Abstract
Background
Plasma lipid levels as well as coronary artery disease (CAD) have been shown to be highly heritable with estimates ranging from 40%–60%. However, top variants detected by large-scale genome-wide association studies (GWAS) explain only a fraction of the total variance in plasma lipid phenotypes and CAD.
Methods and Results
We performed a conditional and joint association analysis using summary-level statistics from two large GWAS meta-analyses: (1) the Global Lipids Genetics Consortium (GLGC) study, and (2) the Coronary ARtery DIsease Genome-wide Replication and Meta-analysis (CARDIoGRAM) study. There were 100,184 individuals from 46 GLGC studies for plasma lipids, and 22,233 cases and 64,762 controls from 14 studies for CAD. We detected a number of loci where multiple independent SNPs were associated with lipid traits within a locus (12 out of 33 loci for high-density lipoprotein cholesterol [HDL-C], 10 of 35 loci for low-density lipoprotein cholesterol [LDL-C], 13 of 44 loci for total cholesterol [TC], and 8 of 28 loci for triglycerides [TG]), reaching genome-wide significance (P<5×10−8), nearly doubling the heritability explained by GWAS (3.6% to 7.6% for HDL-C, 5.0% to 8.8% for LDL-C, 5.5% to 8.8% for TC, and 5.7% to 8.5% for TG). Multiple SNPs were also associated with CAD (3 of 15 loci, 9.6% to 11.4% of increased heritability).
Conclusion
These results demonstrate that a portion of the missing heritability for lipid traits and CAD can be explained by multiple variants at each locus.
Keywords: Genome-wide association study, Lipids, Genetics, Missing heritability, Coronary artery disease
Introduction
Plasma lipids and lipoproteins are heritable risk factors for coronary artery disease (CAD),1 with heritability estimates ranging from 40%–60% for total cholesterol (TC), LDL-cholesterol (LDL-C), HDL-cholesterol (HDL-C), and triglycerides (TG) and 30%–60% for CAD.2-5 Genome-wide association studies (GWAS) for plasma lipids and CAD have successfully identified at least 95 gene regions for plasma lipids and 46 for CAD.1,6 Despite the success of GWAS, single nucleotide polymorphisms (SNPs) at these loci explain only a modest proportion of the heritability - 20% to 25% of the heritability for plasma lipids, and less than 10% of the heritability of CAD.1 These observations have led to the general question of how to account for the unexplained heritability.
Typically GWAS uses a single-locus test, in which each variant is tested individually for association with a specific phenotype and the best SNP at each locus is reported. A single SNP may not capture the overall amount of variation at a locus because there may be multiple causal variants at a locus. If individual level genotype data are available, we can detect additional SNPs using conditional analysis; however, conditional analysis is often infeasible when many studies have contributed results to a large-scale meta-analysis.
Recently, Yang et al. developed a conditional and joint association analysis tool that leverages summary-level statistics and estimated linkage disequilibrium (LD) from a reference sample with individual-level genotype data.7 In the current study, we applied this approach to lipid traits studied by the Global Lipids Genetics Consortium (GLGC) as well as to the dichotomous trait of CAD studied by the Coronary ARtery DIsease Genome-wide Replication and Meta-analysis (CARDIoGRAM) Consortium.5,6
Methods
GWAS Summary-level Statistics for Lipids and Coronary Artery Disease
We obtained the summary-level statistics (rsID, effect allele, the other allele, frequency of the effect allele, effect size, standard error, P value and sample size) from the GLGC meta-analysis study for lipids6 and from the CARDIoGRAM meta-analysis study for CAD.5 These studies included up to 100,184 individuals from 46 studies for lipids, and 22,233 cases and 64,762 controls for CAD, respectively.
