Abstract
Large-scale cancer data sets such as The Cancer Genome Atlas (TCGA) allow researchers to profile tumors based on a wide range of clinical and molecular characteristics. Subsequently, TCGA-derived gene expression profiles can be analyzed with the Connectivity Map (CMap) to find candidate drugs to target tumors with specific clinical phenotypes or molecular characteristics. This represents a powerful computational approach for candidate drug identification, but due to the complexity of TCGA and technology differences between CMap and TCGA experiments, such analyses are challenging to conduct and reproduce. We present Cancer in silico Drug Discovery (CiDD; scheet.org/software), a computational drug discovery platform that addresses these challenges. CiDD integrates data from TCGA, CMap and Cancer Cell Line Encyclopedia (CCLE) to perform computational drug discovery experiments, generating hypotheses for the following three general problems: 1) determining whether specific clinical phenotypes or molecular characteristics are associated with unique gene expression signatures, 2) finding candidate drugs to repress these expression signatures, and 3) identifying cell lines that resemble the tumors being studied for subsequent in vitro experiments. The primary input to CiDD is a clinical or molecular characteristic. The output is a biologically annotated list of candidate drugs and a list of cell lines for in vitro experimentation. We applied CiDD to identify candidate drugs to treat colorectal cancers harboring mutations in BRAF. CiDD identified EGFR and proteasome inhibitors, while proposing five cell lines for in vitro testing. CiDD facilitates phenotype-driven, systematic drug discovery based on clinical and molecular data from TCGA.
Keywords: Colorectal cancer, drug screening, next-generation sequencing, systems biology, gene expression, Connectivity Map
Introduction
Selection of targeted therapies for cancer drug development has traditionally been based on the presence or absence of specific somatic mutations and this has been shown to be an effective strategy to improve patient outcomes (1–4). However, a large number of targeted drugs and other compounds that have anti-tumor properties have not been linked to specific mutations, or biomarkers, that could be used to predict their selective efficacy (5). Although next-generation sequencing (NGS) allows researchers to rapidly and comprehensively profile tumor mutations, the vast majority of these data have not been useful in the clinical setting since only a small number of mutations have been used to inform prognosis or guide therapeutic decisions (6–8).
Several computational approaches exist and have been implemented to predict the functional impact of mutations, and even to predict whether a specific mutation is a driver of the carcinogenesis process, based on several factors such as evolutionary conservation, predicted effects on protein structure and observed recurrence in existing cancer data sets (9–11). However, these computational predictions provide little insight into how cellular processes are altered as a consequence of the mutations. One strategy to assess whether or not specific mutations are influential on cellular processes is to determine whether or not a mutation induces a signature of gene expression changes (12). Gene expression signatures associated with an individual mutation could then be examined to characterize its cellular impact (13) and the signature could be used as a target for candidate drug therapies (14). We have developed the Cancer in silico Drug Discovery (CiDD) platform for the purposes of characterizing tumors with specific mutations, or more generally tumors with specific clinicopathological or molecular characteristics, based on their putative effects on gene expression, and to identify candidate drugs to treat these tumors.
Here, we describe the general framework and integrated data sets of this novel platform. CiDD has been designed to generate hypotheses for the following three general problems: 1) to determine if particular clinical or molecular characteristics are associated with unique gene expression signatures; 2) to find candidate drugs to treat specific tumor subgroups based on these expression changes; and 3) to identify cell lines that resemble the tumors being studied for subsequent in vitro experimentation. In addition, to illustrate the use of CiDD, we have applied it to a clinically relevant context in cancer drug development. We report the in silico identification of candidate drug therapies for colorectal cancers (CRCs) harboring the BRAF V600E mutation. Approximately 10% of CRCs harbor the BRAF V600E mutation, which confers a poor prognosis and presents a therapeutic challenge (4,15). We describe the analyses performed with CiDD that have identified novel targets for BRAF mutant CRCs and drugs such as EGFR inhibitors that have already shown activity at the pre-clinical level in targeting this tumor subtype (4).
Materials and Methods
CiDD is a systematic drug discovery platform that integrates and analyzes large-scale cancer data sets with the primary goal of identifying candidate drugs and cell lines to be validated experimentally in vitro (see Figure 1). The core data sets used by CiDD include The Cancer Genome Atlas (TCGA), the Connectivity Map (CMap) and the Cancer Cell Line Encyclopedia (CCLE). CiDD is purely computational and depends on publicly available clinical and experimental datasets, as well as annotation databases. CiDD is written in Python, has R package dependencies and is command-line driven allowing it to be integrated into bioinformatics pipelines. The software and code are freely available at http://scheet.org/software.
