Abstract
Dihydropteroate synthase (DHPS) mutations in Pneumocystis jiroveci have been associated epidemiologically with resistance to sulfamethoxazole (SMX). Since P. jiroveci cannot be cultured, inherent drug resistance cannot be measured. This study explores the effects of these mutations in a tractable model organism, Saccharomyces cerevisiae. Based on the sequence conservation between the DHPS enzymes of P. jiroveci and S. cerevisiae, together with the structural conservation of the three known DHPS structures, DHPS substitutions commonly observed in P. jiroveci were reverse engineered into the S. cerevisiae DHPS. Those mutations, T597A and P599S, can occur singly but are most commonly found together and are associated with SMX treatment failure. Mutations encoding the corresponding changes in the S. cerevisiae dhps were made in a yeast centromere vector, p414FYC, which encodes the native yeast DHPS as part of a trifunctional protein that also includes the two enzymes upstream of DHPS in the folic acid synthesis pathway, dihydroneopterin aldolase and 2-amino-4-hydroxymethyl dihydropteridine pyrophosphokinase. A yeast strain with dhps deleted was employed as the host strain, and transformants having DHPS activity were recovered. Mutants having both T597 and P599 substitutions had a requirement for p-aminobenzoic acid (PABA), consistent with resistance being associated with altered substrate binding. These mutants could be adapted for growth in the absence of PABA, which coincided with increased sulfa drug resistance. Upregulated PABA synthesis was thus implicated as a mechanism for sulfa drug resistance for mutants having two DHPS substitutions.
Dihydropteroate synthase (DHPS) is an important target for antifolate compounds, such as sulfones and sulfonamides, in both prokaryotic and eukaryotic microbes. Organisms treated with sulfa drugs include pathogens, such as Pneumocystis jiroveci, Mycobacterium leprae, Neisseria meningitidis, and Plasmodium falciparum. Numerous studies have reported that mutations in the primary sequence of dhps are associated with sulfa drug resistance (7, 10, 12, 18, 20, 22, 23, 32, 36, 38, 39). The P. jiroveci mutant alleles map to a highly conserved sequence in DHPS that is associated with direct substrate contact (1, 3, 14), although there is some controversy over whether these influence drug treatment outcomes (2, 6, 15, 18, 19, 24, 26, 28, 33, 34). In vitro culture of P. jiroveci has not yet been possible, nor has heterologous complementation of P. jiroveci DHPS.
For other pathogens, the association between the efficacy of antifolate drugs and genetic mutations in the dihydrofolate reductase gene (dhfr) and dhps has been demonstrated using model systems (11, 29, 31, 40). This would be particularly useful for P. jiroveci, since there is no direct evidence that mutant alleles identified in P. jiroveci dhps cause sulfa drug resistance.
We have sought to better understand the relationship between DHPS changes and sulfa drug resistance by employing a Saccharomyces cerevisiae model system. S. cerevisiae is a good model for studying P. jiroveci DHPS because (i) both organisms are fungi, (ii) both have trifunctional dihydroneopterin aldolase (DHNA)-2-amino-4-hydroxymethyl dihydropteridine pyrophosphokinase (PPPK)-DHPS enzymes, (iii) the DHPS amino acid sequence alignments show 30% identity and 62% similarity and DHNA-PPPK-DHPS (folic acid synthesis [FAS]) alignments show 37% identity and 64% similarity, (iv) the organisms have identical sequences in the regions associated with sulfamethoxazole (SMX) resistance (T597, R598, and P599 in S. cerevisiae; T517, R518, and P519 in P. jiroveci) (Fig. 1); (v) the conservation rates of these residues across 75 published DHPS species are 82, 80, and 99%, respectively, and (vi) the residues map to the same region in the three known structures of DHPS that have been solved to date (1, 3, 14).
FIG. 1.
Alignment of DHPS sequences by using ClustalW. Displayed are sequences from E. coli (Ec), Mycobacterium tuberculosis (Mt), and Staphylococcus aureus (Sa), whose structures have been solved, aligned to those of S. cerevisiae (Sc) and P. jiroveci (Pj). The asterisks indicate totally conserved amino acids, the colons indicate very high conservation, and the dots indicate weak conservation. Amino acids associated with SMX resistance in P. jiroveci DHPS (T→A and P→S) are highlighted and in boldface. Dashes represent gaps introduced by the alignment.
Based on these high levels of identity and similarity, we employed an allelic replacement strategy that utilized an S. cerevisiae dhps knockout strain that was complemented by a vector-encoded FAS, the product of the S. cerevisiae FOL1 gene. Residues in the wild-type FAS were mutated to give the changes associated with SMX resistance. We report here the phenotypic analysis and SMX resistance of various dhps alleles implicated from epidemiological studies with SMX resistance.
