Abstract
Human bestrophin 1 (hBest1) is a calcium-activated chloride channel from the retinal pigment epithelium, where it can suffer mutations associated with vitelliform macular degeneration, or Best disease. We describe the structure of a bacterial homolog (KpBest) of hBest1 and functional characterizations of both channels. KpBest is a pentamer that forms a five-helix transmembrane pore, closed by three rings of conserved hydrophobic residues, and has a cytoplasmic cavern with a restricted exit. From electrophysiological analysis of structure-inspired mutations in KpBest and hBest1, we find a subtle control of ion selectivity in the bestrophins, including reversal of anion/cation selectivity, and dramatic activation by mutations at the exit restriction. A homology model of hBest1 shows the locations of disease-causing mutations and suggests possible roles in regulation.
Keywords: Calcium-activated chloride channel, Crystal structure, Macular degeneration, Single-wavelength anomalous diffraction (SAD), Sodium channel
The human BEST1 gene encodes a protein (bestrophin-1, hBest1) that is highly expressed in retinal pigment epithelium (1-4). Over 120 distinct mutations in hBest1 have been identified that result in multiple retinal degeneration disorders (5-11), notably vitelliform macular dystrophy or Best disease. Functionally, hBest1 was identified as a Cl− channel that can be activated by Ca2+ (8, 12, 13), and most of the disease-causing mutations in hBest1 are point mutations that cause channel dysfunction (8, 12, 14-16). Thus, understanding the structure of hBest1 channel holds tremendous value from both biological and biomedical perspectives.
The bestrophin family identified by hBest1 is distributed widely, with representatives in most metazoan animals, including four in humans, and also in other eukaryotes and in prokaryotes (7, 8). The animal bestrophins are characterized by a highly conserved N-terminal domain that includes four predicted transmembrane helices (TMs) and very diverse C-terminal domains that may be involved in protein-protein interactions (8, 12, 15, 17). Bacterial bestrophins lack the variable C-terminal domain, and they are more divergent in the transmembrane portion. Using a structural genomics approach, we identified a homolog from Klebsiella pneumoniae (KpBest) that could be produced by recombinant expression for structural and functional characterization. In our ultimate structure-based sequence alignment, KpBest shares 14% identity to hBest1 (Fig. S1).
Initial crystals of detergent-solubilized KpBest diffracted poorly; however, constructs from a truncation series did yield suitable crystals. The initial structure was solved from one of these, grown from a solution containing zinc acetate, was solved at 2.9Å resolution by single-wavelength anomalous diffraction (SAD) at the zinc K-edge resonance. Improved diffraction was obtained after further truncation, removing a total of 11 residues from the C-terminus, and the structure was further refined at 2.3Å resolution (Tables S1 and S2). The building and refinement of the structural model were facilitated by the presence of five-fold non-crystallographic symmetry. The refined model comprises ordered residues from 22, 23 or 24 through 285 or 289 in different protomers plus Zn2+ ions and water molecules. Residues 264-271 are also disordered in protomer E.
Bestrophins have been predicted by different groups to form dimers, tetramers, or pentamers (12, 18). Here, we found that KpBest forms a stable pentamer (Figs. 1A, 1B, and S2) with large inter-subunit contacts (23,473 Å2 total buried surface area). The electrostatic potential surface is largely negative on the extracellular surface, neutral in the transmembrane region, and positive at the cytoplasmic membrane surface (Figs.1C and 1D). Consistent with the experimentally determined topology of hBest1 (19), each protomer has four transmembrane helices and the N- and C-termini both reside on the cytoplasmic side (Figs. 1E and 1F). Extracellular inter-helix loops TM1-TM2 (12 residues) and TM3-TM4 (3 residues) are short, whereas the intracellular connection between TM2 and TM3 is long (105 residues) and forms a separate cytoplasmic domain of five helices from this connection (Fig. 1E, α3-α7) plus C-terminal helix α10 (red in Fig. 1F). The TM2 helices line the putative ion-conducting pore through the membrane, and they continue as long and curved, but uninterrupted helices α2 (light blue in Fig. 1F). In contrast, TM3/α8 and TM4/α9 are connected to the cytoplasmic domain by extended segments (4 and 12 residues, respectively). The α9-α10 connection is a loop structure that includes a conserved carboxylate segment (EDDDDFE) in eukaryotes. The α7 helices (light yellow in Fig. 1F) and cytoplasmic portions of α2 line a cytoplasmic cavern inside the pore.
