Abstract
Escherichia coli HKY28, a ceftazidime-resistant strain isolated from a urine specimen in Japan, produced an inhibitor-sensitive AmpC β-lactamase variant. The deduced amino acid sequence of the enzyme contained a number of substitutions and a tripeptide deletion (Gly286-Ser287-Asp288) compared with the sequence of native AmpC of E. coli. When the deletion was reverted by a 9-base insertion at the relevant site of ampC in the clone, the typical inhibitor-resistant phenotype of AmpC was restored, while at the same time the levels of resistance to ceftazidime, cefpirome, and cefepime were reduced eightfold or more. Molecular modeling studies indicated that a structural change took place in the H-10 helix as a result of the deletion, and this change caused an alteration of the substrate binding site, leading to a unique phenotype analogous to that of inhibitor-sensitive class A extended-spectrum β-lactamases. The degree of inhibition was greater with sulbactam and tazobactam than with clavulanic acid. To our knowledge, this is the first report to have characterized an E. coli ampC that encodes chromosomal AmpC β-lactamase sensitive to the available β-lactamase inhibitors.
The principal and most prevalent mechanism of resistance to β-lactam agents among pathogenic gram-negative bacteria is the production of β-lactamases (3, 17). One approach to overcoming the problem has been the development of β-lactams resistant to the hydrolytic activities of these enzymes. The other has been the development of β-lactamase inhibitors, which protect β-lactams from hydrolysis by β-lactamases when the inhibitors are used in combination with β-lactams (28). At present, three β-lactamase inhibitors, clavulanic acid, sulbactam, and tazobactam, are available for clinical use in combination with a number of penicillins. These inhibitors mainly target Ambler class A β-lactamases and inactivate their active-site serines, thus potentiating the actions of β-lactamase-sensitive compounds. Clavulanic acid and sulbactam are generally not effective in inhibiting the activities of AmpC β-lactamases, although some are known to be moderately inhibited by tazobactam (4, 14).
In 1994, we isolated an Escherichia coli clinical strain, HKY28, which produced a chromosomal AmpC β-lactamase that had an inhibitor-sensitive and extended-spectrum activity profile similar to those of class A extended-spectrum β-lactamases (ESBLs). However, the results of PCR experiments with representative TEM- and SHV-derived ESBLs and CTX-M-type β-lactamases were negative. In the present study we conducted genetic, biochemical, and molecular modeling analyses of this unique AmpC β-lactamase variant.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
E. coli HKY28 was isolated from a culture of urine from an inpatient in Japan in 1994. E. coli XL1-Blue (Stratagene, La Jolla, Calif.) was used as the recipient strain for plasmids. E. coli BMH71-18mutS and E. coli MV1184 (Takara Bio Inc., Ohtsu, Japan) were used as the hosts in a site-directed mutagenesis experiment. Plasmid vectors pBCKS+ (Stratagene) and pKF18k (Takara Bio) were used for the cloning and site-directed mutagenesis experiments, respectively. For enzyme purification, ampC-deficient E. coli CS14-2 (7) was used as the host to avoid background AmpC production. Bacteria were grown in Luria-Bertani (LB) broth supplemented with the appropriate antibiotics, unless specified otherwise.
Antibiotics and susceptibility testing.
The following β-lactam antibiotics and β-lactamase inhibitors were obtained from the indicated sources: aztreonam, Eizai Co., Ltd., Tokyo, Japan; ampicillin, amoxicillin, and cefminox, Meiji Seika Kaisha, Ltd., Tokyo, Japan; cefepime, Bristol Pharmaceuticals K. K., Tokyo, Japan; cefmetazole and chloramphenicol, Sankyo Co., Ltd., Tokyo, Japan; cefotaxime and cefpirome, Aventis Pharma, Ltd., Tokyo, Japan; cefoxitin and imipenem, Banyu Pharmaceutical Co., Ltd., Tokyo, Japan; ceftazidime and clavulanic acid, GlaxoSmithKline K. K., Tokyo, Japan; cephaloridine and moxalactam, Shionogi & Co., Ltd., Osaka, Japan; sulbactam, Pfizer Pharmaceuticals Inc., Tokyo, Japan; and tazobactam, Taiho Pharmaceutical Co., Ltd., Tokyo, Japan.
MICs were determined by the agar dilution method by the protocol recommended by the National Committee for Clinical Laboratory Standards (18).
PCR amplification.
To amplify broad-spectrum β-lactamase genes from HKY28, PCR analysis was performed with sets of primers for various β-lactamases, including TEM- and SHV-derived ESBLs as well as CTX-M-1-, CTX-M-2-, and CTX-M-9-type β-lactamases, as described previously (27).
Transfer of ceftazidime resistance.
Conjugation experiments were conducted with E. coli CSH2 as the recipient by broth mating and filter mating methods (7). Transconjugants were selected on LB agar supplemented with rifampin (50 μg/ml), nalidixic acid (50 μg/ml), and ceftazidime (4 μg/ml).
Cloning and sequencing of β-lactamase gene.