MATERIALS AND METHODS
E. coli strains, growth media, and transformation.
The bacterial strain employed for molecular cloning and plasmid amplification was Escherichia coli strain MC1061 [araD139 Δ(araABC-leu)7679 galU galK ΔlacX74 rpsL hsdR (rK− mK−) mcrB]. The growth medium utilized was 1× YT (0.5% [wt/vol] yeast extract, 0.8% [wt/vol] tryptone, 0.5% [wt/vol] NaCl). Cells were made competent by calcium chloride treatment, and transformants were selected on 1× YT + 100 μg of ampicillin/ml.
Yeast strains, growth media, and transformation.
The S. cerevisiae dhps knockout strain used in this study, EHY1, has been described previously (4, 5, 16). EHY1 (MATa leu2-3,112 trp1 ura3-52 tup1 DHPS::LEU2) is a genomic dhps knockout strain. The authenticity of the EHY1 knockout strain was confirmed by molecular approaches and phenotypic analyses and was consistent with a specific dhps deletion. The auxotrophic requirements and complementation of EHY1 were previously described (4). Media for the growth of yeast cells included YEPD (1% [wt/vol] yeast extract [Difco], 2% [wt/vol] peptone, 2% [wt/vol] d-glucose). Synthetic minimal medium (MM0) was 0.67% yeast nitrogen base containing ammonium sulfate minus folic acid minus p-aminobenzoic acid (PABA) (Bio 101) supplemented with 1% (wt/vol) d-glucose and 0.2% (wt/vol) uracil.
Yeast transformants were obtained by the lithium acetate transformation procedure (13), followed by growth and selection on complete synthetic medium minus tryptophan. Complete synthetic medium was 0.67% yeast nitrogen base (Difco) containing ammonium sulfate supplemented with 2% d-glucose, amino acids, and adenine as described in the Cold Spring Harbor manual on yeast genetics (8). This medium contained PABA (200 ng/ml) and folate (2 ng/ml). Before being tested for SMX susceptibility, transformants were passaged on MM0 six times.
Synthesis of mutant alleles implicated with sulfa drug resistance.
The Quickchange XL kit (Stratagene) was used to generate four mutant constructs from the wild-type DHPS using oligonucleotides listed in Table 1. The wild-type construct was designated TRP (T597, P599). The mutants were A597, P599 (designated ARP); T597, S599 (designated TRS); and a double mutant, A597, S599 (designated ARS). The fourth allele has not been reported clinically and was derived from the ARS allele by a conservative change of A597 to V597. This yielded V597, S599 (designated VRS). This clone arose spontaneously during the mutagenesis procedure and was kept for its intrinsic interest. In order to validate the synthesis of the mutant allele sequence, each construct was analyzed by DNA sequencing using a Big Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer). Each of the mutant constructs generated was transformed into EHY1 and plated onto complete synthetic medium minus tryptophan, and complementing clones were selected.
TABLE 1.
Oligonucleotides used for site-directed mutagenesis
| Clone | Substitution | Oligonucleotide no. | Direction of priming | Sequence (5′ → 3′)a |
|---|---|---|---|---|
| ARS | A597, S599 | 146337 | → | GGAGGGTGTTCTGCCAGGTCTAACTCTATTCAGGC |
| 146338 | ← | GCCTGAATAGAGTTAGACCTGGCAGAACACCCTCC | ||
| TRS | T597, S599 | 146339 | → | GGAGGGTGTTCTACCAGGTCTAACTCTATTCAGGC |
| 146340 | ← | GCCTGAATAGAGTTAGACCTGGCAGAACACCCTCC | ||
| ARP | A597, P599 | 146341 | → | CGTTGGAGGGTGTTCTGCCAGGCCTAACTCTATTC |
| 146342 | ← | GAATAGAGTTAGGCCTGGCAGAACACCCTCCAACG |
Nucleotide substitutions are in boldface.
Phenotypic analysis of S. cerevisiae transformants.
Each transformant was precultured in YEPD broth at 30°C. The cultures were harvested in log-phase growth, washed with phosphate-buffered saline (PBS; 8% [wt/vol] NaCl, 0.2% [wt/vol] KCl, 1.44% [wt/vol] Na2HPO4, 0.24% [wt/vol] KH2PO4, pH 7.4), and normalized to a cell density of ∼6 × 106 CFU/ml (A595 = 0.1). A series of twofold dilutions was made in a 96-well microtiter plate. One microliter of the dilution series for each clone was spotted on solidified MM0. Alternatively, plates were supplemented with various combinations of the folate biosynthetic pathway end products, including methionine (20 μg/ml), adenine (20 μg/ml), histidine (20 μg/ml), TMP (20 μg/ml), and formyl-tetrahydrofolate (20 μg/ml). In addition, there was a detailed analysis of the affects of PABA that included supplementing MM0 with PABA to achieve a final concentration of 0 to 10,000 ng/ml. The plates were then incubated at 30°C for 3 to 5 days.
Determination of SMX resistances of prototrophs by drug diffusion assays.
