Abstract
Salmonella genomic island 1 (SGI1) harbors a multidrug resistance (MDR) gene cluster which is a complex class 1 integron. Variant SGI1 MDR gene clusters conferring different MDR profiles have also been identified in several Salmonella enterica serovars and classified as SGI1-A to -F. A retrospective study was undertaken to characterize MDR regions from serovar Agona strains harboring SGI1 isolated from poultry in Belgium between 1992 and 2002. A total of 171 serovar Agona strains, displaying resistance to at least one antibiotic, were studied for the presence of SGI1. SGI1 was detected in 94 serovar Agona strains. The most prevalent variant was SGI1-A (85%), which harbors within the SGI1 complex class 1 integron a common region (CR1) containing orf513, a putative transposase gene, adjacent to the dfrA10 trimethoprim resistance gene. A new variant SGI1 named SGI1-G was identified in two strains. It consisted of the pse-1 gene cassette, as in SGI1-B, but with additional insertion of the orf513/dfrA10 region structure. Seven strains displaying the typical SGI1 MDR profile (Ap Cm Ff Sm Sp Su Tc) showed genetic variation at the 3′ end of SGI1. These strains harbored the insertion of the CR1 containing orf513 as in SGI1-A, -D, and -G. However, downstream the right end of CR1, they presented different 7.4- to 8.5-kb deletions of the SGI1 3′ end that extended to the chromosomal genes yieE and yieF. These results suggest a possible role of CR1 in deletion formation, as has been reported for some insertion sequences. Pulsed-field gel electrophoresis analysis showed that all the serovar Agona SGI1-carrying strains belonged to a single clone. Thus, SGI1 is largely encountered in serovar Agona strains isolated from poultry in Belgium, the most prevalent variant being SGI1-A. SGI1 MDR region undergoes recombinational events resulting in a diversity of MDR gene clusters.
Multidrug-resistant (MDR) Salmonella enterica serovar Typhimurium phage type DT104 emerged during the last decade as a global health problem because of its involvement in diseases in animals and humans (12, 18, 19, 27, 28, 31, 32). MDR DT104 strains are commonly resistant to ampicillin (Apr), chloramphenicol (Cmr) and florfenicol (Ffr), streptomycin (Smr) and spectinomycin (Spr), sulfonamides (Sur), and tetracyclines (Tcr).
This MDR profile is conferred by an MDR gene cluster included in a chromosomal genomic island called Salmonella genomic island 1 (SGI1) (5, 6). The 43-kb SGI1 is located between the thdF and int2 genes of the chromosome of S. enterica serovar Typhimurium DT104. The int2 gene is part of a retron sequence which has been reported to date only in serovar Typhimurium strains (5, 6). In other S. enterica serovars SGI1 is located between thdF and the yidY gene (4, 5, 15, 22). All of the antibiotic resistance genes are located near the 3′ end of SGI1 and are part of a complex class 1 integron that belongs to the In4 group (4). Class 1 integrons contain a 5′-conserved segment (5′-CS) which consists of the intI1 gene encoding the site-specific integrase and the associated attI1 site, the primary site of recombination, and the 3′-CS of variable length but generally consisting of qacEΔ1, which encodes low-level resistance to some antiseptics; the sul1 gene, which encodes sulfonamide resistance; and orf5, a gene of unknown function (16). One or more gene cassettes consisting of the coding region(s) and the downstream 59-base element (59-be), which is responsible for recognition and mobilization of cassettes, are found between the 5′-CS and 3′-CS (16). Transposon Tn402 is a mobile class 1 integron that contains the 5′-CS and a transposition module (tni region) consisting of four genes required for transposition (8). In addition, Tn402 is bound by inverted repeats (IRs) of 25 bp, IRi at the integrase end and IRt at the tni end. Several class 1 integrons appear to have originated from a Tn402-like ancestor by incorporation of the common part of the 3′-CS including qacEΔ1, sul1, and orf5. Most of these integrons, though still bound by IRi and IRt, have lost part or all of the tni module and are deemed defective transposon derivatives (8). The In4 group have a 3′-CS that includes a copy of IS6100 but no transposition genes, and most members are bound by IRi and IRt (24, 26). The MDR gene cluster of SGI1 is bound by IRi and IRt and thus can be considered a complex In4-type integron (4). Further, the MDR region is surrounded by 5-bp direct repeats, strongly suggesting it was integrated by a transposition event (4, 24, 26). Interestingly, in SGI1 there is a duplication of the 5′-CS, each one followed by a gene cassette. The first cassette carries the aadA2 gene, which confers resistance to streptomycin and spectinomycin, and a 3′-CS with a truncated sul1 (sul1Δ) gene. The second cassette contains the β-lactamase gene pse-1, which confers resistance to ampicillin and a 3′-CS with a complete sul1 gene conferring resistance to sulfonamides. Flanked by the two cassettes are the floR gene, which confers cross-resistance to chloramphenicol and florfenicol, and the tetracycline resistance genes tetR and tet(G) (1, 3, 7). Variant MDR gene clusters of SGI1 have recently been described in serovars Typhimurium DT104, Agona, and Albany (4, 9, 15). In some, there is only one 5′-CS and, hence, a single gene cassette, either aadA2 (SGI1-C) or pse-1 (SGI1-B), and floR and tet(G) are not present. In two other variants, SGI-A and SGI1-D, the 3′-CS, designated 3′-CS1, is followed by a 2,154-bp common region (CR) originally described in In6 and In7 (23, 30). Adjacent to the CR is a unique region that includes dfrA10 (trimethoprim resistance) as in In7 (23). Following the unique region is a second partial copy of the 3′-CS, designated 3′-CS2. The CR itself contains a gene, designated orf513, whose putative product, Orf513, due to its weak similarity to the transposase of IS801, is thought to be a putative transposase (4, 25). The CR described above, designated CR1, has been described in other class 1 integrons always in association with antibiotic resistance genes (25). Orf513 exhibits 66% identity to OrfA and 55% identity to Orf2, two proteins whose genes, like orf513 in CR1, are found adjacent to antibiotic resistance genes (4, 10, 25). The orfA gene was originally found adjacent to a floR gene in a plasmid isolated from E. coli in cattle (10), and orf2 is found adjacent to tet(G) in SGI1 and some variants (4, 5, 11). An examination of the downstream sequences revealed a 27-bp conserved region that defines the right-hand boundary of related CRs (25). Thus, each CR, CR1, CR2 (orfA), and CR3 (orf2) encodes a putative transposase which is postulated to interact with the 27-bp boundary sequence and mobilize antibiotic resistance genes (25).