The basic recombinant DNA manipulations were carried out as described by Sambrook et al. (24). The genomic DNA of HKY28 was prepared and digested with EcoRI. The resultant fragments were ligated with plasmid vector pBCKS+, and electrocompetent E. coli XL1-Blue was transformed with these recombinant plasmids. Transformants were selected for resistance to chloramphenicol (30 μg/ml) and ceftazidime (4 μg/ml). For determination of the MICs and use of the transformants for site-directed mutagenesis, the ampC gene of HKY28 was amplified with oligonucleotide primers ampC-U (5′-CGG AAT TCG GTT TTC TAC GGT CTG GC-3′) and ampC-L (5′-CGG GAT CCG ATG ACA GCA AGG AAA AG-3′), which contained EcoRI and BamHI cleavage sites (indicated in boldface), respectively, at their 5′ ends, by using Pyrobest DNA polymerase (Takara Bio). The EcoRI-BamHI fragment containing the ampC gene of E. coli HKY28 was ligated with pBCKS+ to yield pBE28W, which was then used to transform E. coli XL1-Blue and E. coli CS14-2. The coding sequences of the cloned fragments were determined by using custom sequencing primers as well as a BigDye Terminator Cycle Sequencing Ready Reaction kits and an ABI 3100 DNA sequencer (Applied Biosystems, Foster City, Calif.). The enzymes used for gene manipulations were purchased from Nippon Gene Co. Ltd. (Tokyo, Japan) or New England Biolabs, Inc. (Beverly, Mass.).
Reversion of AmpC deletion.
Site-directed mutagenesis was performed to revert the 9-nucleotide deletion in the cloned ampC gene of E. coli HKY28 corresponding to a tripeptide deletion at positions 286 to 288 in AmpC. The reagents and strains contained in the Mutan-Express Km mutagenesis kit (Takara Bio) were used according to the procedures based on the oligonucleotide-directed dual Amber method (9) provided by the manufacturer. The following mutagenic primer containing the 9-nucleotide insertion (in boldface) was used: 5′-CCA GTG CAA TTT TAT TGT CAC TGC CGT TAA TGA TGA TGT CAG G-3′. After mutagenesis, the EcoRI-BamHI fragment containing the revertant ampC was ligated with pBCKS+ to yield pBE28R, which was then used to transform E. coli XL1-Blue and E. coli CS14-2.
Enzyme purification.
E. coli CS14-2 harboring pBE28W or pBE28R was cultured overnight in 2 liters of LB broth supplemented with 30 μg of chloramphenicol per ml. Cells were harvested by centrifugation and washed with and then suspended in 3 ml of 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer (pH 6.0). The cells were frozen and thawed twice and were then ultracentrifuged at 100,000 × g for 4 h at 4°C. For gel filtration, the supernatant containing β-lactamase was chromatographed through a HiLoad 16/60 Superdex 200 prepgrade (Pharmacia Biotech, Uppsala, Sweden) column preequilibrated with 50 mM MOPS buffer (pH 6.0). For cation-exchange chromatography, fractions with activity were then applied to a HiTrap SP HP column (Pharmacia Biotech) preequilibrated with the same buffer. The enzymes were eluted with a linear gradient of 0 to 0.5 M NaCl in the same buffer. The purity of the enzymes was checked by a sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Enzyme assays.
Purified AmpC enzymes were assayed against various β-lactam substrates at 37°C in 50 mM phosphate buffer (pH 7.0) by using an autospectrophotometer (V-550; Nihon Bunko Ltd., Tokyo, Japan). The specific activity of the enzymes was defined as the activity that hydrolyzed 1 μmol of cephaloridine per min. Km and kcat values were obtained by a direct-weight fit to the Michaelis-Menten equation by using KaleidaGraph software (Hulinks, Tokyo, Japan). The concentrations of inhibitors giving a 50% reduction in hydrolysis of cephaloridine (IC50) were measured after 10 min of preincubation of the enzymes with the inhibitors at 37°C and cephaloridine as the substrate at 1 mM. The affinities of the enzymes for the inhibitors (Kis) were measured by competition procedures with cephaloridine in the same buffer with no preincubation of the enzyme or the inhibitor. To determine the isoelectric points, 10 μl of enzyme solution was loaded onto an Immobiline DryStrip (pH 3 to 10 and 6 to 11; Pharmacia Biotech), and electrophoresis was carried out with an IPGphor electrophoresis system (Pharmacia Biotech).
Modeling of substrate-enzyme complex structures.
The crystal structure of the AmpC β-lactamase (Protein Data Bank accession number 2BLS) was used as the reference to build a model of the AmpC enzyme of E. coli HKY28. The tripeptide at the H-10 helix was deleted by the loop search method of the Homology module installed in Insight II software (version 2000; Molecular Simulations Inc., San Diego, Calif.). An initial structure of the enzyme was optimized by use of molecular dynamics calculations at 298 K by the cell multipole method, a distance-dependent dielectric constant, and a time step of 1 fs for 100 ps by sampling the conformation every 1 ps by use of Discover 3 software (version 98.0; Molecular Simulations Inc.). One hundred conformations were minimized until the final root-mean square deviation became less than 0.1 kcal/mol/Å, and the lowest energy conformation was selected for the substrate-docking study. The substrates were roughly docked into the ligand-binding cleft with the guidance of a hydrogen bond of a β-lactam carbonyl oxygen at the oxyanion hole as well as a hydrogen bond of the carboxylate oxygen with Tyr150 (12). The initial complex model was minimized, and then the substrate-binding site was covered by water molecules (sphere thickness, 20 Å). The structure consisted of the substrate and the residues within 10 Å from the substrate, which were energy optimized in the presence of the water molecules by the molecular dynamics and minimization procedure described above. The lowest-energy structures were selected as energy-refined complex models.