In order to evaluate the relative sensitivity or resistance of each DHPS allele to SMX, drug diffusion assays were performed with PABA supplementing the molten medium at a final concentration ranging from 0 to 10,000 ng/ml. The assays were performed in petri dishes 86 mm in diameter containing 25-ml aliquots of MM0 solidified with 1.5% agar (Merck). A 6.5-mm-diameter hole was made in the center of each agar plate. SMX (25 μl at 100 mg/ml dissolved in dimethyl sulfoxide [DMSO]) was loaded into the center and allowed to diffuse through the plate for >2 h. In control plates, 25 μl of DMSO was also loaded into the center and allowed to diffuse. Each experiment was performed in triplicate. Each clone was precultured in YEPD broth at 30°C. The cultures were harvested in log-phase growth, washed with PBS, and normalized to an A595 of 0.1. A stainless-steel wire (2-mm diameter) was used to inoculate each clone radially from the center of the agar plate. The plates were then incubated at 30°C. Growth was evaluated after 3 to 5 days.
Determination of SMX resistances of prototrophs in broth cultures.
In order to accurately compare the growth rates (generation times) of the clones, the growth of broth cultures in MM0 supplemented with 200 ng of pABA/ml (wt/vol) (MM200) was monitored periodically over 180 h by turbidity measurements at A595. Each clone was precultured in YEPD broth at 30°C. The cultures were harvested in log-phase growth, washed with PBS, and normalized to an A595 of 0.1. These cells were then used to seed the growth cultures at an A595 of 0.02. The MM200 broth was supplemented with SMX at 0 to 1,200 μg/ml. Replicate experiments were prepared in 96-well plates (2.2 ml deep), and the cultures were finally aliquoted into four standard-size microtiter plates (Costar 3603) to a final volume of 150 μl. The plates were then incubated in a 30°C humidified incubator and shaken at 1,000 rpm (with a 3-mm displacement). The turbidity was measured using a Multiskan Ascent microplate reader (Thermo Labsystems), and the data were plotted using Graphpad PRISM software (version 3).
Determination of SMX resistance of naïve transformants in broth cultures.
In order to investigate whether elevated PABA levels could account for the increased resistance observed in the adapted strains (prototrophs), the MICs of the naïve transformants were determined in broth cultures. The medium for determination of the MIC was supplemented with PABA at levels up to 1,000 ng/ml. SMX was then added in a range of concentrations (0 to 2,000 μg/ml). After the addition of PABA and SMX to the medium, the pH was adjusted to 7.1 to permit the solubilization of SMX up to 2,000 μg/ml and to eliminate pH differences caused by the SMX and PABA. The upper limit of solubility of SMX is 2,000 μg/ml in MM0 at pH 7.1. The DMSO concentration was constant at (0.5%). Fresh (naïve) transformants were precultured in MM300 containing 2% glucose. Cells were harvested in mid-log-phase growth, washed twice with 50 ml of PBS, and normalized to an A595 of 1.0. Each strain (5 μl of cells) was then seeded into 100 μl of medium in a 96-well plate (Costar 3596). The cultures were grown at 30°C for up to 2 weeks. The seeding density for this experiment was 2.5-fold higher than that of the liquid growth assays, which permitted limited growth of ARS in MM0 (data not shown). The experiments were set up using a Qiagen Rapidplate liquid-handling robot. Each experiment was performed in triplicate.
RESULTS
Construction of yeast strains with altered DHPS.
The yeast strain EHY1 had its chromosomal sequences encoding DHPS deleted so that it no longer synthesized folates. However, EHY1 could grow in the absence of added folate when transformed with p414FYC, a centromere plasmid that encoded the yeast DHNA-PPPK-DHPS trifunctional enzyme (FAS) under the control of its native promoter (16). p414FYC derivatives, altered only by mutations that produced amino acid substitutions at T597 and P599 within DHPS to T597 and S599, A597 and P599, A597 and S599, or V597 and S599 were prepared using site-directed mutagenesis, confirmed by nucleotide sequence analysis, and transformed into the EHY1 strain. Transformants were selected on yeast minimal medium in the absence of added folate and tryptophan. Transformants were obtained for each of the five constructs, indicating functional complementation by each altered DHPS enzyme. For convenience, we shall refer to the five respective transformants as TRP, TRS, ARP, ARS, and VRS.
PABA requirement of mutant transformants.