Recently, SGI1 has also been identified in other serovar Typhimurium phage types, i.e., DT120, and in other S. enterica serovars, i.e., Agona, Paratyphi B, and Albany, indicating the horizontal transfer potential of SGI1 (4, 5, 11, 15, 22). SGI1-carrying MDR serovar Agona, Paratyphi B, and Albany strains were isolated from different animal species such as poultry in Belgium, tropical fish from Singapore, and food fish imported from Thailand to France, respectively (4, 5, 11, 15, 22).
Since 1992 an increasing number of MDR serovar Agona strains exhibiting different antibiotic resistance profiles have been isolated from poultry in Belgium. Some recent isolates were studied and reported to carry SGI1 or SGI1 variants (4, 5). In the present work, we conducted a retrospective study to identify and characterize SGI1 in all serovar Agona strains that were resistant to at least one antibiotic and were isolated from poultry in Belgium from 1992 to 2002. The aim of this study was thus to determine what was the most prevalent SGI1 variant, to identify possible new SGI1 variants, and to determine at the epidemiological level whether these strains were clonal or not.
MATERIALS AND METHODS
Bacteria, media, and antibiotic susceptibility testing.
One hundred and seventy-one serovar Agona strains used in this study, displaying resistance to at least one antibiotic, were isolated from poultry in Belgium between 1992 and 2002. These strains and control strains serovar Typhimurium DT104 BN9181 and serovar Paratyphi B 44 and E. coli strain TOP10 (Invitrogen) used in cloning experiments were grown at 37°C in brain heart infusion broth or agar plates. The strains were tested for their antibiotic susceptibility by the disk-diffusion assay on Mueller-Hinton plates. Susceptibility was tested using disks containing the following antibiotics: ampicillin (10 μg), chloramphenicol (30 μg), florfenicol (30 μg), streptomycin (10 IU), spectinomycin (100 μg), sulfonamides (200 μg), tetracyclines (30 IU), and trimethoprim (Tms) (5 μg). Susceptibilities to cephalothin (Cfs) (30 μg), ceftriaxone (Cros) (30 μg) and ceftiofur (Xnls) (30 μg) were also tested. All antibiotic disks except for florfenicol were purchased from Bio-Rad (Marnes-la-Coquette, France). Florfenicol disks and the drug itself were obtained from Schering-Plough Animal Health (Kenilworth, N.J.). The MICs of florfenicol and chloramphenicol were determined by the standard agar doubling dilution method (21). The MIC breakpoints for chloramphenicol and florfenicol were defined by the Comité de l'Antibiogramme de la Société Française de Microbiologie (21) or by the manufacturer, i.e., susceptible (MIC ≤ 8 μg/ml), intermediate (MIC = 16 μg/ml), or resistant (MIC ≥ 32 μg/ml).
PCR mapping, cloning, and sequencing.
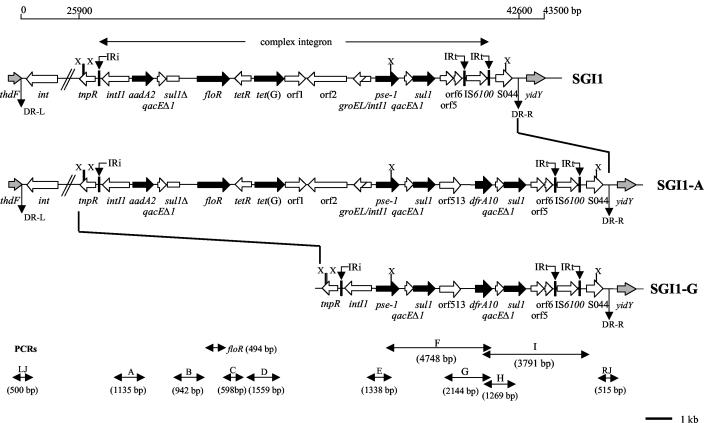
Detection of SGI1 and its location were performed using primers corresponding to left and right (with or without retron) junctions in the chromosome as described previously (Table 1; Fig. 1) (4, 15, 22). PCR mapping of the typical antibiotic resistance genes associated with SGI1 was performed using conditions and primers described previously (11, 15, 22) (Table 1) (Fig. 1).
TABLE 1.
Primers used for PCR in this study
| Primer | Gene | Amplificationa | Size (bp) | Nucleotide sequence (5′-3′) |
|---|---|---|---|---|
| U7-L12 | thdF | Left junction | 500 | ACACCTTGAGCAGGGCAAG |
| LJ-R1 | int | AGTTCTAAAGGTTCGTAGTCG | ||
| 104-RJ | S044 | Right junction | TGACGAGCTGAAGCGAATTG | |
| C9-L2 | int2 | 515 | AGCAAGTGTGCGTAATTTGG | |
| 104-D | yidY | 500 | ACCAGGGCAAAACTACACAG | |
| cmP01 | floR | floR | 494 | TTTGGWCCGCTMTCRGACb |
| cml15 | floR | SGAGAARAAGACGAAGAAGb | ||
| int1 | intI1 | A | 1,135 | GCTCTCGGGTAACATCAAGG |
| aad | aadA2 | GACCTACCAAGGCAACGCTA | ||
| sulTER | sullΔ | B | 942 | AAGGATTTCCTGACCCTG |
| F3 | floR | AAAGGAGCCATCAGCAGCAG | ||
| F4 | floR | C | 598 | TTCCTCACCTTCATCCTACC |
| F6 | tetR | TTGGAACAGACGGCATGG | ||
| tetR | tetR | D | 1,559 | GCCGTCCCGATAAGAGAGCA |
| tetA | tet(G) | GAAGTTGCGAATGGTCTGCG | ||
| int2 | groEL-intI1 | E | 1,338 | TTCTGGTCTTCGTTGATGCC |
| pse1 | pse-1 | CATCATTTCGCTCTGCCATT | ||
| pse-L | pse-1 | F | 4,748 | AATGGCAATCAGCGCTTCCC |
| dfrA10-B | dfrA10 | AACCAACACCACCAATGACA | ||
| Forf513 | orf513 | G | 2,144 | GCAGCACTACCCAGCCTTCA |
| dfrA10-B | dfrA10 | AACCAACACCACCAATGACA | ||
| dfrA10-A | dfrA10 | H | 1,269 | TGTCATTGGTGGTGTTGGTT |
| Rsul1 | sul1 | CGACACCGAGACCAATAGCG | ||
| dfrA10-A | dfrA10 | I | 3,791 | TGTCATTGGTGGTGTTGGTT |
| MDR-B | S044 | GAATCCGACAGCCAACGTTCC | ||
| Fresol | tnpR | TCGGGTCTAGCGGCATTCTT |
See Fig. 1.