Nucleotide sequence accession number.
The nucleotide sequence encoding AmpC characterized in this study appears in the EMBL/GenBank/DDBJ databases under accession number AB108683.
RESULTS
Susceptibility of parental strain.
The MICs of β-lactams for parental strain E. coli HKY28 are shown in Table 1. Strain HKY28 was resistant to ampicillin, amoxicillin, cephaloridine, cefminox, and cefoxitin. It was also resistant to ceftazidime (MIC, 32 μg/ml) but remained susceptible to aztreonam and imipenem. Interestingly, the MIC of ampicillin was reduced by at least 8-fold when it was combined with sulbactam, and the MICs of cefotaxime were reduced by 16- and 8-fold when it was combined with sulbactam and tazobactam, respectively. Addition of sulbactam reduced the MIC of ceftazidime by eightfold. Overall, the reductions in the MICs were the greatest with sulbactam, followed by tazobactam and clavulanic acid.
TABLE 1.
Results of antibiotic susceptibility testing
| β-Lactam | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| E. coli HKY28 | E. coli XL1-Blue(pBE28 W) | E. coli XL1-Blue(pBE28 R) | E. coli XL1-Blue | |
| Amoxicillin | 128 | >128 | >128 | 4 |
| Amoxicillin-clavulanatea | 128 | >128 | >128 | 4 |
| Ampicillin | >128 | >128 | >128 | 2 |
| Ampicillin-sulbactamb | 32 | 64 | >128 | 2 |
| Piperacillin | 8 | 8 | 8 | 0.5 |
| Piperacillin-tazobactamc | 4 | 2 | 4 | 0.5 |
| Cefotaxime | 16 | 32 | 8 | 0.06 |
| Cefotaxime-clavulanatea | 8 | 8 | 2 | 0.06 |
| Cefotaxime-sulbactamb | 1 | 1 | 2 | 0.06 |
| Cefotaxime-tazobactamc | 2 | 4 | 4 | 0.06 |
| Ceftazidime | 32 | 128 | 16 | 0.06 |
| Ceftazidime-clavulanatea | 16 | 32 | 8 | 0.13 |
| Ceftazidime-sulbactamb | 4 | 8 | 8 | 0.06 |
| Ceftazidime-tazobactamc | 16 | 8 | 8 | 0.13 |
| Cephaloridine | 64 | 128 | 128 | 4 |
| Cefminox | 32 | 32 | 32 | 0.5 |
| Cefoxitin | 16 | 32 | >128 | 8 |
| Cefmetazole | 16 | 32 | 128 | 1 |
| Moxalactam | 8 | 4 | 8 | 0.25 |
| Cefpirome | 2 | 4 | 0.03 | 0.015 |
| Cefepime | 2 | 4 | 0.03 | 0.015 |
| Aztreonam | 8 | 16 | 16 | 0.06 |
| Imipenem | 0.13 | 0.13 | 0.13 | 0.13 |
Fixed concentration of clavulanate, 4 μg/ml.
Fixed concentration of sulbactam, 4 μg/ml.
Fixed concentration of tazobactam, 4 μg/ml.
PCR analysis of β-lactamase genes.
By PCR E. coli HKY28 was negative for the genes for the TEM-, SHV-, CTX-M-1-, CTX-M-2- and CTX-M-9-type β-lactamases, which are the prevalent types of ESBLs in Japan.
Transfer of ceftazidime resistance.
The ceftazidime resistance of E. coli HKY28 could not be transferred to recipient E. coli strain CSH2 by conjugation, despite repeated attempts.
Cloning and sequencing of resistance gene.
