman, and Y. Shai. 1995. Interaction of the mammalian antibacterial peptide cecropin P1 with phospholipid vesicles. Biochemistry 34:11479-11488. [DOI] [PubMed] [Google Scholar]
- 25.Ge, Y., D. L. MacDonald, K. J. Holroyd, C. Thornsberry, H. Wexler, and M. Zasloff. 1999. In vitro antibacterial properties of pexiganan, an analog of magainin. Antimicrob. Agents Chemother. 43:782-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh, J. K., D. Shaool, P. Guillaud, L. Ciceron, D. Mazier, I. Kustanovich, Y. Shai, and A. Mor. 1997. Selective cytotoxicity of dermaseptin S3 toward intraerythrocytic Plasmodium falciparum and the underlying molecular basis. J. Biol. Chem. 272:31609-31616. [DOI] [PubMed] [Google Scholar]
- 27.Giacometti, A., O. Cirioni, Y. Gov, R. Ghiselli, D. Del Prete, F. Mocchegiani, V. Saba, F. Orlando, G. Scalise, N. Balaban, and G. Dell'Acqua. 2003. RNA III-inhibiting peptide inhibits in vivo biofilm formation by drug-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:1979-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gottenbos, B., D. W. Grijpma, H. van der Mei, J. Feijen, and H. J. Busscher. 2001. Antimicrobial effects of positively charged surfaces on adhering Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. 48:7-13. [DOI] [PubMed] [Google Scholar]
- 29.Gov, Y., A. Bitler, G. Dell'Acqua, J. V. Torres, and N. Balab. 2001. RNA III-inhibiting peptide (RIP), a global inhibitor of Staphylococcus aureus pathogenesis: structure and function analysis. Peptides 22:1609-1620. [DOI] [PubMed] [Google Scholar]
- 30.Hancock, R. E. 1997. Peptide antibiotics. Lancet 349:418-422. [DOI] [PubMed] [Google Scholar]
- 31.Hancock, R. E., and R. Lehrer. 1998. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 16:82-90. [DOI] [PubMed] [Google Scholar]
- 32.Hariton-Gazal, E., R. Feder, A. Mor, A. Graessmann, A. Brack-Werner, D. Jans, C. Gilon, and A. Loyter. 2001. Targeting of nonkaryophilic cell-permeable peptides into the nuclei of intact cells by covalently attached nuclear localization signals. Biochemistry 41:9208-9214. [DOI] [PubMed] [Google Scholar]
- 33.Heller, W. T., A. J. Waring, R. I. Lehrer, and H. W. Huang. 1998. Multiple states of beta-sheet peptide protegrin in lipid bilayers. Biochemistry 37:17331-17338. [DOI] [PubMed] [Google Scholar]
- 34.Henke, P. K., T. M. Bergamini, S. M. Rose, and J. D. Richardson. 1998. Current options in prosthetic vascular graft infection. Am. Surg. 64:39-45. [PubMed] [Google Scholar]
- 35.Huang, H. W. 2000. Action of antimicrobial peptides: two-state model. Biochemistry 39:8347-8352. [DOI] [PubMed] [Google Scholar]
- 36.Huebner, J., and D. A. Goldman. 1999. Coagulase-negative staphylococci: role as pathogens. Annu. Rev. Med. 50:233-236. [DOI] [PubMed] [Google Scholar]
- 37.Kustanovich, I., D. E. Shalev, M. Mikhlin, L. Gaidukov, A. Mor. 2002. Structural requirements for potent versus selective cytotoxicity for antimicrobial dermaseptin S4 derivatives. J. Biol. Chem. 277:16941-16951. [DOI] [PubMed] [Google Scholar]
- 38.Ladokhin, A. S., M. E. Selsted, and S. H. White. 1997. Sizing membrane pores in lipid vesicles by leakage of co-encapsulated markers: pore formation by melittin. Biophys. J. 72:1762-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy, O. 2000. Antimicrobial proteins and peptides of blood: templates for novel antimicrobial agents. Blood 96:2664-2672. [PubMed] [Google Scholar]
- 40.Linnola, R. 2001. Staphylococcus epidermidis and intraocular lenses. Ophthalmology 108:1518-1519. [DOI] [PubMed] [Google Scholar]
- 41.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 42.Lowy, F. D. 2003. Antimicrobial resistance: the example of Staphylococcus aureus. J. Clin. Investig. 111:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ludtke, S. J., K. He, Y. Wu, and H. W. Huang. 1994. Cooperative membrane insertion of magainin correlated with its cytolytic activity. Biochim. Biophys. Acta 1190:181-184. [DOI] [PubMed] [Google Scholar]
- 44.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 45.Maloy, L. W., and U. P. Kari. 1995. Structure-activity studies on magainins and other host defense peptides. Biopolymers 37:105-122. [DOI] [PubMed] [Google Scholar]
- 46.Marr, K. A. 2000. Staphylococcus aureus bacteremia in patients undergoing hemodialysis. Semin. Dialysis 13:23-29. [DOI] [PubMed] [Google Scholar]
- 47.Merrifield, R. B., E. L. Merrifield, P. Juvvadi, D. Andreu, and H. G. Boman. 1994. Design and synthesis of antimicrobial peptides. Ciba Found Symp. 186:5-20. [PubMed] [Google Scholar]
- 48.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 49.Moll, G. N., S. Brul, W. N. Konings, and A. J. Driessen. 2000. Comparison of the membrane interaction and permeabilization by the designed peptide Ac-MB21-NH2 and truncated dermaseptin S3. Biochemistry 39:11907-11912. [DOI] [PubMed] [Google Scholar]
- 50.Mor, A. 2001. Antimicrobial peptides. In A. Seidel (ed.), The Kirk-Othmer encyclopedia of chemical technology. John Wiley & Sons, New York, N.Y.