degenerate nucleotides: M = A or C, R = A or G, S = C or G, W = A or T.
FIG. 1.
Genetic organization of the MDR gene clusters of SGI1, SGI1-A variant, and new SGI1-G variant. Black and grey arrows correspond to antibiotic resistance genes and chromosomal genes flanking SGI1, respectively. DR-L and DR-R are the left and right direct repeats, respectively, bracketing SGI1. IRi and IRt are 25-bp imperfect IRs defining the left and right ends of complex class 1 integrons. Vertical thick black bars indicate homology regions between SGI1 variants. PCRs used to assess the genetic organization of the antibiotic resistance genes (PCRs floR, A, B, C, D, E, F, G, H, and I) and the SGI1 junctions to the chromosome (PCRs LJ and RJ for left and right junctions, respectively) are indicated. Abbreviation for restriction site: X, XbaI.
To assess a new variant SGI1 MDR gene cluster identified in this study, PCR amplification of a region of the complex class 1 integron was also performed using primer Fresol and primer Rsul of PCR I (Fig. 1). Cloning of this PCR product in plasmid pCR2.1-TOPO was performed using the TOPO TA cloning kit (Invitrogen, Cergy Pontoise, France). Nucleotide sequencing of the insert was achieved by Genome Express (Meylan, France). Sequence analysis was done by using BLAST (available from http://www3.ncbi.nlm.nih.gov/BLAST/).
A set of PCRs was performed to determine the extent of the deletions at the 3′ end of SGI1 in the serovar Agona strains with negative results for the right junction of SGI1. PCR products corresponding to the new right junctions were sequenced.
PFGE.
Strains positive for SGI1 were investigated by pulsed-field gel electrophoresis (PFGE). Preparation of genomic DNA was performed as previously described (11, 15, 22). Genomic DNA, contained in agarose plugs, was digested with BlnI (BioWhttaker, Emerainville, France) or XbaI (Qbiogene, Illkrich, France). Electrophoresis was performed using the CHEF-DR III system (Bio-Rad, Hemel Hempstead, United Kingdom) at 14°C in 0.5× Tris-borate-EDTA running buffer. Electrophoresis conditions were as follows: 0.6 V/cm for 18 h with pulse times of 2.16 to 54.17 s.
Nucleotide sequence accession numbers.
The complete nucleotide sequences of regions resulting from the different SGI1 deletions have been deposited in the GenBank database under accession numbers AY434091, AY434092, and AY434093.
RESULTS
Detection of SGI1 in MDR serovar Agona isolated in Belgium in 1992 to 2002.
One hundred and seventy-one serovar Agona strains resistant to at least one antibiotic and isolated between 1992 and 2002 were analyzed for the presence of SGI1. The SGI1 detection and location based on junction PCRs were realized by using primers corresponding to the left and right junctions of SGI1 in the Salmonella chromosome (Fig. 1). We also used PCR to detect the presence or absence of the retron sequence, which is located downstream of SGI1 in serovar Typhimurium DT104 strains but absent to date in other S. enterica serovars (4, 5, 15, 22). SGI1 was thus detected in 87 serovar Agona strains between the thdF and the yidY genes of the chromosome (Table 2). All strains lacked the retron sequence found downstream of SGI1 in serovar Typhimurium DT104. For seven MDR serovar Agona strains, PCR was positive for the left junction of SGI1 with thdF but was, however, negative for the right junction with yidY (data not shown) (Table 2). Thus, these data indicate that 55% of MDR serovar Agona strains tested harbor SGI1 at the same chromosomal location, i.e., between the thdF and yidY genes, as previously found (5). The earliest MDR serovar Agona strain harboring SGI1 was isolated in 1993 (Table 2). Among 19 serovar Agona strains tested from 1992, none contained SGI1. Seven serovar Agona strains isolated between 2000 and 2002 that were negative for the right junction but positive for the left junction PCR appeared to harbor SGI1 with some genetic variation at its 3′ end.
TABLE 2.