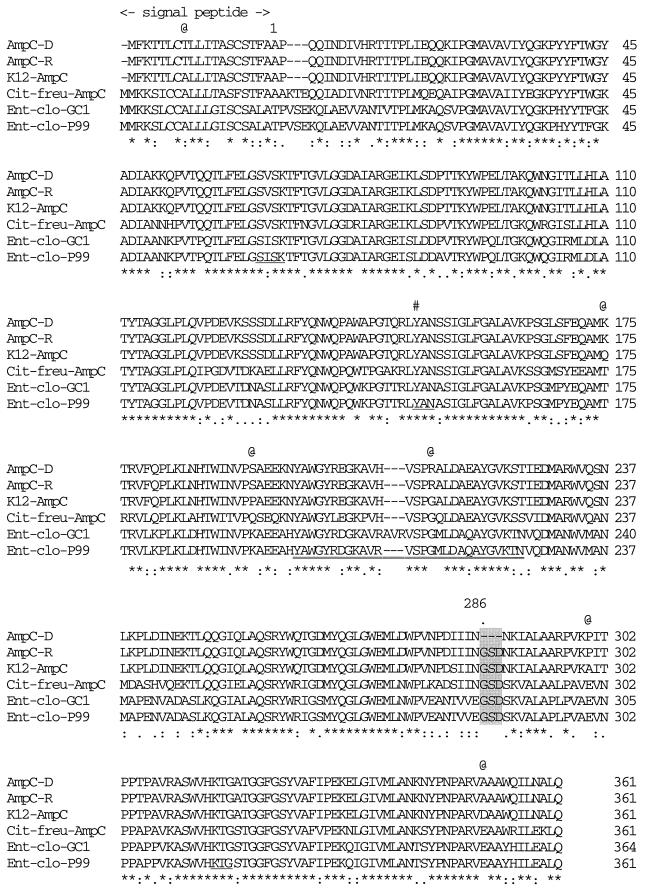
A 6-kb EcoRI fragment containing a ceftazidime resistance determinant was cloned into the vector pBCKS+ and was termed pE753. Nucleotide sequencing analysis revealed a chromosomal locus of E. coli containing ampC flanked by frdD and blc but without any other β-lactamase gene. PCR-generated recombinant plasmid pBE28W containing ampC of E. coli HKY28 was found to possess an ampC gene identical to that of pE753 and conferred resistance to ceftazidime. The deduced amino acid sequence contained seven amino acid substitutions and three amino acid deletions (Gly286, Ser287, and Asp288) of the AmpC product compared with the sequence of E. coli K-12 (10) (Fig. 1). The promoter region of the ampC gene contained three mutations (a C-to-T change at position −73, a C-to-T change at position +6, and a G-to-A change at position +34) and a T insertion between positions −14 and −13 compared with the sequence of the corresponding region of the E. coli K-12 genome. Recombinant plasmid pBE28R, generated by site-directed mutagenesis, was confirmed to possess ampC of E. coli HKY28, except for the insertion of the 9-nucleotide sequence designed to restore the tripeptide deleted from ampC of E. coli HKY28.
FIG. 1.
Predicted amino acid sequence of the AmpC β-lactamase of E. coli HKY28 aligned with that of E. coli K-12 (10). The 3-amino-acid deletion in the HKY28 AmpC is shaded. Underlines, the β-lactamase active site SVSK, the conserved tripeptide KTG, and the class C motif YXN; #, position of Tyr150; @, positions of the amino acid substitutions observed between the AmpCD of strain HKY28 and the AmpC of strain K-12; numbers on the right, numbers of amino acid residues from the N terminus of each mature protein; ✻, amino acid residues conserved among the six AmpC-type enzymes; colons and dots, amino acid substitutions that result in homologous amino acid residues; Cit-freu, Citrobacter freundii; Ent-clo, E. cloacae; double underline, AmpC Ω-loop domain.
Susceptibilities of clones to β-lactams.
Both E. coli XL1-Blue harboring pBE28W (the HKY28 clone) and that harboring pBE28R (the revertant clone) displayed resistance or reduced susceptibilities to all β-lactams except cefpirome, cefepime, and imipenem; but the degree of resistance varied significantly between the two clones. The cefotaxime and ceftazidime MICs were fourfold or more higher for the HKY28 clone than for the revertant clone. The cefpirome and cefepime MICs were 64-fold higher for the HKY28 clone than for the revertant clone. On the other hand, the degree of resistance to cefoxitin and cefmetazole conferred by the revertant clone was significantly higher than that conferred by the HKY28 clone. When various β-lactam-β-lactamase inhibitor combinations were tested, the piperacillin, cefotaxime, and ceftazidime MICs for the HKY28 clone were reduced by up to 16-fold. The degree of reduction was the greatest when sulbactam was used as the inhibitor. The reductions in the MICs of the three inhibitors for the revertant clone were fourfold or less.
Isoelectric focusing.
The isoelectric points were estimated to be 9.9 for the HKY28 AmpC (AmpCD) and 9.8 for the revertant AmpC (AmpCR). When the crude extract of E. coli HKY28 was subjected to analytical isoelectric focusing, only one band corresponding to AmpCD was visualized with nitrocefin, confirming that AmpCD is the only β-lactamase produced by E. coli HKY28 (data not shown).
Enzyme assays.
The specific activites of AmpCD and AmpCR were 88 and 220 U/mg of protein, respectively. The kinetic parameters (Km and kcat) and hydrolytic efficiencies (kcat/Km) of AmpCD and AmpCR against various β-lactams are given in Table 2. The kcat values of AmpCD were greater than those of AmpCR for cefpirome and cefepime but lower for the rest of the substrates tested. However, for all substrates with the exception of cefotaxime, AmpCD exhibited lower Km values than AmpCR. This difference was approximately 100-fold for ceftazidime, and overall, AmpCD showed a 2.5-fold greater hydrolytic efficiency for ceftazidime than AmpCR, despite the much poorer kcat. AmpCD exhibited much lower Km values and higher kcat values for cefpirome and cefepime than AmpCR, resulting in approximately 40- and 20-fold greater hydrolytic efficiencies, respectively.
TABLE 2.