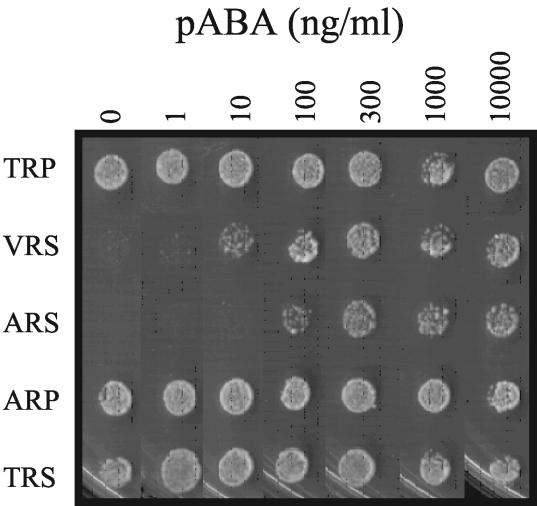
The generation times of transformants expressing each mutant DHPS gene were compared to determine if there were defects caused by the mutant DHPS enzymes. Since SMX resistance assays must be performed in the presence of low levels of PABA (27), the transformants were adapted to low-PABA-folate medium by serial passaging six times on MM0. The growth rates of the strains were compared on minimal medium supplemented with various levels of PABA ranging from 0 to 10,000 ng/ml (Fig. 2). A significant defect was noted with respect to a requirement for PABA by the mutants ARS and VRS. The TRP (wild-type), ARP, and TRS transformants exhibited no significant PABA requirement, while the double mutants, ARS and VRS, exhibited significant PABA requirements, with VRS having less PABA dependence. This indicated that the combined changes of T597 and P599 caused a change in PABA binding or condensation with 2-amino-4-hydroxy-6-hydroxymethyl-dihydropteridine-pyrophosphate (H2PtPP).
FIG. 2.
Growth of transformants on minimal medium supplemented with various levels of PABA.
SMX resistance of prototrophs by drug diffusion assays.
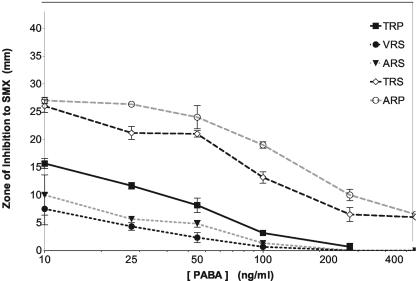
We evaluated the relative resistance of each transformant to SMX, using drug diffusion assays following adaptation to PABA-free minimal medium (MM0). MM0 was used initially to avoid the competitive effect of PABA with the sulfa drugs. The results of the replicates of drug diffusion assays in the presence of various levels of PABA are shown in Fig. 3. In agreement with previous studies (9), PABA and SMX were shown to compete so that the degree of inhibition by SMX for each strain decreased with increasing concentrations of PABA. The smallest inhibitory zones were observed with VRS and ARS, indicating that the double substitutions confer the greatest SMX resistance, and ARP and TRS, with single-amino-acid substitutions, were more sensitive to growth inhibition than the wild type.
FIG. 3.
Determination of SMX resistance by agar drug diffusion assay. SMX was solubilized in DMSO (100 mg/ml), and 25 μl was applied to the centers of agar plates. Additional plates were supplemented with increasing concentrations of PABA. Transformants were harvested in log-phase growth, washed in PBS, and normalized to 6 × 106 CFU/ml. Each strain was then streaked radially from the center of the plate. The plates were grown for 3 to 5 days, and the zones of inhibition for each isolate, TRP, VRS, ARS, TRS, and ARP, were then scored. The results are averages of triplicate experiments; error bars represent the standard deviations of the means.
All transformants retained at least some SMX sensitivity, probably reflecting the fact that DHPS is essential, and it might not be possible to diverge to retain DHPS function and have no sensitivity to SMX. It was possible to show that the inhibition of VRS and ARS, as well as TRP, converged to zero, or no inhibition (complete resistance), when the PABA concentration was >300 ng/ml, while the single-mutant alleles (ARP and TRS) were still inhibited with PABA concentrations as high as 500 ng/ml (Fig. 3). By drug diffusion assays on solidified media, the pattern of resistance to the action of SMX was VRS > ARS > TRP > TRS > ARP (most resistant to least resistant).
SMX resistances of prototrophs in broth cultures.
In order to precisely analyze whether the growth rate influenced drug resistance, growth assays were performed in MM200 broth supplemented with increasing concentrations of SMX. This analysis produced MICs (Table 2) that agreed with those in the drug diffusion assays (Fig. 3). VRS was highly resistant to SMX, as strong growth was observed at the highest SMX concentration tested, suggesting that the MIC was significantly greater than 1,200 μg/ml (i.e., ≫1,200 μg/ml). TRP and ARS were significantly (but not completely) inhibited at 1,200 μg/ml, suggesting that the MICs were slightly greater than 1,200 μg of SMX/ml (i.e., >1,200 μg/ml). By contrast, the single mutants appeared to be considerably more sensitive (Table 2).
TABLE 2.