Characteristics of the 94 serovar Agona SGI1-carrying strains tested in this study
| Yr | No. of strains | MDR resistance profile | MIC (μg/ml) of:
|
SGI1 junctions PCRs
|
Resistance gene cluster PCRsa
|
Variant genomic island name | PFGE patternb
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Florfenicol | Chloramphenicol | Left | Right
|
A | B | C | D | E | F | G | H | I | floR | XbaI | BlnI | |||||
| With retron | Without retron | |||||||||||||||||||
| 1993 | 1 | Ap Cm Ff Sm Sp Su Tc Tm | 64 | 256 | + | − | + | + | + | + | + | + | + | + | + | + | + | SGI1-A | A | A1 |
| 1994 | 3 | Ap Cm Ff Sm Sp Su Tc Tm | ≥64 | ≥256 | + | − | + | + | + | + | + | + | + | + | + | + | + | SGI1-A | A | A1 |
| 1995 | 7 | Ap Cm Ff Sm Sp Su Tc Tm | ≥64 | ≥256 | + | − | + | + | + | + | + | + | + | + | + | + | + | SGI1-A | A | A1 |
| 1996 | 7 | Ap Cm Ff Sm Sp Su Tc Tm | ≥64 | ≥256 | + | − | + | + | + | + | + | + | + | + | + | + | + | SGI1-A | A | A1 |
| 1 | Ap Su Tm | 8 | 8 | + | − | + | − | − | − | − | − | + | + | + | + | − | SGI1-G | A | A2 | |
| 1 | Ap Cm Ff Sm Sp Su Tc | 32 | >256 | + | − | + | + | + | + | + | + | − | − | − | − | + | SGI1 | A | A3 | |
| 1997 | 11 | Ap Cm Ff Sm Sp Su Tc Tm | ≥64 | ≥256 | + | − | + | + | + | + | + | + | + | + | + | + | + | SGI1-A | A | A1 |
| 1 | Ap Cm Ff Sm Sp Su Tc | 64 | 256 | + | − | + | + | + | + | + | + | − | − | − | − | + | SGI1 | A | A3 | |
| 1 | Sm Sp Su | 8 | 8 | + | − | + | + | − | − | − | − | − | − | − | − | − | SGI1-C | A | A4 | |
| 1998 | 3 | Ap Cm Ff Sm Sp Su Tc Tm | ≥64 | ≥256 | + | − | + | + | + | + | + | + | + | + | + | + | + | SGI1-A | A | A1 |
| 1999 | 2 | Ap Cm Ff Sm Sp Su Tc Tm | ≥64 | ≥256 | + | − | + | + | + | + | + | + | + | + | + | + | + | SGI1-A | A | A1 |
| 2000 | 9 | Ap Cm Ff Sm Sp Su Tc Tm | ≥64 | ≥256 | + | − | + | + | + | + | + | + | + | + | + | + | + | SGI1-A | A | A1 |
| 1 | Ap Su Tm | 8 | 8 | + | − | + | − | − | − | − | − | + | + | + | + | − | SGI1-G | A | A2 | |
| 1 | Sm Sp Su (Tc) | 8 | 8 | + | − | + | + | − | − | − | − | − | − | − | − | − | SGI1-C | A | A4 | |
| 2 | Ap Cm Ff Sm Sp Su Tc | 64 | 256 | + | − | − | + | + | + | + | + | − | − | − | − | + | SGI1-AΔ3R | A | A3 | |
| 2001 | 13 | Ap Cm Ff Sm Sp Su Tc Tm | ≥64 | ≥256 | + | − | + | + | + | + | + | + | + | + | + | + | + | SGI1-A | A | A1 |
| 1 | Ap Cf Cm Cro Ff Sm Sp Su Tc Tm Xnl | 64 | 256 | + | − | + | + | + | + | + | + | + | + | + | + | + | SGI1-A | A | A1 | |
| 1 | Ap Cm Ff Sm Sp Su Tc | 64 | 256 | + | − | − | + | + | + | + | + | − | − | − | − | + | SGI1-AΔ2R | A | A2 | |
| 3 | Ap Cm Ff Sm Sp Su Tc | 64 | 256 | + | − | − | + | + | + | + | + | − | − | − | − | + | SGI1-AΔ2R | A | A3 | |
| 2002 | 23 | Ap Cm Ff Sm Sp Su Tc Tm | ≥64 | ≥256 | + | − | + | + | + | + | + | + | + | + | + | + | + | SGI1-A | A | A1 |
| 1 | Ap Cm Ff Sm Sp Su Tc | 64 | >256 | + | − | + | + | + | + | + | + | − | − | − | − | + | SGI1 | A | A3 | |
| 1 | Ap Cm Ff Sm Sp Su Tc | 64 | >256 | + | − | − | + | + | + | + | + | − | − | − | − | + | SGI1-AΔ1R | A | A3 | |
MDR profiles.
SGI1-positive serovar Agona strains showed five different MDR profiles (Table 2). They consisted of the following: Ap Cm Ff Sm Sp Su Tc Tm (81 strains), Ap Cm Ff Sm Sp Su Tc (9 strains), Sm Sp Su (2 strains), Ap Su Tm (2 strains), and Ap Cf Cm Cro Ff Sm Sp Su Tc Tm Xnl (1 strain). The serovar Agona strains resistant to florfenicol showed the same level of resistance to florfenicol as previously reported for serovars Typhimurium DT104, Paratyphi B, and Albany harboring SGI1 (MIC of florfenicol, 64 μg/ml) (Table 2) (11, 15, 22).
MDR gene clusters.
PCR mapping of the typical antibiotic resistance genes associated with SGI1-A is schematized in Fig. 1. PCRs A to I and floR were performed on genomic DNAs extracted from SGI1-positive serovar Agona strains. Eighty-one strains displaying the MDR profile Ap Cm Ff Sm Sp Su Tc Tm yielded fragments A to I and floR of the sizes expected from the control serovar Agona strain 1169SA97 harboring SGI1-A described previously (data not shown) (Table 2) (4). Thus, these PCR mapping results indicated most serovar Agona strains harbor the variant MDR gene cluster of SGI1-A.
Two serovar Agona strains (1233SA96 and 959SA97) isolated in 1996 and 1997 showed the typical MDR profile of serovar Typhimurium DT104 harboring SGI1 (Ap Cm Ff Sm Sp Su Tc) with lack of resistance to trimethoprim (Table 2). PCRs A to E and partial floR results were positive and yielded fragments of expected sizes as for serovar Typhimurium DT104 control strain BN9181 (data not shown). However, PCR fragments F to I specific for the orf513/dfrA10 region were not obtained. The absence of this region was further assessed by PCR with a forward primer of fragment F and a reverse primer of fragment I. This PCR result was positive and yielded a 4.4-kb fragment as with DNA from serovar Typhimurium DT104 control strain BN9181 (data not shown). These results indicated that these two serovar Agona strains harbor SGI1 with the classical MDR gene cluster described for serovar Typhimurium DT104 (1, 3, 5, 6, 7).
Two others serovar Agona strains (47SA97 and 278SA00) isolated in 1997 and 2000 were only resistant to streptomycin or spectinomycin and sulfonamides (Table 2). PCR mapping results of these strains were only positive for fragment A specific for the aadA2 gene cassette. This SGI1 variant containing a single aadA2 gene cassette was previously characterized for strain 47SA97 and was named SGI1-C (4). Thus, the other serovar Agona strain isolated in 2000 was also shown to harbor SGI1-C (4).