Kinetic activity of AmpCD and AmpCR
| Substrate | AmpCD
|
AmpCR
|
||||
|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) | |
| Cephaloridine | 100 ± 10 | 64 ± 4 | 6.4 × 105 | 780 ± 40 | 300 ± 10 | 3.9 × 105 |
| Ampicillin | 6.2 ± 1.6 | 0.43 ± 0.06 | 7.0 × 104 | 13 ± 3 | 4.0 ± 0.3 | 3.2 × 105 |
| Cefoxitin | 1.2 ± 0.3 | 0.043 ± 0.006 | 3.8 × 104 | 3.9 ± 0.1 | 0.35 ± 0.01 | 9.1 × 104 |
| Ceftazidime | 5.7 ± 0.8 | 0.084 ± 0.006 | 1.5 × 104 | 550 ± 10 | 3.5 ± 0.1 | 6.4 × 103 |
| Cefotaxime | 31 ± 8 | 0.37 ± 0.02 | 1.2 × 104 | 13 ± 2 | 1.2 ± 0.1 | 9.7 × 104 |
| Cefpirome | 21 ± 1 | 1.5 ± 0.1 | 7.1 × 104 | 120 ± 20 | 0.21 ± 0.03 | 1.8 × 103 |
| Cefepime | 49 ± 5 | 1.0 ± 0.1 | 2.1 × 104 | 200 ± 40 | 0.21 ± 0.02 | 1.1 × 103 |
The IC50s of the β-lactamase inhibitors for AmpCD and AmpCR and the Ki values of the enzymes against the inhibitors are listed in Table 3. AmpCD exhibited approximately 5- to 10-fold lower Ki values than AmpCR against all three inhibitors. Tazobactam was the best inhibitor and had the lowest IC50 for AmpCD.
TABLE 3.
IC50s and Ki values of β-lactamase inhibitors for AmpCD and AmpCR
| β-Lactamase | Clavulanic acid
|
Sulbactam
|
Tazobactam
|
|||
|---|---|---|---|---|---|---|
| IC50 (μM) | Ki (μM) | IC50 (μM) | Ki (μM) | IC50 (μM) | Ki (μM) | |
| AmpCD | 19 ± 1 | 320 ± 30 | 3.9 ± 0.2 | 9.2 ± 0.2 | 1.4 ± 0.1 | 8.7 ± 2.4 |
| AmpCR | 140 ± 20 | 4,100 ± 1,600 | 24 ± 4 | 780 ± 150 | 25 ± 1 | 1,100 ± 120 |
Molecular modeling study.
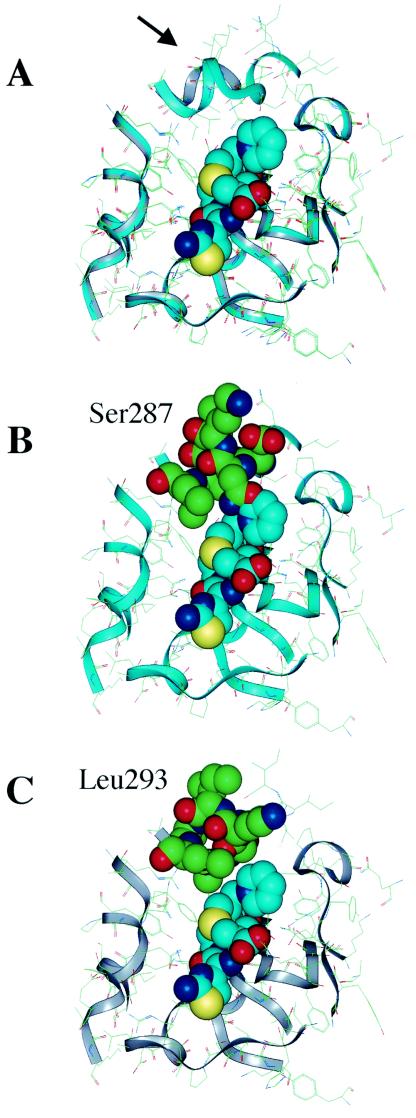
A molecular modeling study was conducted to elucidate the mechanism for the lower Km of AmpCD for ceftazidime (Fig. 2). In the AmpC of E. coli K-12, the tripeptide Gly286-Ser287-Asp288 loops out in the direction of ceftazidime (Fig. 2B). Conversely, the tripeptide deletion in AmpCD creates an open site in the vicinity of the R-2 side chain of ceftazidime (Fig. 2C). Similar models were obtained for cefpirome and cefepime (data not shown).
FIG. 2.
Optimized ribbon structures of ceftazidime docked in the active site of HKY28 AmpC (gray) compared with that of E. coli K-12 AmpC (light blue) (10). (A) The two structures are superimposed. The Gly286-Ser287-Asp288 deletion in the HKY28 AmpC creates an open space at the top of the binding site (arrow) that allows the accommodation of the R-2 side chain of ceftazidime in the E. coli K-12 AmpC and the R-2 side chain of ceftazidime collides with Ser287(B), but in the HKY28 AmpC (C) it comes near Leu293 but does not make direct contact.