Fig. 1. Crystal structure of KpBest.
(A & B) Ribbon diagram of the KpBest pentamer with each protomer colored differently, (A) as viewed from outside the membrane and (B) as viewed from the side (rotated 90° through × axis). (C & D) Electrostatic potential at the molecular surface viewed as in A & B, respectively). The contour level is at ±5 kT/e red for negative potential and blue for positive potential. Membrane boundaries in B & D were calculated by OPM server. (E) 2D topology of a protomer, colored spectrally from dark blue at its N-terminal segment to red at its C-terminal segment. (F) Ribbon diagram of a protomer. Colored as in E.
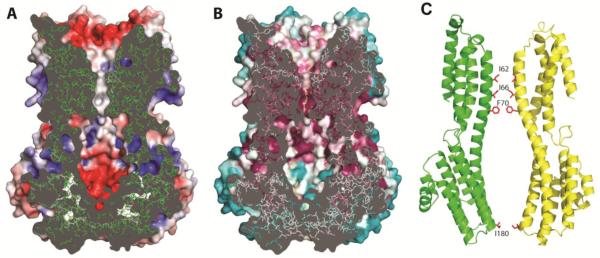
An apparent ion conduction pathway is at the center of the KpBest pentamer. A funnel-shaped electronegative vestibule, penetrating midway into the membrane, precedes a hydrophobic five-helix transmembrane pore (Fig. 2A). The pore is followed by a cytoplasmic cavern and a central exit restriction. The pore and upper parts of the cavern are highly conserved, whereas outer surfaces and lower parts of the cytoplasmic domain are not (Fig. 2B). Overall, the ion permeation pathway has a flower-vase shape, with one restriction (radius < 2.0 Å) from three rings of TM2 residues (I62, I66 and F70) at the pore and another (I180, radius = 1.1 Å) at the start of cytoplasmic helix α7 (Fig. 2C). Therefore, the structure of KpBest predicts two distinct permeation restrictions in the ion passageway, providing a vital clue for the functional mechanism of bestrophin channels. Notably, all four residues located at the predicted restrictions are highly conserved and/or disease related in hBest1: I76, F84 and I205 (cf. KpBest I62, F70, and I180, respectively) are identical (Fig. S1), while point mutation of either F80 or I205 (cf. KpBest I66 and I180, respectively) causes retinal disorders (6, 20, 21).
Fig. 2. Structure of the ion conducting pathway through KpBest.
(A) Cross-section through the pore center. The model is viewed as in 1D, with the electrostatic potential shown on exposed surfaces of the molecular envelope. (B) Cross-section as in A, but colored by Consurf sequence conservation. The calculation used 150 prokaryotic homologs with 95% maximal and 35% minimal sequence identities compared to KpBest. . (C) Ribbon diagram of two oppositely facing (144°) protomers of a KpBest pentamer are shown with the extracellular side on the top. The side chains of critical residues are red.
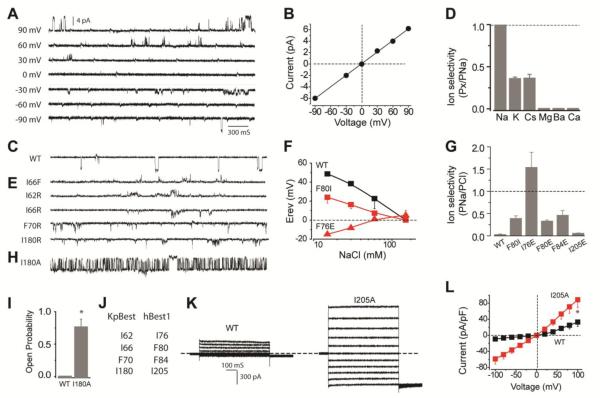
Although eukaryotic bestrophins are known as Ca2+-activated Cl− channels, the function of KpBest had not been previously examined. To test its function, purified KpBest was fused into planar lipid bilayer with 150 mM NaCl in both the trans (internal) and cis (external) solutions. Applying a range of transmembrane potentials resulted in well-resolved unitary currents with a linear single-channel I-V relationship (Figs. 3A and 3B), confirming that KpBest is indeed an ion channel. Ca2+ was not required for KpBest activation, as might have been expected given that KpBest lacks the C-terminal domain that contains putative Ca2+ binding sites in eukaryotic bestrophins (7, 13, 22). Strikingly, with 150 mM NaCl on the cis side and no NaCl on the trans side, inward single channel currents were recorded (Fig. 3C), demonstrating that KpBest is a cation channel that conducts Na+, unlike Cl−-conducting eukaryotic bestrophins. To fully assess KpBest ion selectivity, reversal potentials under various bionic conditions were measured. KpBest is permeable to monovalent cations with rank order Na+ > K+ ≈ Cs+, but not to bivalent cations Mg2+, Ca2+ or Ba2+ (Fig. 3D). It is noteworthy that ion channels in the same family can have reversed charge selectivity, as exemplified by TMEM16 Ca2+-activated channels: TMEM16A and 16B are anion channels, while TMEM16F conducts cations (23).