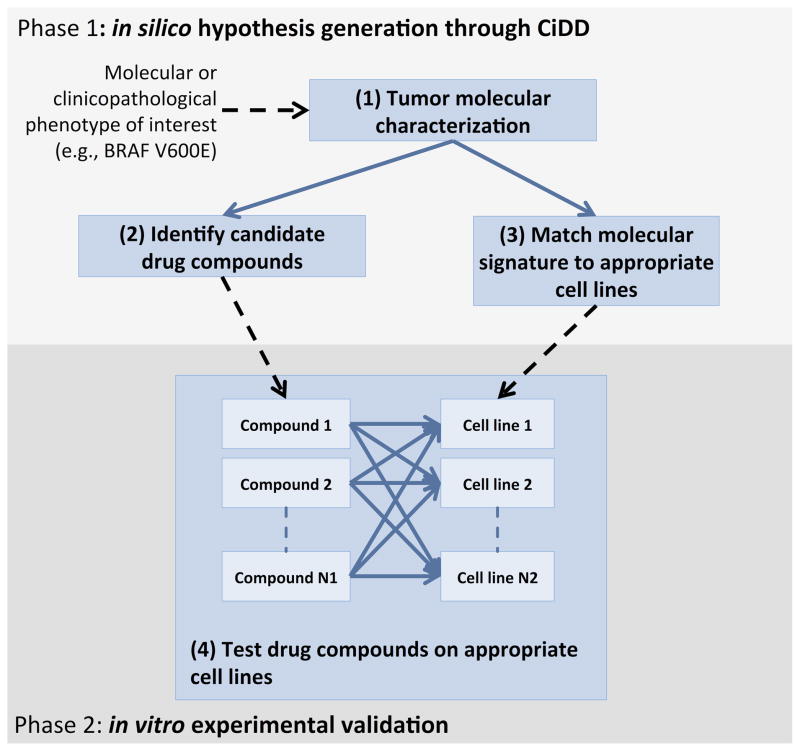
Figure 1.
A CiDD analysis produces a list of candidate drugs to treat tumors with the molecular or clinicopathological phenotype of interest and a list of cell lines that are representative of the phenotype of interest.
Data assembly
Required experimental data sets for performing CiDD analyses are TCGA (16) and CMap (14). CCLE (17) is required to identify cell-lines for subsequent experimentation. TCGA includes clinical, mutation and gene expression data for thousands of samples across multiple cancer types. CiDD provides commands to download, query and analyze these data. CMap is a collection of gene expression data for cell lines treated with small molecules paired with pattern-matching algorithms that attempt to identify biologically functional connections between drugs and gene expression profiles (14). CiDD utilizes CMap build 02, which contains more than 7,000 expression profiles representing the effects of 1,309 compounds. CCLE provides molecular profiles for 947 cancer cell lines which include DNA copy number, gene expression and DNA mutation data (17).
The experimental data from CMap consists of rank-based gene expression values from the Affymetrix HG-U133A microarray. Thus, CMap is designed for the analysis of Affymetrix gene expression data only, which hinders using CMap with gene expression data collected from non-Affymetrix platforms. To overcome this limitation, CiDD transforms bulk-downloaded CMap data from Affymetrix probe-based rank values to Entrez gene-based ranks. Gene-based ranks are determined by taking the mean probe rank for each gene, sorting the mean rank values and then assigning a rank for each gene based on the sorted values. This allows results from RNA sequencing and Agilent microarray technologies, such as those provided by TCGA, to be analyzed with the drug-perturbed data of CMap in a standardized way at the gene level. A similar strategy has been applied in the R package gCMAP (18) that allows users to query CMap using Affymetrix probe identifiers or gene symbols. Gene expression signatures derived from both Agilent microarrays and RNA sequencing have identified validated candidate drugs when analyzed with the Affymetrix-based drug signatures of CMap (19–21) demonstrating the feasibility of a cross-platform approach.
CiDD also uses annotation datasets, which include the Molecular Signatures Database (MSigDB) (13) for characterizing gene sets and drug databases including DrugBank (22), Matador (23) and KEGG Drug (24) for annotating candidate drugs. These databases provide information such as drug pharmacology, gene and pathway targets to make the CiDD’s drug reports more informative. Public data from TCGA are automatically downloaded by CiDD, while data from CMap, CCLE and MSigDB require registration at their respective websites prior to downloading. Upon download, CiDD automatically prepares and manages data sets for drug discovery analyses. Descriptions of these data sets are provided in Supplementary Methods.