Reference Samples for Linkage Disequilibrium
We used the European ancestry individual-level genotype of 9,796 individuals and phenotype data of the Atherosclerosis Risk in Communities Study (ARIC) cohort8 as a reference sample. ARIC represents a large population-based cohort and this cohort contributed to the GLGC and CARDIoGRAM meta-analyses. SNP quality control was performed, excluding SNPs with missingness >2%, minor allele frequency (MAF) <0.01 or Hardy-Weinberg equilibrium (HWE) P value <1×10−6. Among a total of 805,437 genotyped SNPs, 617,428 SNPs were retained in the ARIC data. We discarded samples with missingness >3% and one of each pair of samples with an estimated genetic relatedness >0.25. A total of 8,682 individuals of European ancestry in the ARIC cohort were included for LD calculation. The SNP data for ARIC were phased by MaCH and imputed into the HapMap Phase 2 CEU panel by minimac, the same panel that was used for the initial GWAS.9,10 We used the best guess genotypes of the imputed SNPs and excluded imputed SNPs with HWE P value <1×10−6, imputation quality Rsq <0.3 or MAF <0.01 and retained 2,490,789 SNPs in the ARIC cohort.
Conditional and Joint GWAS Analysis
We performed a stepwise model selection procedure to select independently associated SNPs using the GCTA tool available online (http://www.complextraitgenomics.com/software/gcta/massoc.html) for each lipid trait and CAD. Briefly, the procedure begins with the most significant SNP with P<5×10−8 in the single-SNP meta-analysis and tests all the remaining SNPs conditional on the selected SNP(s) in the model. It then selects the SNP with the minimum conditional P value and fits all the selected SNPs in a model, dropping the SNP with the largest P value >5×10−8. The algorithm iterates until no SNP is added to or removed from the model. The joint effects of all selected SNPs are estimated after the model has been optimized. We define a locus as a chromosomal region at which adjacent pairs of associated SNPs are less than 1 megabase (Mb) distant. Details about the conditional and joint analysis are fully described in ref. 7.
Estimation of the Variance Explained by the Joint Association
We calculated the variance explained using the following equation where βM and βJ are effect sizes in standard deviation units obtained from the original meta-analysis and the joint analysis, respectively.11
Variance explained was calculated for (1) the top SNPs from original meta-analysis, and (2) the top original SNPs plus the additionally associated SNPs found from the conditional and joint analysis.
Replication Analysis for Lipid Traits
We replicated the variance explained by all jointly associated SNPs detected from the GLGC data using 7,312 individuals from the Malmö Diet and Cancer (MDC) cohort as an independent sample in two models: Method A) A multiple regression of the SNPs selected from the discovery set (GLGC); Method B) A replication analysis using the SNPs with their effect sizes estimated from the discovery sample (GLGC) to predict the phenotype in MDC. In Method A, two predictors were created in the MDC cohort by PLINK12 for each lipid trait, one based on all additionally associated SNPs or its proxy SNPs (r2>0.8) and the other based on the GWAS top SNPs only, and the observed lipid phenotypes were regressed on the predictors. In Method B, we created two predictors in the MDC cohort but with SNP effects estimated from the GLGC dataset and regressed the observed lipid phenotypes on the predictors. In both methods, adjusted R2 values of MDC were compared with the explained variances for lipid traits in GLGC. Also, we checked the independency of multiple associated SNPs within a locus by comparing β from the model using multiple SNPs and β from the model using each SNP within each locus that has the largest number of multiple associated SNPs for each trait.
Informed Consent and Institutional Review Board Approval
Most of the analyses utilized summary statistics from prior publications. For genetic association analyses in the MDC cohort using de-identified genotype and phenotype data, each participant had provided written informed consent, and approval was given by the institutional review board at Partners Healthcare.
Results
Lipid Phenotypes
Using summary statistics of ∼2.5 million SNPs from the GLGC meta-analysis of 100,184 individuals for four lipid fractions along with SNP LD estimated in 8,682 unrelated European-Americans selected from the ARIC cohort study (See Methods), we identified 62, 61, 68, and 41 jointly associated SNPs for each lipid trait (HDL-C, LDL-C, TC, and TG) with P<5×10−8 (Supplemental Tables 1-4), respectively. When compared with previous results conducted by conventional conditional analysis in the original GLGC study, we could detect more additionally associated SNPs with each trait (11 vs. 29 for HDL-C, 12 vs. 26 for LDL-C, 12 vs. 24 for TC, and 9 vs. 13 for TG, Supplemental Table 5).