DISCUSSION
E. coli HKY28 produced an AmpC β-lactamase which conferred resistance to ceftazidime and reduced susceptibility to cefotaxime (MICs, 32 and 16 μg/ml, respectively). This resistance was significantly compromised by the β-lactamase inhibitors sulbactam and tazobactam and to some extent by clavulanic acid. This was an uncommon finding, since E. coli rarely acquires resistance to ceftazidime solely by the production of chromosomal β-lactamase. Also, the AmpC β-lactamase, which belongs to Ambler class C β-lactamases, is not usually inhibited well by β-lactamase inhibitors. We therefore investigated the AmpC β-lactamase of the strain.
When the ampC gene was cloned and expressed in E. coli XL1-Blue, it conferred resistance to ceftazidime and cefotaxime, and the resistance could be reversed by any of the three commercially available β-lactamase inhibitors. Sulbactam and tazobactam were much more potent inhibitors in terms of lowering the MICs than clavulanic acid, a distinct profile compared with those of class A ESBLs, which are generally inhibited well by any of the three inhibitors (4).
Sequencing of the entire ampC structural gene of E. coli HKY28 revealed the presence of seven amino acid alterations and a tripeptide deletion at positions 286 to 288 corresponding to Gly-Ser-Asp in the deduced amino acid sequence of AmpC (Fig. 1). None of the substituted residues has been implicated in playing a functional role in the hydrolysis of β-lactams (23, 25). On the other hand, residues 287 to 289, which overlap the residues deleted from AmpCD, is known to be positioned in close proximity to R-2 substituents of β-lactams (16). The levels of resistance to ceftazidime and cefotaxime were reduced by 4-fold or more, while those of newer oxyiminocephalosporins, such as cefepime and cefpirome, were also reduced by 64-fold for the revertant clone producing AmpCR. The three β-lactamase inhibitors no longer reversed resistance to cefotaxime and ceftazidime in the revertant clone. AmpCR has a G214R substitution in the so-called Ω loop, and this substitution may have some influence on the expansion of substrate specificity, especially for cephamycins such as cefoxitin and cefmetazole. The kinetic values of AmpCR for broad-spectrum cephalosporins, including cefotaxime, ceftazidime, cefepime, and cefpirome, as well as cephamycins, such as cefoxitin, indicate that AmpCR certainly has some unusual properties. Some of the five amino acid substitutions found in AmpCR might contribute to such a phenotype. In addition, the three amino acid deletions at the H-10 domain observed in AmpCD might provide this enzyme with a special characteristic, such as enhanced susceptibility to β-lactamase inhibitors and an augmented ability to hydrolyze ceftazidime, cefepime, and cefpirome. However, the deletion might result in a decrease in the ability to hydrolyze cephamycins.
The results of the kinetics studies were very much in accordance with the susceptibility profiles. AmpCD generally exhibited lower Km values than AmpCR against all substrates tested except cefotaxime. These reductions in Km values were accompanied by compromised kcat values, with the exception of those for cefpirome and cefepime. AmpCD showed both lower Km values and greater kcat values for these two agents, resulting in 40- and 20-fold better hydrolytic efficiencies, respectively, compared with those of AmpCR. The kinetic data for cefotaxime did not correlate well with the MICs. A similar observation was reported for an atypical AmpC of an Enterobacter cloacae clinical isolate lacking 6 amino acids at positions 289 to 294, located adjacent to the deletion identified in AmpCD (2). By consideration of the fact that these data were obtained for two clones which differed only by the presence and the absence of the 3 amino acids in AmpC, one possibility is that AmpCD is unstable.
The results of inhibition studies confirmed the role of the Gly286-Ser287-Asp288 deletion in the increased sensitivity of AmpCD to all three commercially available β-lactamase inhibitors. The tripeptide deletion in AmpCD was shown to lower the Ki values against the inhibitors by approximately 10- to 100-fold. In terms of IC50s, sulbactam and tazobactam were potent inhibitors of AmpCD, whereas clavulanic acid only mildly inhibited the enzyme. The AmpC β-lactamase of E. cloacae P99 is inhibited well by tazobactam but is inhibited only modestly by sulbactam and is hardly inhibited at all by clavulanic acid (4). In this respect, AmpCD is an AmpC β-lactamase that is unusually sensitive, especially to sulbactam.
Gly286-Ser287-Asp288 is located in the H-10 helix of AmpC (16). While the functional roles of these residues in the catalytic mechanism have not been clearly elucidated, Asp288 of the E. coli AmpC has been suggested to play a role in recognizing the carboxylate group of β-lactams (23, 25). In native AmpC, Ser287 forms hydrogen bonds with Asn346 and Arg349 (23), but these bonds are lost in AmpCD, along with the deletion of Asp288. The result of the molecular modeling study provided a structural explanation for the lowered Km of AmpCD for ceftazidime, as shown in Fig. 2. In the E. coli K-12 AmpC, the tripeptide Gly286-Ser287-Asp288 impeded access of ceftazidime to the active site of the enzyme, resulting in high Km values, whereas the tripeptide deletion in AmpCD was found to provide an open site where the R-2 side chain of ceftazidime could readily be accommodated. This explains the significantly lower Km for AmpCD compared with that for AmpCR.