CiDD workflow
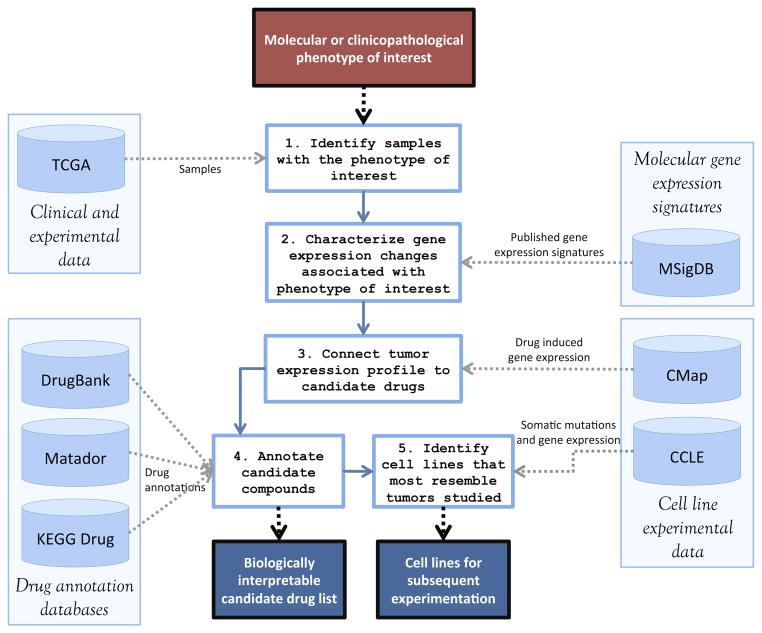
A common workflow using CiDD is illustrated in Figure 2. Initially, a CiDD project based on a TCGA cancer type is created and clinical, mutation and gene expression data for TCGA samples are automatically downloaded. For an analysis, CiDD first identifies TCGA samples for use in computational experiments based on user-defined clinicopathological phenotypes or molecular characteristics, such as specific gene mutations, microsatellite instability status, tumor stage, or a variety of other patient or tumor characteristics reported through TCGA projects. Based on the defined phenotype, CiDD identifies 2 classes of samples to compare. For a mutation-based phenotype, CiDD establishes one class containing samples with a defined mutation or set of mutations and a second class containing samples that are wild-type for the genes of interest. For a clinical phenotype, the user specifies both classes explicitly, such as two classes corresponding to microsatellite instable and stable tumors. CiDD attempts to identify a gene expression signature that is associated with the defined patient or tumor characteristic. If a gene expression signature exists for the phenotype of interest, that signature is characterized with MSigDB gene sets and the signature is used to identify candidate drugs through pattern-matching algorithms proposed by CMap. Subsequently, CiDD characterizes candidate drugs using databases such as DrugBank, Matador and KEGG Drug. Finally, CiDD identifies candidate cell lines on which to test the drugs in vitro by analyzing data from CCLE. The primary results of a CiDD execution are a biologically annotated candidate drug list and candidate cell lines for subsequent drug experimentation.
Figure 2.
A CiDD workflow shows the 5 main steps of an analysis with their data set dependencies. Input to this workflow includes point mutations (such as BRAF V600E) or other molecular and clinical phenotypes of interest paired with a cancer type (e.g., CRC). The primary output includes a candidate drug list that has been annotated with drug databases and a list of cell lines for subsequent experimentation.
Gene signature identification
TCGA provides gene expression data from Agilent microarrays, Illumina GA RNA sequencing and Illumina HiSeq RNA sequencing. The data type to analyze can be specified as a parameter to CiDD. By default, CiDD will choose the technology that provides data for the largest number of samples with the phenotype of interest. Using the R package Limma (25) which is designed for both microarray and RNA sequencing differential expression analyses, CiDD identifies up- and down-regulated genes. CiDD characterizes these results with biological pathways by performing gene set tests using the piano Bioconductor package (26), while using gene sets defined by MSigDB.
Generation of a k-top scoring pairs (k-TSP) classifier
For generating a classifier that is robust across gene expression technologies, CiDD takes a non-parametric approach to classification and adopts an extension of the top scoring pairs (TSP) method (27). Using the R package ktspair (28), CiDD generates a k-TSP classifier for predicting the status of the phenotype of interest on independent samples. The k-TSP algorithm is described in Supplementary Methods.