Fig. 3. Ionic conductance measurements of KpBest and hBest1.
(A) Representative families of single KpBest currents recorded from planar lipid bilayers at different voltages (150 mM NaCl in both trans and cis solutions). (B) Single KpBest channel current-voltage relationship. (C) Current trace of wild-type single KpBest channels (150 mM NaCl in cis and 0 NaCl in trans solutions). (D) Relative cation permeability; n = 3 for each point. (E) Current traces of mutant KpBest channels (same condition as C). (F) Changes in wild-type and mutant hBest1 channel reversal potentials (Erev) as a function of extracellular NaCl concentration (150 mM NaCl in internal); n=3−4 for each point. (G) Relative permeability (PNa/PCl) of hBest1 wild-type and mutant variants; n=12−15 for each bar. (H) Current trace of I180A single KpBest channels (same condition as C). (I) Wild-type and I180A single KpBest channel open probabilities. (J) Critical residues in KpBest and hBest1. (K) Exemplar whole-cell currents of wild type (left) and I205A (right) hBest1 in HEK 293 cells. (L) Population steady-state current-voltage relationships; n= 3−6 for each point. * P<0.05 when compared to wild-type using two-tailed unpaired Student's t test.
Despite extensive studies on the ion conducting pores of eukaryotic bestrophins, including hBest1 and mouse bestrophin-2 (mBest2) (15, 24-26), the molecular basis for ion selection in these channels is not known. Our KpBest model predicts three critical residues (I62, I66 and F70) at the first permeation restriction (Fig. 2C) that likely control ion selectivity. To test this hypothesis, we first examined I66, because it is the only residue among the three that is different in KpBest compared to anion-conducting bestrophin channels (where this residue is F) (Fig. 3J and S1) (12, 15, 24-27). In bilayer experiments, KpBest I66F showed outward current with 150 mM NaCl on the cis side and no NaCl on the trans side, indicating that this mutant channel conducts Cl- rather than Na+ (Fig. 3E, top). Remarkably, a modest single amino-acid substitution at the first predicted permeation restriction flips the cation/anion selectivity of KpBest, suggesting a key role in ion selectivity. To further test this premise in the eukaryotic counterpart, WT and F80I mutant hBest1 (corresponding to KpBest I66) were transfected into HEK 293 cells, and their reversal potentials were determined in whole-cell voltage clamp experiments. Consistent with the impact of the I66F substitution on switching ion selectivity of KpBest1 towards Cl−, hBest1 F80I was much less permeable to Cl− compared to WT (PNa/PCl = 0.39 for F80I, compared to PNa/PCl = 0.03 for the WT, Figs. 3F and 3G).