While inducible chromosomal AmpC β-lactamases are known to confer resistance to oxyiminocephalosporins and cephamycins by mutations in their regulator genes that lead to derepressed production of the enzymes in many species of gram-negative bacteria, only a few AmpC enzymes with altered substrate specificities have been reported to date (17). The extended-spectrum AmpC produced by E. cloacae GC1 contained a tripeptide insertion of a tandem repeat, Ala211-Val212-Arg213, in the Ω loop (6, 20). It was suggested that the conformational flexibility in the expanded Ω loop facilitates hydrolysis of oxyiminocephalosporins (6). It is noteworthy that an AmpC with extended resistance to cefepime and cefpirome was recently described from an E. cloacae clinical isolate, as mentioned above (2). A deletion of 6 amino acids (Ser, Lys, Val, Ala, Leu, and Ala) from positions 289 to 294 was likely responsible for the extension of the spectrum of activity. The enzyme showed approximately 10 times higher hydrolytic efficiency for the oxyiminocephalosporins than the P99 β-lactamase did, mostly due to lower Km values. This amino acid deletion is in close proximity to that in the AmpCD studied here, both of which are located in the H-10 helix. Therefore, it is not surprising that the two enzymes share similar kinetic characteristics.
E. coli is known to constitutively produce only an insignificant amount of chromosomal AmpC β-lactamase, due to relatively weak promoter activity and the presence of a transcriptional attenuator (11, 13). However, occasional isolates produce large amounts of the enzyme and become resistant to various β-lactams, including ceftazidime. This overproduction could result from gene amplification (8) or the acquisition of a stronger promoter region (21, 22); but most commonly it results from mutations that take place in the promoter region at positions such as −42, −32, and +24, which lead to enhanced transcription of ampC (5, 19). These modifications in transcription typically lead to moderately elevated ceftazidime MICs (13). The nucleotide sequence of the promoter and attenuator regions of the ampC gene of E. coli HKY28 revealed the presence of three mutations (a C-to-T change at position −73, a C-to-T change at position +6, and a G-to-A change at position +34) and a T insertion between positions −14 and −13. The first two mutations have not been implicated as a cause of increased ampC transcription, while a nucleotide insertion between −35 and −10 hexamers is known to enhance AmpC transcription, possibly by bringing the distance between the hexamers to the optimal 17 bp (5, 11). Therefore, it is likely that the insertion of a T residue between positions −14 and −13 caused the hyperproduction of AmpCD, explaining in part the ceftazidime resistance displayed by E. coli HKY28.
Class A ESBLs are inhibited well by the β-lactamase inhibitors clavulanic acid and sulbactam, a characteristic that serves to differentiate them from other β-lactamases, including AmpC (15). The Ki values of class A enzymes for the inhibitors are in the nanomolar range, but those of the AmpCD enzyme reported in the present study are in the micromolar range. Anyway, AmpCD acquired considerable sensitivity to inhibition by sulbactam and tazobactam but acquired sensitivity to inhibition by clavulanic acid to a much lower degree, as it extended its spectrum to cephalosporins, including ceftazidime. A few other studies have also reported on the isolation of E. coli strains displaying similar inhibitor-sensitive phenotypes, but their mechanisms remain to be described (1, 26). It would be interesting to know if they produce AmpC variants with characteristics similar to those of the AmpC described in the present study.
Acknowledgments
We are grateful to Kevin Young for kindly providing E. coli CS14-2. We also thank Kumiko Kai for expert technical assistance.
This work was supported by Grant-in-Aid for Young Scientists (B) no. 14771358 from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Biochemical analysis of this work was supported in part by grants H12-Shinkou-19 and H12-Shinkou-20 from the Ministry of Health, Labor and Welfare of Japan.
REFERENCES
- 1.Babini, G. S., F. Danel, S. D. Munro, P. A. Micklesen, and D. M. Livermore. 1998. Unusual tazobactam-sensitive AmpC β-lactamase from two Escherichia coli isolates. J. Antimicrob. Chemother. 41:115-118. [DOI] [PubMed] [Google Scholar]
- 2.Barnaud, G., R. Labia, L. Raskine, M. J. Sanson-Le Pors, A. Philippon, and G. Arlet. 2001. Extension of resistance to cefepime and cefpirome associated to a six amino acid deletion in the H-10 helix of the cephalosporinase of an Enterobacter cloacae clinical isolate. FEMS Microbiol. Lett. 195:185-190. [DOI] [PubMed] [Google Scholar]
- 3.Bush, K. 2001. New β-lactamases in gram-negative bacteria: diversity and impact on the selection of antimicrobial therapy. Clin. Infect. Dis. 32:1085-1089. [DOI] [PubMed] [Google Scholar]