For the loci where the increasing alleles of at least two SNPs were negatively correlated, some associated variants were undetected in the original GWAS. For example, rs180349 and rs3741298 at the APOA1-C3-A4-A5 locus on chromosome 11 did not exhibit a significant association with HDL-C in single-SNP meta-analyses (P value from the single-SNP meta-analysis [PM]=8.67×10−3 and 4.12×10−4, respectively), but both SNPs reached genome-wide significance when fitted jointly (Supplemental Table 1). In addition, the significance and effect size of the leading SNP at the locus also increased (PM=2.94×10−42 to PJ=1.84×10−73 for rs964184) (Supplemental Table 1). There were 12 of 33 HDL-C, 10 of 35 LDL-C, 13 of 44 TC, and 8 of 28 TG loci harboring more than two associated SNPs, with the maximum number of 9, 9, 6, and 7 SNPs at a locus, respectively. The lead SNPs (33 for HDL-C, 35 for LDL-C, 44 for TC, and 28 for TG) explained 3.6%, 5.0%, 5.5%, and 5.7% of phenotypic variance, respectively. These values were almost doubled (7.6% for HDL-C, 8.8% for LDL-C, 8.8% for TC, and 8.5% for TG) when all jointly associated SNPs (62 for HDL-C, 61 for LDL-C, 68 for TC, and 41 for TG) were taken into account.
Coronary Artery Disease
Using summary statistics of ∼2.5 million SNPs from the CARDIoGRAM meta-analysis of 22,233 cases and 64,762 controls for CAD along with the same reference SNP data from the ARIC cohort described above, we identified 18 jointly associated SNPs for CAD with P<5×10−8. Of these SNPs associated with CAD, 3 of 15 loci represent multiple associated SNPs within a single locus (Figure 1, Supplemental Table 6).
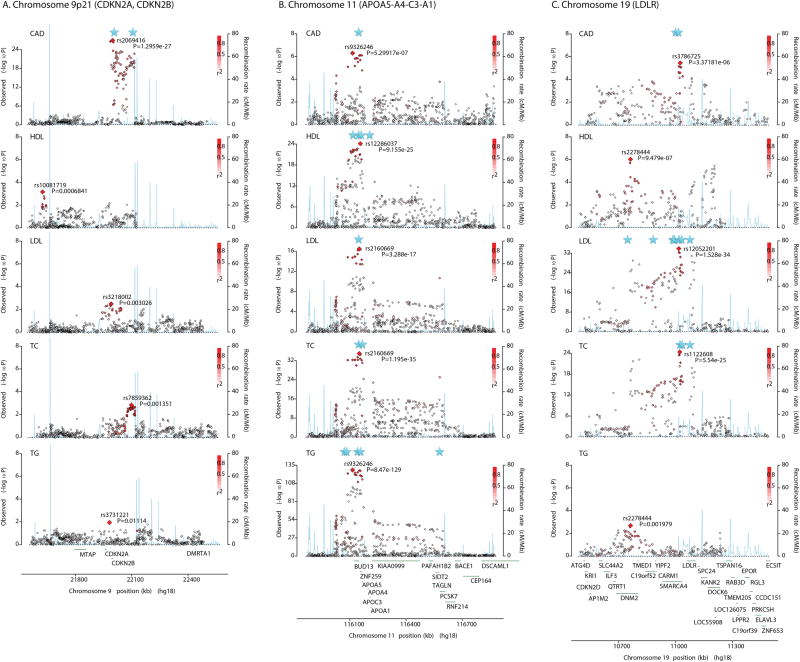
Figure 1.