MICs of SMX for yeast isolates grown in MM200
| DHPS sequence | Isolate | MIC (μg/ml) |
|---|---|---|
| A597, P599 | ARP | 150 |
| T597, S599 | TRS | 600 |
| A597, S599 | ARS | >1,200 |
| T597, P599 | TRP | >1,200 |
| V597, S599 | VRS | >>1,200 |
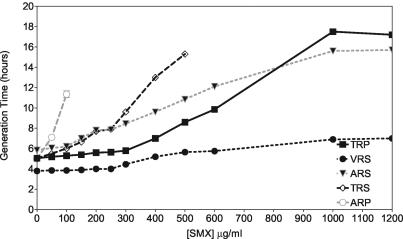
The growth rates of the adapted strains were also measured in the presence of SMX and are shown in Fig. 4. In the absence of SMX, VRS grew faster than all the other transformants, with a generation time of 4 h. TRP, TRS, and ARP had similar generation times of ∼5 h, and ARS had the longest generation time, almost 6 h. With increasing concentrations of SMX, the generation times of all transformants increased (Fig. 4), but the effect on the generation time varied for each transformant. The single mutant ARP became inhibited most quickly, followed by the other single mutant, TRS. The resistance pattern in Fig. 4 precisely matched that shown in Table 2. Likewise, the consistency of profiles held with the other transformants. At SMX concentrations of ≤600 μg/ml, TRP had a shorter generation time than the double mutant ARS. However, at SMX concentrations of >800 μg/ml, the double mutant ARS grew faster than TRP (Fig. 4). Therefore, ARS was more resistant than TRP, as it had a shorter generation time and the MIC for it was higher than that of TRP at high SMX concentrations (data deduced from Fig. 4; cf. Table 2).
FIG. 4.
Generation time of adapted transformants supplemented with increasing concentrations of SMX in MM200. The results are averages of four growth experiments.
VRS, by comparison to all other transformants tested, was the most resistant to SMX in MM100 (data not shown) and MM200. The MIC was measured as ≫1,200 μg/ml (Table 2), but it is obvious from Fig. 4 that at that level of SMX, it had a generation time of ∼7 h.
SMX resistances of naïve transformants in broth cultures.
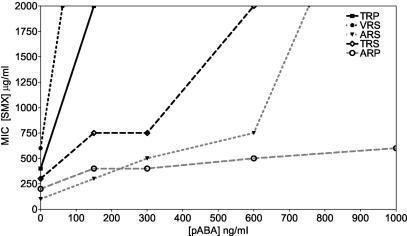
The MIC of SMX for each allele was plotted against the PABA concentration. The data indicated that the resistance of the single mutant ARP did not change significantly when the PABA concentration was elevated. By contrast, the MICs of all other mutants increased significantly with added PABA. The rate of change differed for each clone, indicating that the resistance of each allele was modulated by PABA to various degrees. Specifically, the MIC for naïve ARS was lower than those for TRS and the wild type (TRP) for all conditions tested. This indicated that PABA was not the only factor that led to the resistance of the passaged ARS prototroph (Fig. 3). The MIC for VRS was observed to be highest, 2,000 μg/ml, when the PABA concentration was 100 ng/ml, reflecting the lower requirement of VRS for PABA relative to the ARS mutant. The MIC for TRP was 2,000 μg/ml when the PABA concentration was 150 μg/ml.
DISCUSSION
Mutations in the DHPS gene that are implicated in sulfa drug resistance were reverse engineered into vector-encoded S. cerevisiae FAS (DHNA-PPPK-DHPS) and then transformed into an S. cerevisiae strain having a dhps deleted, effectively resulting in allelic replacement of the sequences encoding DHPS. The two mutations found in the P. jiroveci DHPS gene from patients for whom sulfa prophylaxis failed were sufficient to cause a PABA auxotrophic requirement, as well as SMX resistance, when engineered into S. cerevisiae DHPS (Fig. 1). We conclude from this that such amino acid substitutions probably exert their effects either by reducing the affinity of DHPS for sulfa drugs and for PABA or by affecting the catalysis of condensation of sulfa drugs and PABA with H2PtPP. We have also determined that the requirement for exogenous PABA is much greater in primary transformants. After six passages, yeast cells adapted to abrogate this PABA requirement. The mechanism of this adaptation will be the subject of a future publication (P. Iliades, M. M. Barraclough, S. R. Meshnick, and I. G. Macreadie, unpublished data). This adaptive mechanism may involve increased biosynthesis of PABA. Upregulated PABA synthesis has been suggested as a mechanism for drug-resistant isolates of N. meningitides (17, 21, 37). In a yeast model, it has been established that overexpression of the yeast PABA synthase gene, ABZ1, results in resistance to sulfa drugs (9). In order to ascertain whether adaptation (which possibly alters PABA synthesis) was the only parameter affecting resistance in our model, we determined the MICs for naïve strains in a range of PABA concentrations. These data indicated that PABA can confer increased SMX resistance on mutant strains. However, the most striking observation was the divergent resistance pattern among alleles (Fig. 5). If PABA was the only factor effecting resistance, the change in resistance would be expected to be uniform for each mutant. The divergent resistance profiles observed demonstrate that PABA levels can modulate the resistance of each mutant to various degrees. The ARS mutant showed significantly lower resistance than the ARS prototroph relative to the wild type, indicating that PABA may not be the only parameter implicated in the increased resistance of the adapted strains.
FIG. 5.
MICs for naïve transformants growing in MM0 supplemented with increasing concentrations of SMX and PABA. The results are averages of three experiments. Note that the higher seeding density for this experiment permitted the growth of all naïve transformants.