New variant SGI1 MDR gene cluster.
Two SGI1-positive serovar Agona strains (712SA96 and 169SA00) isolated in 1996 and 2000 displayed the MDR profile Ap Su Tm, which did not correspond to any variant SGI1 MDR gene cluster previously described (Table 2) (4, 9). All PCRs were negative except PCRs F to I (data not shown) (Fig. 1). Thus, PCR mapping of the antibiotic resistance genes showed the presence of the pse-1 gene cassette and the orf513 and dfrA10 genes. To further assess the pse-1 gene cassette region in this complex integron, an additional PCR amplification using primer Fresol and primer Rsul of PCR H was performed (Table 1, Fig. 1). A fragment of approximately 3.7 kb was obtained, and its nucleotide sequence was determined. Sequence analysis showed 100% identity with the nucleotide sequences of the tnpR gene and the pse-1 gene cassette from the serovar Typhimurium DT104 SGI1 MDR gene cluster (5). This MDR gene cluster thus shows the same genetic organization as in the SGI1-B variant with a complete integrase gene intI1, the pse-1 gene cassette and the 3′-CS of the integron (4). However, it constitutes a new SGI1 variant by the additional insertion of the orf513/dfrA10 region (Fig. 1); we propose the name SGI1-G for this new SGI1 variant, according to the previously proposed nomenclature (4, 15).
SGI1 3′-end deletions.
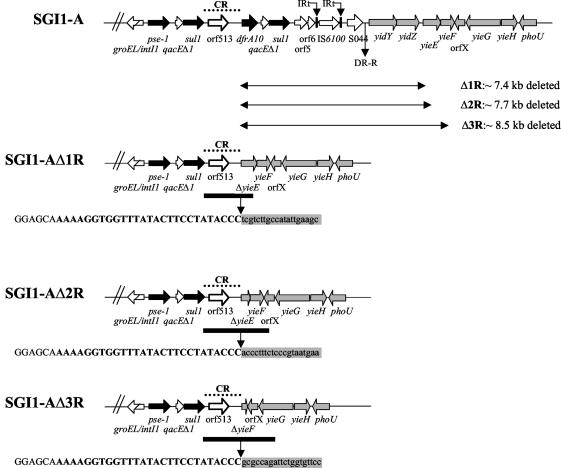
Seven serovar Agona strains (1690SA00, 1691SA00, 507SA01, 519SA01, 773SA01, 781SA01, and 1254SA02) isolated between 2000 and 2002 were negative for the right junction PCR of SGI1 with the yidY gene of the chromosome (Table 2). These strains showed the typical MDR profile Ap Cm Ff Sm Sp Su Tc from serovar Typhimurium DT104 harboring SGI1. PCRs A to E and floR were positive, whereas PCRs F to I were negative (data not shown) (Fig. 1). A set of further PCR mappings was done to try to determine the genetic variation, possibly involving deletions, at the SGI1 3′ end of these strains. PCR mapping of the SGI1 3′ end showed for each strain the presence of the complete pse-1 gene cassette and a specific orf513 fragment of the expected size (data not shown). PCR amplifications corresponding to dfrA10, qacEΔ1, sul1, orf5, orf6, IS6100, and S044 were not obtained (Fig. 2). Thus, these data indicated deletions of the respective genes downstream of orf513. Further PCR mapping of the chromosome downstream of orf513 showed that in these seven strains, the yidY, yidZ, and yieE genes were absent. However, the yieF gene was detected in all strains except in two (1690SA00 and 1691SA00) isolated in 2000 (data not shown). Genes downstream of yieF were detected in all strains. A small insertion of about 500 bp between the yieF and yieG genes was found in all strains which was not present in the genome sequence of serovar Typhimurium LT2 (GenBank accession number NC_003197) (Fig. 2). Thus, PCR mapping results indicated that three different deletions had occurred in these strains indicated as SGI1-AΔ1R, SGI1-AΔ2R, and SGI1-AΔ3R in Table 1. A PCR product of about 3.5 kb between the end of the sul1 gene downstream of the pse-1 gene cassette to the yieG gene of the chromosome was amplified and sequenced for strain 1690SA00 (GenBank accession number AY434092). A PCR product of shorter size ending in the insertion between the yieF and yieG genes of the chromosome was amplified and sequenced for strain 507SA01 (Fig. 2) (GenBank accession number AY434091). A PCR product of about 2.8 kb was obtained for strain 1254SA02 that corresponds to a fragment between the beginning of the CR1 to the yieE gene of the chromosome and was also sequenced (GenBank accession number AY434093). Sequence analysis showed that for all strains SGI1 deletions that extended to chromosomal genes started downstream of the CR1 of 2,154 bp that includes orf513 (25). For the sequenced strain 1254SA02, the deletion extended from the end of the CR1 to bp 64 of the yieE gene of the chromosome, which represents an approximately 7.4-kb deleted fragment from SGI1-A and the chromosome (Fig. 2). For the sequenced strain 507SA01, the deletion ended at bp 362 of the yieE gene of the chromosome, which represents a 7.7-kb deletion. For strain 1690SA00, the deletion was approximately 8.5 kb long to bp 408 of the yieF gene of the chromosome. Thus, we named these SGI1 deletion variants SGI1-AΔ1R (strain 1254SA02), SGI1-AΔ2R (strains 507SA01, 519SA01, 773SA01, and 781SA01), and SGI1-AΔ3R (strains 1690SA00 and 1691SA00) (Table 2; Fig. 2). Sequence analysis of the insertion of about 500 bp between yieF and yieG genes in these seven serovar Agona strains, relative to the genome of serovar Typhimurium LT2 (GenBank accession number NC_003197), revealed the presence of an open reading frame of 426 bp named orfX, with absence of homology to any known proteins (Fig. 2). Sequence alignment of CR1 from strains 1690SA00 and 507SA01 with the previously sequenced CR1 from SGI1-D of serovar Agona strain 953SA98 (GenBank accession number AY049746) showed two and six nucleotide substitutions, respectively. The nucleotide sequence of the CR1 from strain 1254SA02 was 100% identical to that of the CR1 from SGI1-D. The deduced amino acid sequence of orf513 from strain 1690SA00 was 100% identical to that of strain 953SA98 harboring SGI1-D (4). For strain 507SA01, all six nucleotide substitutions in orf513 led to amino acid changes in the deduced protein. The Orf513 protein from strain 507SA01 was thus 98.8% identical to that from the others.