Regional association plots of three loci with significant joint associations for CAD. A, CDKN2A and CDKN2B on chromosome 9p21. B, APOA5-A4-C3-A1 on chromosome 11. C, LDLR on chromosome 19. SNPs are plotted as red diamonds with –log10 P (y-axis) from the meta-analysis result for each trait. Significant joint association is marked as blue star corresponding to genomic position (x-axis) (See Supplemental Tables 1-6 for details). Color scheme illustrates linkage disequilibrium r2 based the 1000 Genomes European Ancestry (CEU) data. Genes are shown at the bottom of each plot. Note that scale of y-axis is different from each figure.
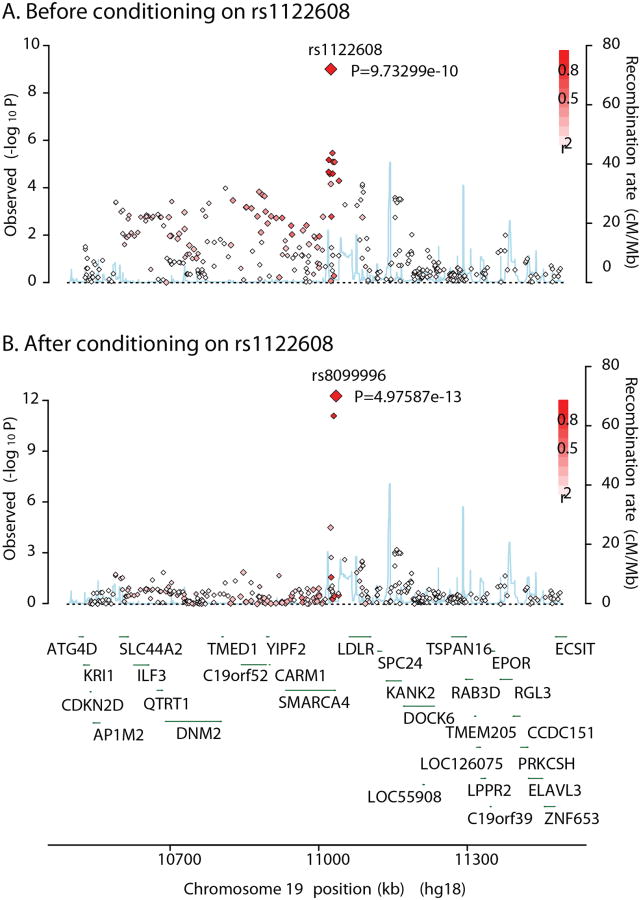
We found a significant joint association with CAD in the LDLR region, where multiple common variants for LDL-C and rare mutations in familial hypercholesterolemia have been previously reported.13 Two SNPs, rs8099996 and rs1122608, which are 11,024 bp apart, were retained in the stepwise model selection as jointly associated SNPs with PJ<3.5×10−11 (Supplemental Table 6). The secondary SNP (rs8099996; PM=0.67) was masked by the primary SNP (rs1122608; PM=9.7×10−10) in single-SNP analyses but it appeared significant (rs8099996; P=5.0×10−13) in conditional analysis on the primary SNP (Figure 2). This region was also significant for LDL-C and TC (Figure 1C). While the 9p21 CDKN2A and CDKN2B region was only significant for CAD in joint association analysis, the APOA5-A4-C3-A1 gene cluster locus was significant for four lipid traits as well as CAD (Figures 1A-1B). When they were fitted jointly, their effects, as well as statistical significance, were substantially increased compared to those in single-SNP analyses. The 15 leading SNPs explained 9.6% of phenotypic variance. The three additional SNPs detected by the joint analysis accounted for 1.8% of the variance explained.
Figure 2.
Example of a locus with a secondary joint association signal conditioning on a primary signal. A, SNPs are plotted as red diamonds with −log10 P (y-axis) from the meta-analysis result for CAD. The rs1122608 SNP is a primary signal at this LDLR locus. B, P values of the same SNP sets, which were calculated from conditional analysis on rs1122608, are plotted.