Importantly, our findings that the double-mutant constructs were more resistant than the wild-type and single-mutant alleles are consistent with the epidemiological data that indicate that double-mutant alleles predominate in frequency over the single mutants (6, 18). If the same is true in P. jiroveci, then the evolution of the double mutation occurred despite negative selection for the first mutation.
Finally, we have found a mutant that was more resistant to SMX than constructs with changes found naturally. The VRS mutant, with the novel T597V and P599S DHPS substitutions, was robust, having the lowest PABA dependence (of the double mutants), the shortest generation times, and the highest SMX resistance. Clearly there is an important interplay that appears to operate between PABA requirements and SMX resistance. This mutant has fortunately not been observed clinically, suggesting that in P. jiroveci it is disadvantageous or, conversely, it might be just a matter of time before it appears.
The construction of S. cerevisiae strains with altered DHPS sequences opens many new approaches to examining resistance to sulfa drugs. It will now be possible to more precisely assay the whole process of the emergence of sulfa drug resistance in a model system that may have implications for understanding drug resistance in infectious organisms, using sophisticated tools associated with S. cerevisiae genetics and molecular biology. For example, using S. cerevisiae FAS mutants (30) showed that sulfa drugs are converted to sulfa-dihydropteroate (DHP) adducts that are growth inhibitory. Thus, sulfa drugs exert an effect by more than one mechanism. It will be interesting to compare the roles of SMX-DHP and the depletion of folate production with different DHPS enzymes. We predict that VRS and ARS synthesize DHP in preference to sulfa-DHP, and the ability of the double mutants to do this is due to their discrimination between PABA and sulfa drugs based on the binding affinity of DHPS. In addition, there may be other factors affecting sulfa drug resistance, such as the scavenging of nutrients (25, 35), PABA levels (9), and folic acid levels (4).
Acknowledgments
We thank Neil McKern and Tim Adams for their helpful suggestions in the preparation of the manuscript.
This work was supported by the National Institutes of Health through the University of North Carolina at Chapel Hill grant no. 1 RO1AI46966-01A1 awarded to Steven Meshnick.
The findings, opinions, and recommendations expressed here are those of the authors and not necessarily those of the University of North Carolina at Chapel Hill or the National Institutes of Health.
REFERENCES
- 1.Achari, A., D. O. Somers, J. N. Champness, P. K. Bryant, J. Rosemond, and D. K. Stammers. 1997. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat. Struct. Biol. 4:490-497. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, W., S. Meshnick, and P. Kazanjian. 2000. Pneumocystis carinii mutations associated with sulfa and sulfone prophylaxis failures in immunocompromised patients. Microbes Infect. 2:61-67. [DOI] [PubMed] [Google Scholar]
- 3.Baca, A. M., R. Sirawaraporn, S. Turley, W. Sirawaraporn, and W. G. Hol. 2000. Crystal structure of Mycobacterium tuberculosis 7,8-dihydropteroate synthase in complex with pterin monophosphate: new insight into the enzymatic mechanism and sulfa-drug action. J. Mol. Biol. 302:1193-1212. [DOI] [PubMed] [Google Scholar]
- 4.Bayly, A. M., J. M. Berglez, O. Patel, L. A. Castelli, E. G. Hankins, P. Coloe, C. Hopkins Sibley, and I. G. Macreadie. 2001. Folic acid utilisation related to sulfa drug resistance in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 204:387-390. [DOI] [PubMed] [Google Scholar]
- 5.Bayly, A. M., and I. G. Macreadie. 2002. Cytotoxicity of dihydropteroate in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 213:189-192. [DOI] [PubMed] [Google Scholar]
- 6.Beard, C. B., J. L. Carter, S. P. Keely, L. Huang, N. J. Pieniazek, I. N. Moura, J. M. Roberts, A. W. Hightower, M. S. Bens, A. R. Freeman, S. Lee, J. R. Stringer, J. S. Duchin, C. del Rio, D. Rimland, R. P. Baughman, D. A. Levy, V. J. Dietz, P. Simon, and T. R. Navin. 2000. Genetic variation in Pneumocystis carinii isolates from different geographic regions: implications for transmission. Emerg. Infect. Dis. 6:265-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks, D. R., P. Wang, M. Read, W. M. Watkins, P. F. Sims, and J. E. Hyde. 1994. Sequence variation of the hydroxymethyldihydropterin pyrophosphokinase:dihydropteroate synthase gene in lines of the human malaria parasite, Plasmodium falciparum, with differing resistance to sulfadoxine. Eur. J. Biochem. 224:397-405. [DOI] [PubMed] [Google Scholar]