As all three critical residues in the first permeation restriction are hydrophobic, we next examined whether KpBest ion selectivity could be altered by individually substituting those residues with positively charged arginine (R), which in principle might favor negatively charged Cl-. When purified KpBest I62R, I66R and F70R mutants were tested in bilayer experiments, I62R (but not I66R or F70R) conducted Cl− rather than Na+ (Fig. 3E). Following the same logic, the equivalent residues on hBest1 (I76, F80 and F84, respectively) were individually mutated to negatively charged glutamic acid (E) to test whether the ion selectivity of hBest1 could be altered towards Na+. Consistent with the KpBest results, only I76E flipped the ion selectivity to Na+ (PNa/PCl = 1.54, Figs. 3F and 3G), although F80E and F84E were also less permeable to Cl− than WT (PNa/PCl = 0.33 and 0.46, respectively, Fig. 3G). None of the mutations significantly affected current amplitude (Fig. S3). Notably, our results are in accord with previous reports: the rectification of mBest2 can be altered in opposite directions by replacing F80 (equivalent to KpBest I66) with either R or E (26), and the corresponding F81E mutation of Drosophila melanogaster bestrophin-1 (dBest1) flips the cation/anion selectivity (28). Taken together, using our KpBest model as a guide, we have identified three residues that control the ion selectivity in bestrophins.
The dramatic change in ion selectivity from substitutions at the first hydropobic residue of the pore (I62R in KpBest and I76E in hBest1) suggesting a particularly critical role for this position. Interestingly, the expression level of I62R in E. coli was much lower compared to that of WT KpBest (about 1/14, Fig. S4), suggesting that I62 is important for the expression/assembly of the channel. Consistent with this idea, mutating the equivalent mBest2 I76 to C/L/V resulted in no currents (26). More amazingly, the simple swapping of the non-identical central pore residues (KpBest I66 and hBest1 F80) significantly alters ion selectivity without affecting expression levels. The hydrophobic character of the five-helix transmembrane pore of bestrophins in reminiscent of that in the SLAC1 channel (29) where the anion selectivity series, like that observed for mBest2 (24, 25) and also for TMEM16A (30, 31), is inversely related to the hydration energy of monovalent anions (32).
The second restriction in the permeation pathway of KpBest, at the base of the cytoplasmic cavity where I180 residues interact (Fig. 2C), suggests a possible permeation gate. As I180 produces the narrowest bottleneck in the ion permeation pathway, we reasoned that it may control the overall permeability of the channel, thereby acting as an activation gate. To test this idea, we replaced the bulky isoleucine alanine and tested the purified I180A mutant channels in bilayer experiments. Indeed, I180A channels conducted unitary currents with the same amplitude (6 pA) as WT, but with a markedly enhanced open probability (Fig. 3H and 3I). Substituting I180 on KpBest with R which has a longer side-chain than A yielded mutant channels that still conduct Na+ (Fig. 3E) with a low open probability similar to WT KpBest. These results indicate I180 residues act as an activation gate. To test whether this scenario also applied to hBest1, the comparable I205A/E mutants were generated and subjected to whole-cell voltage clamp. Phenocopying their respective KpBest counterparts, hBest I205A displayed significantly larger currents compared to WT, and lost inward rectification indicating a change in channel gating (Figs. 3K and 3L), while I205E barely affected channel ion selectivity (PNa/PCl = 0.06, Fig. 3G). Overall, these results demonstrate that the gate predicted by our KpBest model (KpBest I180 and hBest1 I205) controls permeability/gating, but not selectivity, of the channels. Notably, hBest1 I205T is a disease causing mutation with significantly decreased Cl− conductance (6), reinforcing the functional significance of I205.