FIG. 2.
Schematic view of the 3′ end of SGI1-AΔRs characterized in this study. SGI1-A 3′ end is shown at the top. Black and grey arrows correspond to antibiotic resistance genes and chromosomal genes found downstream of SGI1, respectively. Right-hand boundary sequences are indicated under each SGI1-AΔR. Uppercase letters represent the CR1 sequences. Nucleotides in boldface letters indicate the 27-bp potential recognition site of Orf513. Nucleotides in lowercase letters on a grey background represent chromosomal gene sequences found downstream of SGI1. The extents of deletions are indicated by arrows. Locations of sequenced regions are indicated by thick black bars, and locations of the CR1 sequences are indicated by dotted lines.
Macrorestriction analysis by PFGE.
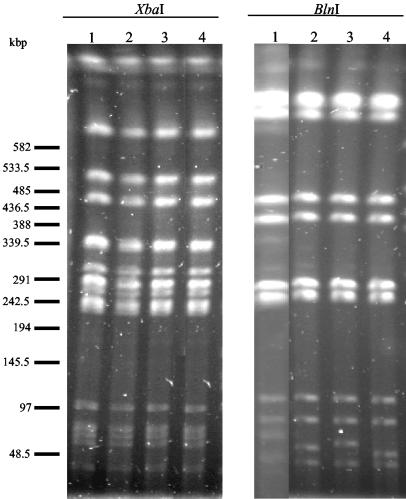
Macrorestriction analysis by PFGE of the SGI1 carrying serovar Agona strains DNAs cut by XbaI showed that all strains had identical XbaI PFGE patterns (Table 2; Fig. 3). Four BlnI PFGE patterns (A1 to A4) were obtained and corresponded to only one band difference each in the bottom of the gel. These different BlnI patterns correlated well with the type of SGI1 variant; i.e., pattern A1 correlated well with SGI1-A, pattern A2 correlated well with SGI1-G and SGI1-AΔRs, pattern A3 correlated well with SGI1 and SGI1-AΔRs, and pattern A4 correlated well with SGI1-C. Thus, these results indicate that all serovar Agona SGI1-carrying strains from this study are clonally related.
FIG. 3.
Macrorestriction analysis by PFGE of genomic DNAs cut by XbaI or BlnI of serovar Agona strains harboring SGI1-A (lanes 1), SGI1 (lanes 2), SGI1-G (lanes 3), and SGI1-C (lanes 4).
DISCUSSION
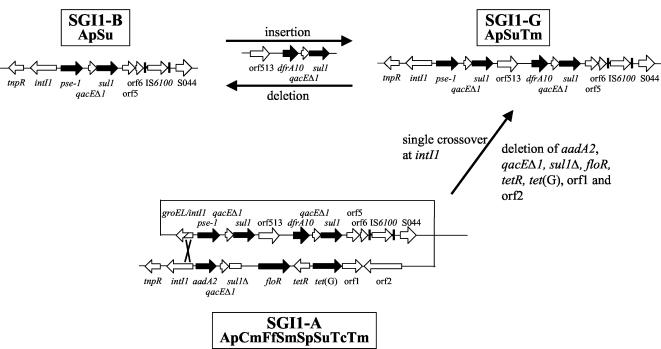
SGI1 is the first genomic island containing an MDR gene cluster identified in S. enterica. Among MDR serovar Typhimurium DT104 strains, most of them show the Ap Cm Ff Sm Sp Su Tc resistance profile and harbor the classical MDR gene cluster from SGI1. Acquisition of SGI1 may have been an important trait in the worldwide epidemic of the MDR serovar Typhimurium DT104 clone causing disease in animals as well as in humans. SGI1 has been further identified in other S. enterica serovars, such as serovar Agona strains isolated from poultry in Belgium and serovars Paratyphi B and Albany strains isolated from fish in Asia (4, 5, 11, 15, 22). The identification of SGI1 at the same chromosomal location in several S. enterica serovars indicates horizontal transfer of this island and a SGI1 site-specific insertion in the chromosome. SGI1-carrying MDR serovar Agona strains isolated from poultry in Belgium between 1992 and 2002 represent 55% of serovar Agona strains tested (94 SGI1-positive serovar Agona strains). It appears from the present study that the emergence of SGI1 in serovar Agona strains from poultry in Belgium started in 1993. The most prevalent SGI1 variant identified in these strains was SGI1-A, which accounts for 85% of the total SGI1-positive strains. This SGI1-A variant, which confers additional Tmr, relative to that of SGI1 mostly encountered in MDR serovar Typhimurium DT104 strains, carries in its MDR gene cluster the additional structure orf513/dfrA10 downstream of the pse-1 gene cassette 3′-CS1 (4). A new SGI1 variant identified in this study, named SGI1-G, consists of the complex integron structure that includes only the β-lactamase pse-1 gene cassette associated with the orf513/dfrA10 region downstream of the 3′-CS1. The generation of this variant SGI1-G could be explained by the insertion of the orf513/dfrA10 structure via homologous recombination between the duplicated qacEΔ1/sul1 genes of this variant (Fig. 4) (4, 25). This hypothesis is supported by a recent study of Partridge et al. (25) who showed that an artificially excised circular molecule which consists of the orf513/dfrA10 region from In34 could recombine with the 3′-CS in another class 1 integron. Another explanation could be that SGI1-G originated from SGI1-A by a single crossover between the copies of intI1 with loss of the aadA2, qacEΔ1, sul1, floR, tetR, tet(G), orf1, and orf2 region (Fig. 4).
FIG. 4.
Schematic diagram of the generation of variant SGI1-G from SGI1-A or SGI1-B.