- 8.Burke, D., D. Dawson, and T. Stearns. 2000. Cold Spring Harbor Laboratory course manual: methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 9.Castelli, L. A., N. P. Nguyen, and I. G. Macreadie. 2001. Sulfa drug screening in yeast: fifteen sulfa drugs compete with p-aminobenzoate in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 199:181-184. [DOI] [PubMed] [Google Scholar]
- 10.Demanche, C., J. Guillot, M. Berthelemy, T. Petitt, P. Roux, and A. E. Wakefield. 2002. Absence of mutations associated with sulfa resistance in Pneumocystis carinii dihydropteroate synthase gene from non-human primates. Med. Mycol. 40:315-318. [DOI] [PubMed] [Google Scholar]
- 11.Ferlan, J. T., S. Mookherjee, I. N. Okezie, L. Fulgence, and C. H. Sibley. 2001. Mutagenesis of dihydrofolate reductase from Plasmodium falciparum: analysis in Saccharomyces cerevisiae of triple mutant alleles resistant to pyrimethamine or WR99210. Mol. Biochem. Parasitol. 113:139-150. [DOI] [PubMed] [Google Scholar]
- 12.Fermer, C., B. E. Kristiansen, O. Skold, and G. Swedberg. 1995. Sulfonamide resistance in Neisseria meningitidis as defined by site-directed mutagenesis could have its origin in other species. J. Bacteriol. 177:4669-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gietz, R. D., and R. H. Schiestl. 1991. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast 7:253-263. [DOI] [PubMed] [Google Scholar]
- 14.Hampele, I. C., A. D'Arcy, G. E. Dale, D. Kostrewa, J. Nielsen, C. Oefner, M. G. Page, H. J. Schonfeld, D. Stuber, and R. L. Then. 1997. Structure and function of the dihydropteroate synthase from Staphylococcus aureus. J. Mol. Biol. 268:21-30. [DOI] [PubMed] [Google Scholar]
- 15.Helweg-Larsen, J., T. L. Benfield, J. Eugen-Olsen, J. D. Lundgren, and B. Lundgren. 1999. Effects of mutations in Pneumocystis carinii dihydropteroate synthase gene on outcome of AIDS-associated P. carinii pneumonia. Lancet 354:1347-1351. [DOI] [PubMed] [Google Scholar]
- 16.Iliades, P., J. Berglez, S. R. Meshnick, and I. G. Macreadie. 2003. Promoter strength of folic acid synthesis genes affects sulfa drug resistance in Saccharomyces cerevisiae. Microb. Drug Resist. 9:249-256. [DOI] [PubMed] [Google Scholar]
- 17.Ivler, D., J. M. Leedom, A. W. Mathies, Jr., J. C. Fremont, L. D. Thrupp, B. Portnoy, and P. F. Wehrle. 1965. Correlates of sulfadiazine resistance in meningococci isolated from civilians. Antimicrob. Agents Chemother. 5:358-365. [PubMed] [Google Scholar]
- 18.Kazanjian, P., W. Armstrong, P. A. Hossler, W. Burman, J. Richardson, C. H. Lee, L. Crane, J. Katz, and S. R. Meshnick. 2000. Pneumocystis carinii mutations are associated with duration of sulfa or sulfone prophylaxis exposure in AIDS patients. J. Infect. Dis. 182:551-557. [DOI] [PubMed] [Google Scholar]
- 19.Kazanjian, P., A. B. Locke, P. A. Hossler, B. R. Lane, M. S. Bartlett, J. W. Smith, M. Cannon, and S. R. Meshnick. 1998. Pneumocystis carinii mutations associated with sulfa and sulfone prophylaxis failures in AIDS patients. AIDS 12:873-878. [DOI] [PubMed] [Google Scholar]
- 20.Kun, J. F., L. G. Lehman, B. Lell, R. Schmidt-Ott, and P. G. Kremsner. 1999. Low-dose treatment with sulfadoxine-pyrimethamine combinations selects for drug-resistant Plasmodium falciparum strains. Antimicrob. Agents Chemother. 43:2205-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landy, M., N. W. Larkum, E. J. Oswald, and F. Steightoff. 1943. Increased synthesis of p-aminobenzoic acid associated with the development of sulfonamide resistance in Staphylococcus aureus. Science 97:265-267. [DOI] [PubMed] [Google Scholar]
- 22.Lane, B. R., J. C. Ast, P. A. Hossler, D. P. Mindell, M. S. Bartlett, J. W. Smith, and S. R. Meshnick. 1997. Dihydropteroate synthase polymorphisms in Pneumocystis carinii. J. Infect. Dis. 175:482-485. [DOI] [PubMed] [Google Scholar]
- 23.Ma, L., and J. A. Kovacs. 2001. Genetic analysis of multiple loci suggests that mutations in the Pneumocystis carinii f. sp. hominis dihydropteroate synthase gene arose independently in multiple strains. Antimicrob. Agents Chemother. 45:3213-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma, L., J. A. Kovacs, A. Cargnel, A. Valerio, G. Fantoni, and C. Atzori. 2002. Mutations in the dihydropteroate synthase gene of human-derived Pneumocystis carinii isolates from Italy are infrequent but correlate with prior sulfa prophylaxis. J. Infect. Dis. 185:1530-1532. [DOI] [PubMed] [Google Scholar]