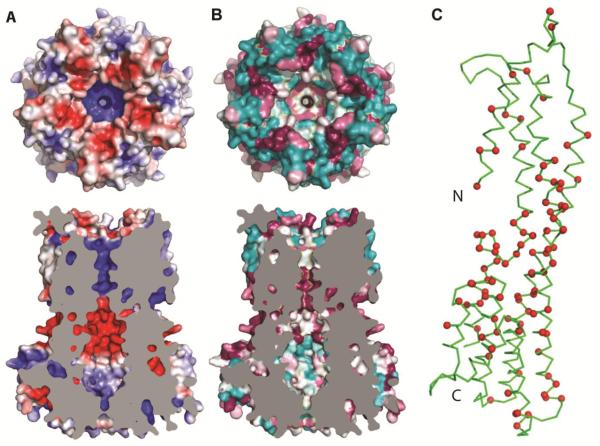
To understand characteristics of hBest1, including the disposition of disease-causing mutations, we generated a homology model for its transmembrane portion based on the KpBest structure (Fig.4). Seemingly consistent with the anion selectivity, the transmembrane pore and the lower restriction of hBest1 are both positively charged (Fig. 4A), contrasting with the negative interior of the KpBest permeation pathway. The conservation pattern among eukaryotic bestrophins (Fig. 4B) is roughly similar to that of prokaryotic relatives (Fig. 3B), although the cytoplasmic exterior is less variable. Disease-causing mutations in hBest1 are abundant (http://www-huge.uni-regensburg.de/BEST1_database/home.php?select_db=BEST1) and have been clustered into hot spots in the sequence (Fig. S1) (8, 33). When plotted onto the three-dimensional model (Figs. 4C and S5), the hot spots segregate into a cavity-lining set from hot spot 2 along α2 and a juxtamembrane set from hot spots 3 and 4 at the top of α7 and at the α9-α10 carboxylate loop, which is more prominent electrostatically in hBest1 (Fig. S5A) than in KpBest (Fig. 1D). The carboxylate loop could conceivably play a role in regulation by Ca2+. Hot spot 1 is along the N-terminal segment that is disordered in KpBest, but presumably it too would be alongside other hot spots in the juxtamembrane region in hBest1.
Fig. 4. Homology model of hBest1.
(A) Top, electrostatic potential at the extracellular surface of the hBest1 homology model. Viewed and drawn as for 1C. Bottom, cross-section through the homology model of hBest1. Viewed and drawn as for 2A, except that cut surface is plain grey. (B) Top, top view as in A, but colored by surface conservation. This calculation used 150 homologs with 95% maximal and 35% minimal sequence identities compared to hBest1. Bottom, cross-section viewed and drawn as in 2B but having a plain grey cut surface. (C) Ribbon diagram of a protomer from the hBest1 homology pentamer. The Cα positions of residues that are sites of disease-causing point mutations are marked in red.
Supplementary Material
Acknowledgments
We thank Youzhong Gao and Oliver Clarke for help in protein production and crystallography, Wenjun Xie for help in planar lipid experiments, and John Schwanof and Randy Abramowitz at National Synchrotron Light Source (NSLS) beamlines X4A and X4C and Frank Murphy at the Advanced Photon Source beamline 24-ID-E for their assistance in data collection. This work was supported in part by NIH grant GM095315 and GM107462 to W.A.H. X4 beamlines are supported by the New York Structural Biology Center at the National Synchrotron Light Source (NSLS) of Brookhaven National Laboratory, a DOE facility. The data reported in this paper are tabulated in the Supplementary Material and deposited to the Protein Data Bank with access codes listed in Table S1.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online.
References
- 1.Marmorstein AD, Marmorstein LY, Rayborn M, Wang X, Hollyfield JG, Petrukhin K. Bestrophin, the product of the Best vitelliform macular dystrophy gene (VMD2), localizes to the basolateral plasma membrane of the retinal pigment epithelium. Proc Natl Acad Sci U S A. 2000;97:12758–12763. doi: 10.1073/pnas.220402097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marquardt A, Stohr H, Passmore LA, Kramer F, Rivera A, Weber BH. Mutations in a novel gene, VMD2, encoding a protein of unknown properties cause juvenile-onset vitelliform macular dystrophy (Best's disease) Hum Mol Genet. 1998;7:1517–1525. doi: 10.1093/hmg/7.9.1517. [DOI] [PubMed] [Google Scholar]
- 3.Petrukhin K, J. Koisti M, Bakall B, Li W, Xie G, Marknell T, Sandgren O, Forsman K, Holmgren G, Andreasson S, Vujic M, Bergen AA, McGarty-Dugan V, Figueroa D, Austin CP, Metzker ML, Caskey CT, Wadelius C. Identification of the gene responsible for Best macular dystrophy. Nat Genet. 1998;19:241–247. doi: 10.1038/915. [DOI] [PubMed] [Google Scholar]