Over the past 10 years, a number of complex class 1 integrons have been described as containing CR1 that includes orf513 (2, 4, 13, 25, 29, 30, 35, 36). These complex integrons contain the 5′-CS and two 3′-CSs designated 3′-CS1 and 3′-CS2. 5′-CS (intI1 gene, attI) and 3′-CS1 (qacEΔ1 and sul1 genes) constitute a typical class 1 integron with one or more gene cassettes containing a typical 59-be. Following the 3′-CS1 is the CR1 of 2,154 bp that contains orf513 previously known as orf341 (34, 35). Downstream of the CR1 is a unique region that includes antibiotic resistance genes such as catA2, dfrA10, dfrA19, blaDHA-1, blaCTX-M-9, blaCTX-M-2, blaCMY-9, and qnr, which are not present as gene cassettes as they are in integrons (2, 4, 13, 25, 29, 30, 34, 35, 36). These resistance genes are indeed not followed by the 59-be present in class 1 integrons. Downstream of the unique region is the 3′-CS2 of a typical class 1 integron (qacEΔ1 and sul1 genes). SGI1-A, -D, and -G variant MDR gene clusters present the same organization as complex In4-like integrons (4). As seen in the present study, in contrast to SGI1-carrying serovar Typhimurium DT104 strains, the CR1 that includes orf513 is dominantly prevalent in SGI1-carrying Agona strains (SGI1-A, -D, -G, -AΔ1R, -AΔ2R, and -AΔ3R). The role of the putative transposase-recombinase Orf513 has not yet been experimentally determined (4, 10, 25). In the early 1990s, it was speculated that antibiotic resistance genes could have been inserted as gene cassettes, using a 59-be, with the subsequent loss of this element (13, 23, 25, 30). Nowadays, with the increasing number of complex integrons described, it appears that Orf513 may recognize a 27-bp segment well conserved at the 3′ end of the CR1 where Orf513 will thus catalyze recombinational events to incorporate antibiotic resistance genes adjacent to this recognition site (2, 13, 25, 29).
Three different deletions at the 3′ end of SGI1 downstream of the CR1 are described in the present study (i.e., SGI1-AΔ1R, -AΔ2R, -AΔ3R). These deletions of different sizes (7.4, 7.7, and 8.5 kb) start at the precise right-hand end of the CR1, downstream from the 27-bp putative recognition site of Orf513, and extend further downstream of SGI1 to proximate chromosomal genes. Most likely these deletions originated from SGI1-A according to the high prevalence of SGI1-A. The insertion of the CR1 from the orf513/dfrA10 region downstream from the pse-1 gene cassette when recombined with the SGI1 3′ end could have led to concomitant deletion of downstream chromosomal genes in some strains. Recombinational events at the 27-bp segment involving the putative transposase Orf513 may thus have resulted in deletion of the SGI1-A 3′ end and part of the adjacent chromosomal region. Considering the similarity of Orf513 to the transposase of IS801 (4, 10, 25), our results suggest a possible role of the CR1 in generating deletions, as has been reported for some insertion sequences (ISs), and also raise the question of mobility of the CR1. Indeed, IS elements may mediate deletion events and remove DNA from one or the other side of the elements, as seen with deletions found downstream of the CR1 described in this study (17, 20, 33). However, in the case of IS, deletion formation appears to be directly related to insertion and transposition events (17). The most studied are IS1-mediated deletions that are generated by intramolecular tranposition (17, 20, 33). Another possible explanation in the case of the CR1 that includes orf513 is that a copy of the CR1 moved and inserted in the yieE or yieF genes in strains carrying SGI1-A. Subsequently, a single crossover event between the two CR1 led to the deletion of the internal region. Thus, further studies concerning mobility of the CR1 and recombination events mediated by it, are of interest.
Macrorestrction PFGE analysis showed that all serovar Agona strains carrying SGI1 and variants were clonally related. According to published data, it is likely that horizontal transfer of SGI1 is not a frequent event and that, as in the case of MDR serovar Typhimurium DT104 strains, S. enterica serovars acquiring SGI1 spread clonally in animals and humans (12, 18, 19, 27, 28, 31, 32). Only recently, we have reported the first human case of infection with a serovar Agona strain harboring the variant SGI1-A (14). The patient was probably infected by one of the clonal SGI1-carrying serovar Agona strains from poultry, on the basis of the fact that XbaI and BlnI PFGE patterns of this human strain were identical to those of poultry strains.
In conclusion, SGI1 is largely encountered in clonally related serovar Agona strains isolated from poultry in Belgium, with the most prevalent variant being SGI1-A. SGI1 MDR gene clusters are subjected to recombinational events, which results in a great diversity of MDR gene clusters, such as the new one, SGI1-G, described in this study. The deletions found at the SGI1 3′ end that extend to proximate chromosomal genes also raise new questions about possible recombinational events mediated by the CR1 as well as CR1’s mobility.
Acknowledgments
We thank J. Borlang, C. Mouline, and V. Verberen for expert technical assistance.