- 25.Mberu, E. K., A. M. Nzila, E. Nduati, A. Ross, S. M. Monks, G. O. Kokwaro, W. M. Watkins, and C. Hopkins Sibley. 2002. Plasmodium falciparum: in vitro activity of sulfadoxine and dapsone in field isolates from Kenya: point mutations in dihydropteroate synthase may not be the only determinants in sulfa resistance. Exp. Parasitol. 101:90-96. [DOI] [PubMed] [Google Scholar]
- 26.Mei, Q., S. Gurunathan, H. Masur, and J. A. Kovacs. 1998. Failure of co-trimoxazole in Pneumocystis carinii infection and mutations in dihydropteroate synthase gene. Lancet 351:1631-1632. [DOI] [PubMed] [Google Scholar]
- 27.Milhous, W., N. F. Weatherly, J. H. Bowdre, and R. E. Desjardins. 1985. In vitro activities of and mechanisms of resistance to antifol antimalarial drugs. Antimicrob. Agents Chemother. 27:525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navin, T. R., C. B. Beard, L. Huang, C. del Rio, S. Lee, N. J. Pieniazek, J. L. Carter, T. Le, A. Hightower, and D. Rimland. 2001. Effect of mutations in Pneumocystis carinii dihydropteroate synthase gene on outcome of P. carinii pneumonia in patients with HIV-1: a prospective study. Lancet 358:545-549. [DOI] [PubMed] [Google Scholar]
- 29.Nopponpunth, V., W. Sirawaraporn, P. J. Greene, and D. V. Santi. 1999. Cloning and expression of Mycobacterium tuberculosis and Mycobacterium leprae dihydropteroate synthase in Escherichia coli. J. Bacteriol. 181:6814-6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel, O., J. Satchell, J. Baell, R. Fernley, P. Coloe, and I. G. Macreadie. 2003. Inhibition studies of sulfonamide-containing folate analogs in yeast. Microb. Drug Resist. 9:139-146. [DOI] [PubMed] [Google Scholar]
- 31.Sibley, C. H., V. H. Brophy, S. Cheesman, K. L. Hamilton, E. G. Hankins, J. M. Wooden, and B. Kilbey. 1997. Yeast as a model system to study drugs effective against apicomplexan proteins. Methods 13:190-207. [DOI] [PubMed] [Google Scholar]
- 32.Swedberg, G., S. Ringertz, and O. Skold. 1998. Sulfonamide resistance in Streptococcus pyogenes is associated with differences in the amino acid sequence of its chromosomal dihydropteroate synthase. Antimicrob. Agents Chemother. 42:1062-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi, T., N. Hosoya, T. Endo, T. Nakamura, H. Sakashita, K. Kimura, K. Ohnishi, Y. Nakamura, and A. Iwamoto. 2000. Relationship between mutations in dihydropteroate synthase of Pneumocystis carinii f. sp. hominis isolates in Japan and resistance to sulfonamide therapy. J. Clin. Microbiol. 38:3161-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visconti, E., E. Ortona, P. Mencarini, P. Margutti, S. Marinaci, M. Zolfo, A. Siracusano, and E. Tamburrini. 2001. Mutations in dihydropteroate synthase gene of Pneumocystis carinii in HIV patients with Pneumocystis carinii pneumonia. Int. J. Antimicrob. Agents 18:547-551. [DOI] [PubMed] [Google Scholar]
- 35.Wang, P., R. K. Brobey, T. Horii, P. F. Sims, and J. E. Hyde. 1999. Utilization of exogenous folate in the human malaria parasite Plasmodium falciparum and its critical role in antifolate drug synergy. Mol. Microbiol. 32:1254-1262. [DOI] [PubMed] [Google Scholar]
- 36.Warhurst, D. C. 2002. Resistance to antifolates in Plasmodium falciparum, the causative agent of tropical malaria. Sci. Prog. 85:89-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White, P. J., and D. D. Woods. 1965. The synthesis of p-aminobenzoic acid and folic acid by Staphylococci sensitive and resistant to sulphonamides. J. Gen. Microbiol. 40:243-253. [DOI] [PubMed] [Google Scholar]
- 38.Williams, D. L., L. Spring, E. Harris, P. Roche, and T. P. Gillis. 2000. Dihydropteroate synthase of Mycobacterium leprae and dapsone resistance. Antimicrob. Agents Chemother. 44:1530-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wongsrichanalai, C., A. L. Pickard, W. H. Wernsdorfer, and S. R. Meshnick. 2002. Epidemiology of drug-resistant malaria. Lancet Infect. Dis. 2:209-218. [DOI] [PubMed] [Google Scholar]
- 40.Wooden, J. M., L. H. Hartwell, B. Vasquez, and C. H. Sibley. 1997. Analysis in yeast of antimalaria drugs that target the dihydrofolate reductase of Plasmodium falciparum. Mol. Biochem. Parasitol. 85:25-40. [DOI] [PubMed] [Google Scholar]