- 4.Neussert R, Muller C, Milenkovic VM, Strauss O. The presence of bestrophin-1 modulates the Ca2+ recruitment from Ca2+ stores in the ER. Pflugers Archiv : European journal of physiology. 2010;460:163–175. doi: 10.1007/s00424-010-0840-2. [DOI] [PubMed] [Google Scholar]
- 5.Boon CJ, Klevering BJ, Leroy BP, Hoyng CB, Keunen JE, den Hollander AI. The spectrum of ocular phenotypes caused by mutations in the BEST1 gene. Progress in retinal and eye research. 2009;28:187–205. doi: 10.1016/j.preteyeres.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Davidson AE, Millar ID, Urquhart JE, Burgess-Mullan R, Shweikh Y, Parry N, O'Sullivan J, Maher GJ, McKibbin M, Downes SM, Lotery AJ, Jacobson SG, Brown PD, Black GC, Manson FD. Missense mutations in a retinal pigment epithelium protein, bestrophin-1, cause retinitis pigmentosa. American journal of human genetics. 2009;85:581–592. doi: 10.1016/j.ajhg.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao Q, Hartzell HC, Yu K. Bestrophins and retinopathies. Pflugers Archiv : European journal of physiology. 2010;460:559–569. doi: 10.1007/s00424-010-0821-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartzell HC, Qu Z, Yu K, Xiao Q, Chien LT. Molecular physiology of bestrophins: multifunctional membrane proteins linked to best disease and other retinopathies. Physiol Rev. 2008;88:639–672. doi: 10.1152/physrev.00022.2007. [DOI] [PubMed] [Google Scholar]
- 9.Kramer F, Mohr N, Kellner U, Rudolph G, Weber BH. Ten novel mutations in VMD2 associated with Best macular dystrophy (BMD) Human mutation. 2003;22:418. doi: 10.1002/humu.9189. [DOI] [PubMed] [Google Scholar]
- 10.Kramer F, White K, Pauleikhoff D, Gehrig A, Passmore L, Rivera A, Rudolph G, Kellner U, Andrassi M, Lorenz B, Rohrschneider K, Blankenagel A, Jurklies B, Schilling H, Schutt F, Holz FG, Weber BH. Mutations in the VMD2 gene are associated with juvenile-onset vitelliform macular dystrophy (Best disease) and adult vitelliform macular dystrophy but not age-related macular degeneration. European journal of human genetics : EJHG. 2000;8:286–292. doi: 10.1038/sj.ejhg.5200447. [DOI] [PubMed] [Google Scholar]
- 11.Allikmets R, M. Seddon J, Bernstein PS, Hutchinson A, Atkinson A, Sharma S, Gerrard B, Li W, Metzker ML, Wadelius C, Caskey CT, Dean M, Petrukhin K. Evaluation of the Best disease gene in patients with age-related macular degeneration and other maculopathies. Human genetics. 1999;104:449–453. doi: 10.1007/s004390050986. [DOI] [PubMed] [Google Scholar]
- 12.Sun H, Tsunenari T, Yau KW, Nathans J. The vitelliform macular dystrophy protein defines a new family of chloride channels. Proc Natl Acad Sci U S A. 2002;99:4008–4013. doi: 10.1073/pnas.052692999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao Q, Prussia A, Yu K, Cui YY, Hartzell HC. Regulation of bestrophin Cl channels by calcium: role of the C terminus. The Journal of general physiology. 2008;132:681–692. doi: 10.1085/jgp.200810056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu K, Qu Z, Cui Y, Hartzell HC. Chloride channel activity of bestrophin mutants associated with mild or late-onset macular degeneration. Invest Ophthalmol Vis Sci. 2007;48:4694–4705. doi: 10.1167/iovs.07-0301. [DOI] [PubMed] [Google Scholar]
- 15.Tsunenari T, Sun H, Williams J, Cahill H, Smallwood P, Yau KW, Nathans J. Structure-function analysis of the bestrophin family of anion channels. The Journal of biological chemistry. 2003;278:41114–41125. doi: 10.1074/jbc.M306150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu K, Cui Y, Hartzell HC. The bestrophin mutation A243V, linked to adult-onset vitelliform macular dystrophy, impairs its chloride channel function. Invest Ophthalmol Vis Sci. 2006;47:4956–4961. doi: 10.1167/iovs.06-0524. [DOI] [PubMed] [Google Scholar]