REFERENCES
- 1.Arcangioli, M. A., S. Leroy-Sétrin, J. L. Martel, and E. Chaslus-Dancla. 1999. A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol. Lett. 174:327-332. [DOI] [PubMed] [Google Scholar]
- 2.Arduino, S. M., P. H. Roy, G. A. Jacoby, B. E. Orman, S. A. Pineiro, and D. Centron. 2002. blaCTX-M-2 is located in an unusual class 1 integron (In35) which includes orf513. Antimicrob. Agents Chemother. 46:2303-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolton, L. F., L. C. Kelley, M. D. Lee, P. J. Fedorka-Cray, and J. J. Maurer. 1999. Detection of multidrug-resistant Salmonella enterica serotype typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J. Clin. Microbiol. 37:1348-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, D., A. Cloeckaert, E. Chaslus-Dancla, and M. R. Mulvey. 2002. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob. Agents Chemother. 46:1714-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, D., G. A. Peters, A. Cloeckaert, K. Sidi Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, D. A., G. A. Peters, L. K. Ng, and M. R. Mulvey. 2000. Partial characterization of a genomic island associated with the multidrug resistance region of Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 189:285-291. [DOI] [PubMed] [Google Scholar]
- 7.Briggs, C. E., and P. M. Fratamico. 1999. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, H. J., H. W. Stokes, and R. M. Hall. 1996. The integrons In0, In2, and In5 are defective transposon derivatives. J. Bacteriol. 178:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carattoli, A., E. Filetici, L. Villa, A. M. Dionisi, A. Ricci, and I. Luzzi. 2002. Antibiotic resistance genes and Salmonella genomic island 1 in Salmonella enterica serovar Typhimurium isolated in Italy. Antimicrob. Agents Chemother. 46:2821-2828, 430-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cloeckaert, A., S. Baucheron, G. Flaujac, S. Schwarz, C. Kehrenberg, J.-L. Martel, and E. Chaslus-Dancla. 2000. Plasmid-mediated florfenicol resistance encoded by the floR gene in Escherichia coli isolated from cattle. Antimicrob. Agents Chemother. 44:2858-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cloeckaert, A., K. Sidi Boumedine, G. Flaujac, H. Imberechts, I. D'Hooghe, and E. Chaslus-Dancla. 2000. Occurrence of a Salmonella enterica serovar Typhimurium DT104-like antibiotic resistance gene cluster including the floR gene in S. enterica serovar Agona. Antimicrob. Agents Chemother. 44:1359-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cloeckaert, A., and S. Schwarz. 2001. Molecular characterization, spread and evolution of multidrug resistance in Salmonella enterica serotype Typhimurium DT104. Vet. Res. 32:301-310. [DOI] [PubMed] [Google Scholar]
- 13.Conza, J. R., J. A. Ayala, P. Power, M. Mollerach, and G. Gutkind. 2002. Novel class 1 integron (InS21) carrying blaCTX-M-2 in Salmonella enterica serovar Infantis. Antimicrob. Agents Chemother. 46:2257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doublet, B., P. Butaye, H. Imberechts, J. M. Collard, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Salmonella Agona harboring genomic island 1-A. Emerg. Infect. Dis. 10:756-758. [DOI] [PMC free article] [PubMed]
- 15.Doublet, B., R. Lailler, D. Meunier, A. Brisabois, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2003. Variant Salmonella genomic island 1 antibiotic resistance gene cluster in Salmonella enterica serovar Albany. Emerg. Infect. Dis. 9:585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fluit, A. C., and F. J. Schmitz. 1999. Class 1 integrons, gene cassettes, mobility, and epidemiology. Eur. J. Microbiol. Infect. Dis. 18:761-770. [DOI] [PubMed] [Google Scholar]
- 17.Galas, D. J., and M. Chandler. 1989. Bacterial insertion sequences, p. 109-162. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 18.Glynn, M. K., C. Bopp, W. Dewitt, P. Dabney, M. Mokhtar, and F. J. Angulo. 1998. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N. Engl. J. Med. 338:1333-1338. [DOI] [PubMed] [Google Scholar]
- 19.Hancock, D., T. Besser, J. Gay, D. Rice, M. Davis, and C. Gay. 2000. The global epidemiology of multiresistant Salmonella enterica serovar Typhimurium DT104, p. 217-243. In C. Brown and C. Bolin (ed.), Emerging diseases of animals. American Society for Microbiology, Washington, D.C.
- 20.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Members of the SFM Antibiogram Committee. 2003. Comité de l'antibiogramme de la société française de microbiologie report 2003. Int. J. Antimicrob. Agents 21:364-391. [DOI] [PubMed] [Google Scholar]
- 22.Meunier, D., D. Boyd, M. R. Mulvey, S. Baucheron, C. Mammina, A. Nastasi, E. Chaslus-Dancla, and A. Cloeckaert. 2002. Salmonella enterica serotype Typhimurium DT 104 antibiotic resistance genomic island I in serotype Paratyphi B. Emerg. Infect. Dis. 8:430-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsons, Y., R. M. Hall, and H. W. Stokes. 1991. A new trimethoprim resistance gene, dhfrX, in the In7 integron of plasmid DGO100. Antimicrob. Agents Chemother. 35:2436-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Partridge, S. R., H. J. Brown, H. W. Stockes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Partridge, S. R., and R. M. Hall. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poppe, C., N. Smart, R. Khakhria, W. Johnson, J. Spika, and J. Prescott. 1998. Salmonella typhimurium DT104: a virulent and drug-resistant pathogen. Can. Vet. J. 39:559-565. [PMC free article] [PubMed] [Google Scholar]
- 28.Ribot, E. M., R. K. Wierzba, F. J. Angulo, and T. J. Barrett. 2002. Salmonella enterica serotype Typhimurium DT104 isolated from humans, United States, 1985, 1990, and 1995. Emerg. Infect. Dis. 8:387-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabate, M., F. Navarro, E. Miro, S. Campoy, B. Mirelis, J. Barbe, and G. Prats. 2002. Novel complex sul1-type integron in Escherichia coli carrying blaCTX-M-9. Antimicrob. Agents Chemother. 46:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stokes, H. W., C. Tomaras, Y. Parsons, and R. M. Hall. 1993. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid 30:39-50. [DOI] [PubMed] [Google Scholar]
- 31.Tauxe, R. V. 1999. Salmonella Enteritidis and Salmonella Typhimurium DT104: successful subtypes in the modern world, p. 37-53. In W. M. Scheld, W. A. Craig, and J. M. Hughes (ed.), Emerging infections 3. American Society for Microbiology, Washington, D.C.
- 32.Threlfall, E. J. 2000. Epidemic Salmonella Typhimurium DT104-a truly international multiresistant clone. J. Antimicrob. Chemother. 46:7-10. [DOI] [PubMed] [Google Scholar]
- 33.Turlan, C., and M. Chandler. 1995. IS1-mediated intramolecular rearrangements: formation of excised transposon circles and replicative deletions. EMBO J. 14:5410-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valentine, C. R., M. J. Heinrich, S. L. Chissoe, and B. A. Roe. 1994. DNA sequence of direct repeats of the sul1 gene in plasmid pSa. Plasmid 32:222-227. [DOI] [PubMed] [Google Scholar]
- 35.Villa, L., P. Visca, F. Tosini, C. Pezella, and A. Carattoli. 2002. Composite integron array generated by insertion of an ORF341-type integron within a Tn21-like element. Microb. Drug Res. 8:1-8. [DOI] [PubMed] [Google Scholar]
- 36.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]