- 4.Bush, K., C. Macalintal, B. A. Rasmussen, V. J. Lee, and Y. Yang. 1993. Kinetic interactions of tazobactam with β-lactamases from all major structural classes. Antimicrob. Agents Chemother. 37:851-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caroff, N., E. Espaze, I. Berard, H. Richet, and A. Reynaud. 1999. Mutations in the ampC promoter of Escherichia coli isolates resistant to oxyiminocephalosporins without extended spectrum β-lactamase production. FEMS Microbiol. Lett. 173:459-465. [DOI] [PubMed] [Google Scholar]
- 6.Crichlow, G. V., A. P. Kuzin, M. Nukaga, K. Mayama, T. Sawai, and J. R. Knox. 1999. Structure of the extended-spectrum class C β-lactamase of Enterobacter cloacae GC1, a natural mutant with a tandem tripeptide insertion. Biochemistry 38:10256-10261. [DOI] [PubMed] [Google Scholar]
- 7.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edlund, T., T. Grundstrom, and S. Normark. 1979. Isolation and characterization of DNA repetitions carrying the chromosomal β-lactamase gene of Escherichia coli K-12. Mol. Gen. Genet. 173:115-125. [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto-Gotoh, T., T. Mizuno, Y. Ogasahara, and M. Nakagawa. 1995. An oligodeoxyribonucleotide-directed dual Amber method for site-directed mutagenesis. Gene 152:271-275. [DOI] [PubMed] [Google Scholar]
- 10.Jaurin, B., and T. Grundstrom. 1981. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of β-lactamases of the penicillinase type. Proc. Natl. Acad. Sci. USA 78:4897-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaurin, B., T. Grundstrom, T. Edlund, and S. Normark. 1981. The E. coli β-lactamase attenuator mediates growth rate-dependent regulation. Nature 290:221-225. [DOI] [PubMed] [Google Scholar]
- 12.Kato-Toma, Y., T. Iwashita, K. Matsuda, Y. Oyama, and M. Ishiguro. 2003. pKa measurements from nuclear magnetic resonance of tyrosine-150 in class C β-lactamase. Biochem. J. 371:175-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livermore, D. M. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livermore, D. M. 1993. Determinants of the activity of β-lactamase inhibitor combinations. J. Antimicrob. Chemother. 31(Suppl. A):9-21. [DOI] [PubMed] [Google Scholar]
- 15.Livermore, D. M., and D. F. Brown. 2001. Detection of β-lactamase-mediated resistance. J. Antimicrob. Chemother. 48(Suppl. 1):59-64. [DOI] [PubMed] [Google Scholar]
- 16.Lobkovsky, E., P. C. Moews, H. Liu, H. Zhao, J. M. Frere, and J. R. Knox. 1993. Evolution of an enzyme activity: crystallographic structure at 2-Å resolution of cephalosporinase from the ampC gene of Enterobacter cloacae P99 and comparison with a class A penicillinase. Proc. Natl. Acad. Sci. USA 90:11257-11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medeiros, A. A. 1997. Evolution and dissemination of β-lactamases accelerated by generations of β-lactam antibiotics. Clin. Infect. Dis. 24(Suppl. 1):S19-S45. [DOI] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 2000. Methods for disk antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne Pa.
- 19.Nelson, E. C., and B. G. Elisha. 1999. Molecular basis of AmpC hyperproduction in clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 43:957-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nukaga, M., S. Haruta, K. Tanimoto, K. Kogure, K. Taniguchi, M. Tamaki, and T. Sawai. 1995. Molecular evolution of a class C β-lactamase extending its substrate specificity. J. Biol. Chem. 270:5729-5735. [DOI] [PubMed] [Google Scholar]
- 21.Olsson, O., S. Bergstrom, F. P. Lindberg, and S. Normark. 1983. ampC β-lactamase hyperproduction in Escherichia coli: natural ampicillin resistance generated by horizontal chromosomal DNA transfer from Shigella. Proc. Natl. Acad. Sci. USA 80:7556-7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olsson, O., S. Bergstrom, and S. Normark. 1982. Identification of a novel ampC β-lactamase promoter in a clinical isolate of Escherichia coli. EMBO J. 1:1411-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powers, R. A., E. Caselli, P. J. Focia, F. Prati, and B. K. Shoichet. 2001. Structures of ceftazidime and its transition-state analogue in complex with AmpC β-lactamase: implications for resistance mutations and inhibitor design. Biochemistry 40:9207-9214. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Usher, K. C., L. C. Blaszczak, G. S. Weston, B. K. Shoichet, and S. J. Remington. 1998. Three-dimensional structure of AmpC β-lactamase from Escherichia coli bound to a transition-state analogue: possible implications for the oxyanion hypothesis and for inhibitor design. Biochemistry 37:16082-16092. [DOI] [PubMed] [Google Scholar]
- 26.Wong-Beringer, A., J. Hindler, M. Loeloff, A. M. Queenan, N. Lee, D. A. Pegues, J. P. Quinn, and K. Bush. 2002. Molecular correlation for the treatment outcomes in bloodstream infections caused by Escherichia coli and Klebsiella pneumoniae with reduced susceptibility to ceftazidime. Clin. Infect. Dis. 34:135-146. [DOI] [PubMed] [Google Scholar]
- 27.Yagi, T., H. Kurokawa, N. Shibata, K. Shibayama, and Y. Arakawa. 2000. A preliminary survey of extended-spectrum β-lactamases (ESBLs) in clinical isolates of Klebsiella pneumoniae and Escherichia coli in Japan. FEMS Microbiol. Lett. 184:53-56. [DOI] [PubMed] [Google Scholar]
- 28.Yang, Y., B. A. Rasmussen, and D. M. Shlaes. 1999. Class A β-lactamases-enzyme-inhibitor interactions and resistance. Pharmacol. Ther. 83:141-151. [DOI] [PubMed] [Google Scholar]