For a set of 184 SNPs in the Supplemental Tables 1-4, we evaluated the associations with CAD in CARDIoGRAM meta-analysis. Of these 184 SNPs, 38 SNPs were nominally associated (PM<0.05) and 20 SNPs showed a significant association after the Bonferroni's correction (PM<2.2×10−4) (Supplemental Tables 1-4).
Replication of the Lipid Results in an Independent Sample
We validated the direction of effect in each SNP between GLGC results and MDC results for all the jointly associated variants (Supplemental Figure 1). The multiple regression analysis showed that the prediction R2 values of top primary SNPs were 4.6%, 5.8%, 6.7%, and 5.2%, consistent with the estimate of 3.6%, 5.0%, 5.5%, and 5.7% of variance explained in the discovery sample (GLGC), for HDL-C, LDL-C, TC, and TG, respectively (Supplemental Figure 2: Method A). And the R2 values of the additionally associated SNPs were 5.2%, 3.1%, 2.7%, and 2.2%, in line with the estimate of 4.0%, 3.8%, 3.3%, and 2.8% of variance explained by these SNPs in the discovery sample (GLGC), respectively (Supplemental Figure 2: Method A).
In addition, when we used the SNP effects estimated from the GLGC dataset (the second method), the R2 values of top primary SNPs were 3.4%, 4.6%, 4.6%, and 4.4%, consistent with the estimate in the discovery sample (GLGC), and those of the additionally associated SNPs were 4.1%, 3.1%, 2.6%, and 2.2%, in line with the estimate of those explained by these SNPs in the discovery sample (GLGC), respectively (Supplemental Figure 2: Method B). Therefore, these replication analyses in an independent sample confirmed that additional associated variants could explain approximately 2%–4% of phenotypic variation for each lipid trait.
Supplemental Figure 3 shows that β from the multiple SNP model and β from the single SNP model are consistent for each locus with the largest number of multiple SNPs (A:HDL-C for CETP locus, B:LDL-C for APOE-C1-C2 locus, C:TC for APOE-C1-C2 locus, D:TG for LPL locus), suggesting that the variants from the multiple SNP model are independent.
Discussion
In this study, we detected a number of loci where multiple independent SNPs were associated with lipid traits and accounting for these variants nearly doubled the heritability explained by the previous GWAS results (HDL-C, LDL-C, TC, and TG). In addition, the joint associations of lipid traits were validated in an independent sample.
GWAS results have explained only a fraction of the heritability of complex traits. There has been extensive debate regarding this unexplained heritability, with hypotheses ranging from rare variants to epistasis.14-16 Here, we explored the possibility of multiple independent signals at a given locus as a contributor to the unexplained heritability.
Conditional analysis has been used as a tool to identify secondary association signals at a locus, starting with the top associated SNP, across the whole genome followed by a stepwise procedure of selecting additional SNPs, one by one, according to their conditional P values. However, nearly always, pooled individual level genotype data are unavailable in large-scale meta-analyses. In that sense, the tool we used is useful in terms of saving computational time and cost because it does not require individual genotype data except for the samples used as a LD reference. As a result, the current study clearly indicates that an increased portion of the missing heritability could be explained by the joint influence of multiple variants within a locus, suggesting the importance of digging into known loci to identify causal variants and understand the genetic architecture of complex diseases.