- 51.Mor, A., M. Amiche, and P. Nicolas. 1994. Structure, synthesis, and activity of dermaseptin b, a novel vertebrate defensive peptide from frog skin: relationship with adenoregulin. Biochemistry 33:6642-6650. [DOI] [PubMed] [Google Scholar]
- 52.Mor, A., K. Hani, and P. Nicolas. 1994. The vertebrate peptide antibiotics dermaseptins have overlapping structural features but target specific microorganisms. J. Biol. Chem. 269:31635-31640. [PubMed] [Google Scholar]
- 53.Mor, A., V. H. Nguyen, A. Delfour, S. D. Migliore, and P. Nicolas. 1991. Isolation, amino acid sequence, and synthesis of dermaseptin, a novel antimicrobial peptide of amphibian skin. Biochemistry 30:8824-8830. [DOI] [PubMed] [Google Scholar]
- 54.Mor, A., and P. Nicolas. 1994. Isolation and structure of novel defensive peptides from frog skin. Eur. J. Biochem. 219:145-154. [DOI] [PubMed] [Google Scholar]
- 55.Mor, A., and P. Nicolas. 1994. The NH2-terminal alpha-helical domain 1-18 of dermaseptin is responsible for antimicrobial activity. J. Biol. Chem. 269:1934-1939. [PubMed] [Google Scholar]
- 56.Navon-Venezia, S., R. Feder, L. Gaidukov, Y. Carmeli, and A. Mor. 2002. Antibacterial properties of dermaseptin S4 derivatives with in vivo activity. Antimicrob. Agents Chemother. 46:689-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nicolas, P., and A. Mor. 1995. Peptides as weapons against microorganisms in the chemical defense system of vertebrates. Annu. Rev. Immunol. 49:277-304. [DOI] [PubMed] [Google Scholar]
- 58.Oren, Z., and Y. Shai. 2000. Cyclization of a cytolytic amphipathic alpha-helical peptide and its diastereomer: effect on structure, interaction with model membranes, and biological function. Biochemistry 39:6103-6114. [DOI] [PubMed] [Google Scholar]
- 59.Oren, Z., and Y. Shai. 1998. Mode of action of linear amphipathic alpha-helical antimicrobial peptides. Biopolymers 47:451-463. [DOI] [PubMed] [Google Scholar]
- 60.Patrzykat, A., C. L. Friedrich, L. Zhang, V. Mendoza, and R. E. Hancock. 2002. Sublethal concentrations of pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob. Agents Chemother. 46:605-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Piers, K. L., M. H. Brown, and R. E. Hancock. 1994. Improvement of outer membrane-permeabilizing and lipopolysaccharide-binding activities of an antimicrobial cationic peptide by C-terminal modification. Antimicrob. Agents Chemother. 38:2311-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pouny, Y., D. Rapaport, A. Mor, P. Nicolas, and Y. Shai. 1992. Interaction of antimicrobial dermaseptin and its fluorescently labeled analogues with phospholipid membranes. J. Biol. Chem. 31:12416-12423. [DOI] [PubMed] [Google Scholar]
- 63.Sardelic, F., P. Y. Ao, D. A. Taylor, and J. P. Fletcher. 1996. Prophylaxis against Staphylococcus epidermidis vascular graft infection with rifampicin-soaked, gelatin-sealed Dacron. Cardiovasc. Surg. 4:389-392. [DOI] [PubMed] [Google Scholar]
- 64.Schierholz, J. M., and J. Beuth. 2001. Implant infections: a haven for opportunistic bacteria. J. Hosp. Infect. 49:87-93. [DOI] [PubMed] [Google Scholar]
- 65.Shai, Y. 2002. Mode of action of membrane active antimicrobial peptides. Biopolymers 66:236-248. [DOI] [PubMed] [Google Scholar]
- 66.Shai, Y. 1999. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1462:55-70. [DOI] [PubMed] [Google Scholar]
- 67.Shin, S. Y., J. H. Kang, M. K. Lee, S. Y. Kim, Y. Kim, and K. S. Hahm. 1998. Cecropin A-magainin 2 hybrid peptides having potent antimicrobial activity with low hemolytic effect. Biochem. Mol. Biol. Int. 44:1119-1126. [DOI] [PubMed] [Google Scholar]
- 68.Sokolov, Y., T. Mirzabekov, D. W. Martin, R. I. Lehrer, and B. L. Kagan. 1999. Membrane channel formation by antimicrobial protegrins. Biochim. Biophys. Acta 1420:23-29. [DOI] [PubMed] [Google Scholar]
- 69.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 70.Strahilevitz, J., A. Mor, P. Nicolas, and Y. Shai. 1994. Spectrum of antimicrobial activity and assembly of dermaseptin-b and its precursor form in phospholipid membranes. J. Biol. Chem. 33:10951-10960. [DOI] [PubMed] [Google Scholar]
- 71.Tiller, J. C., C. J. Liao, K. Lewis, and A. M. Klibanov. 2001. Designing surfaces that kill bacteria on contact. Proc. Natl. Acad. Sci. USA 98:5981-5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tossi, A., L. Sandri, and A. Giangaspero. 2000. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 55:4-30. [DOI] [PubMed] [Google Scholar]
- 73.Vieira-da-Motta, O., X. Damasceno, P. Ribeiro, W. Dias da Silva, and E. Medina-Acosta. 2001. RNA III inhibiting peptide (RIP), a global inhibitor of Staphylococcus aureus pathogenesis: structure and function analysis. Peptides 22:1621-1627.11587790 [Google Scholar]
- 74.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]