- 17.Marmorstein AD, Cross HE, Peachey NS. Functional roles of bestrophins in ocular epithelia. Progress in retinal and eye research. 2009;28:206–226. doi: 10.1016/j.preteyeres.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanton JB, Goldberg AF, Hoppe G, Marmorstein LY, Marmorstein AD. Hydrodynamic properties of porcine bestrophin-1 in Triton X-100. Biochim Biophys Acta. 2006;1758:241–247. doi: 10.1016/j.bbamem.2006.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milenkovic VM, Rivera A, Horling F, Weber BH. Insertion and topology of normal and mutant bestrophin-1 in the endoplasmic reticulum membrane. The Journal of biological chemistry. 2007;282:1313–1321. doi: 10.1074/jbc.M607383200. [DOI] [PubMed] [Google Scholar]
- 20.Lotery AJ, Munier FL, Fishman GA, Weleber RG, Jacobson SG, Affatigato LM, Nichols BE, Schorderet DF, Sheffield VC, Stone EM. Allelic variation in the VMD2 gene in best disease and age-related macular degeneration. Invest Ophthalmol Vis Sci. 2000;41:1291–1296. [PubMed] [Google Scholar]
- 21.Maia-Lopes S, Castelo-Branco M, Silva E, Aguirre J, Riveiro-Alvarez R, Trujillo-Tiebas MJ, Ayuso C. Gene symbol: BEST1. Disease: Best macular dystrophy. Human genetics. 2008;123:112. [PubMed] [Google Scholar]
- 22.Kranjc A, Grillo FW, Rievaj J, Boccaccio A, Pietrucci F, Menini A, Carloni P, Anselmi C. Regulation of bestrophins by Ca2+: a theoretical and experimental study. PloS one. 2009;4:e4672. doi: 10.1371/journal.pone.0004672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang H, Kim A, David T, Palmer D, Jin T, Tien J, Huang F, Cheng T, Coughlin SR, Jan YN, Jan LY. TMEM16F forms a Ca2+-activated cation channel required for lipid scrambling in platelets during blood coagulation. Cell. 2012;151:111–122. doi: 10.1016/j.cell.2012.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qu Z, Fischmeister R, Hartzell C. Mouse bestrophin-2 is a bona fide Cl(−) channel: identification of a residue important in anion binding and conduction. The Journal of general physiology. 2004;123:327–340. doi: 10.1085/jgp.200409031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu Z, Hartzell C. Determinants of anion permeation in the second transmembrane domain of the mouse bestrophin-2 chloride channel. The Journal of general physiology. 2004;124:371–382. doi: 10.1085/jgp.200409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu Z, T. Chien L, Cui Y, Hartzell HC. The anion-selective pore of the bestrophins, a family of chloride channels associated with retinal degeneration. J Neurosci. 2006;26:5411–5419. doi: 10.1523/JNEUROSCI.5500-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsunenari T, Nathans J, Yau KW. Ca2+-activated Cl− current from human bestrophin-4 in excised membrane patches. The Journal of general physiology. 2006;127:749–754. doi: 10.1085/jgp.200609527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chien LT, Hartzell HC. Rescue of volume-regulated anion current by bestrophin mutants with altered charge selectivity. The Journal of general physiology. 2008;132:537–546. doi: 10.1085/jgp.200810065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen YH, Hu L, Punta M, Bruni R, Hillerich B, Kloss B, Rost B, Love J, Siegelbaum SA, Hendrickson WA. Homologue structure of the SLAC1 anion channel for closing stomata in leaves. Nature. 2010;467:1074–1080. doi: 10.1038/nature09487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schroeder BC, Cheng T, Jan YN, Jan LY. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008;134:1019–1029. doi: 10.1016/j.cell.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang YD, Cho H, Koo JY, Tak MH, Cho Y, Shim WS, Park SP, Lee J, Lee B, Kim BM, Raouf R, Shin YK, Oh U. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 2008;455:1210–1215. doi: 10.1038/nature07313. [DOI] [PubMed] [Google Scholar]
- 32.Wright EM, Diamond JM. Anion selectivity in biological systems. Physiol Rev. 1977;57:109–156. doi: 10.1152/physrev.1977.57.1.109. [DOI] [PubMed] [Google Scholar]
- 33.Marmorstein AD, Kinnick TR. Focus on molecules: bestrophin (best-1) Experimental eye research. 2007;85:423–424. doi: 10.1016/j.exer.2006.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.