For plasma lipids, we found that many signals at specific loci can explain a large proportion of the variance. From our estimation, 9 SNPs in the CETP locus could explain as much as 2.6% of variance explained in HDL-C, 9 SNPs in the APOE-C1-C2 locus explained 2.1% of that in LDL-C, 6 SNPs in the APOE-C1-C2 locus explained 1.1% of that in TC, 7 SNPs in the LPL locus explained 1.7% in that of TG, and 2 SNPs in the CDKN2A/CDKN2B locus explained 2.8% of that in CAD trait. This is consistent with a previous report that suggests greater heritability of common variants in known loci.17
The variance explained by the top SNPs for each lipid trait from the GLGC dataset was relatively small compared with the one from the original article.6 This could be due to variability in estimates from different studies as well as the different method employed in this analysis compared to that of original GWAS, where only one study (i.e., the Framingham Heart Study) contributed to the estimation of variance explained. Although we showed evidence of multiple associations at several loci using summary statistics of the CARDIoGRAM meta-analysis, recently published data of the CARDIoGRAMplusC4D meta-analysis with 63,746 CAD cases and 130,681 controls based on the Metabochip array might also be useful to find additional associated signals.1
In summary, we detected a number of loci where multiple associated SNPs within a single locus were associated with lipid traits or CAD. For lipid traits, these variants nearly doubled the heritability explained.
Supplementary Material
Acknowledgments
Funding Sources: HT is supported by a grant for studying overseas from Japanese Circulation Society. The Malmö Diet and Cancer study was made possible by grants from the Swedish Cancer Society, the Swedish Medical Research Council, the Swedish Dairy Association, the Albert Påhlsson and Gunnar Nilsson Foundations and the Malmö city council. OM is supported by the European Research Council (StG-282255), the Swedish Heart and Lung Foundation, Swedish Research Council; the Novo Nordisk Foundation, the Skåne University Hospital donation funds; the Medical Faculty, Lund University; the Governmental funding of clinical research within the national health services, the Albert Påhlsson Research Foundation, Region Skåne, the King Gustav V and Queen Victoria Foundation and the Marianne and Marcus Wallenberg Foundation. GMP is supported by Award Number T32HL007208 from the National Heart, Lung, and Blood Institute. SK is supported by a Research Scholar award from the Massachusetts General Hospital (MGH), the Howard Goodman Fellowship from MGH, the Donovan Family Foundation, R01HL107816, and a grant from Fondation Leducq.
Footnotes
Disclosures: None
References
- 1.CARDIoGRAMplusC4D Consortium. Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arsenault BJ, Boekholdt SM, Kastelein JJ. Lipid parameters for measuring risk of cardiovascular disease. Nat Rev Cardiol. 2011;8:197–206. doi: 10.1038/nrcardio.2010.223. [DOI] [PubMed] [Google Scholar]
- 3.Emerging Risk Factors Collaboration. Di Angelantonio E, Sarwar N, Perry P, Kaptoge S, Ray KK, et al. Major lipids, apolipoproteins, and risk of vascular disease. JAMA. 2009;302:1993–2000. doi: 10.1001/jama.2009.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss LA, Pan L, Abney M, Ober C. The sex-specific genetic architecture of quantitative traits in humans. Nat Genet. 2006;38:218–222. doi: 10.1038/ng1726. [DOI] [PubMed] [Google Scholar]
- 5.Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, Ferreira T, Morris AP, Medland SE, Genetic Investigation of ANthropometric Traits (GIANT) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–375. S1–3. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rimm EB, Giovannucci EL, Willett WC, Colditz GA, Ascherio A, Rosner B, et al. Prospective study of alcohol consumption and risk of coronary disease in men. Lancet. 1991;338:464–468. doi: 10.1016/0140-6736(91)90542-w. [DOI] [PubMed] [Google Scholar]
- 9.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linsel-Nitschke P, Götz A, Erdmann J, Braenne I, Braund P, Hengstenberg C, et al. Lifelong reduction of LDL-cholesterol related to a common variant in the LDL-receptor gene decreases the risk of coronary artery disease--a Mendelian Randomisation study. PLoS One. 2008;3:e2986. doi: 10.1371/journal.pone.0002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritchie MD. Using biological knowledge to uncover the mystery in the search for epistasis in genome-wide association studies. Ann Hum Genet. 2011;75:172–182. doi: 10.1111/j.1469-1809.2010.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaitlen N, Kraft P. Heritability in the genome-wide association era. Hum Genet. 2012;131:1655–1664. doi: 10.1007/s00439-012-1199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.