Candidate drug identification
CiDD connects gene expression changes associated with the phenotype of interest with candidate drug compounds that induce a negatively correlated (or “negatively connected”) gene expression profile. CiDD compares the phenotype gene expression changes, termed a query signature, to rank-based gene expression profiles induced by CMap compounds. To compare rank-based gene expression profiles, CiDD implements nonparametric pattern-matching algorithms based on the Kolmogorov-Smirnov statistic as described by Lamb et al (14). An enrichment score ranging from −1 to +1 provides a measure of the negative or positivity connectivity of a drug to the phenotype of interest. A permutation p-value provides a measure of significance for the enrichment scores. These algorithms and the resulting metrics are described in Supplementary Methods.
Cell line identification
CiDD first selects CCLE cell lines based on user-specified tissue types. Then, CiDD optionally identifies cell lines that contain user-specified mutations by interrogating CCLE mutation data derived from either targeted sequencing of common cancer genes or from Oncomap 3.0, which is a SNP array that genotypes samples at known cancer-related sites. Finally, CiDD runs its k-TSP classifier on CCLE gene expression data to predict if a cell line’s gene expression profile is representative of the phenotype being studied. Cell lines that meet these criteria are reported as candidates for use in subsequent drug experiments.
Results
We applied CiDD to identify candidate drugs to treat CRCs harboring BRAF V600E mutations using mutation and RNA-sequencing data from the TCGA colon and rectum projects. We also identified cell lines from CCLE that are representative of colorectal tumors with BRAF mutations, thus making them candidates for in vitro drug testing. We refer to these analyses as the TCGA-derived analyses. The detailed commands to re-run these analyses are provided in Supplementary Methods. We then compared our systematic TCGA-derived analyses generated from CiDD with analyses performed using a previously published gene expression signature for BRAF V600E generated from CRC samples of the PETACC3 (Pan-European Trial Adjuvant Colon Cancer 3) clinical trial (15). We refer to these published gene expression analyses as the PETACC3-derived analyses.
Identification of a BRAF V600E CRC gene expression signature
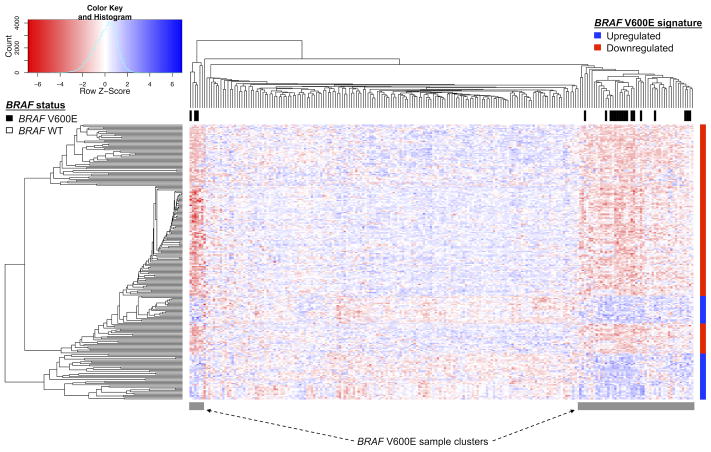
We used CiDD to identify 20 TCGA CRC samples with a BRAF V600E mutation and 149 BRAF wild-type samples with available Illumina GA RNA sequencing data. CiDD identified 63 up-regulated and 170 down-regulated genes (log fold-change >= 2 and Benjamini Hochberg adjusted P-value <= 0.05) that generated a clustering of samples representative of BRAF mutation status as shown in Figure 3.
Figure 3.
CiDD-generated heat map and clustering of BRAF V600E mutated CRCs based on TCGA Illumina GA RNA sequencing data. Differentially expressed genes comparing BRAF V600E and BRAF wildtype samples were identified using the Limma package in R and required to have a Benjamini Hochberg adjusted p-value <= 0.05 and a minimum log fold change >= 2. Hierarchical clustering of the samples and genes were performed using hclust with a “pearson” distance measure in R. The BRAF V600E gene expression signature is represented with the vertical colored bar on the right side of the figure, where red represents down-regulated genes and blue up-regulated genes. BRAF V600E mutant samples all reside within 2 sample clusters of the heatmap, which suggests that the BRAF V600E signature captures the gene expression response of BRAF V600E mutations.
We identified pathways associated with the BRAF signature through CiDD using Wilcoxon-based gene set tests (26). For assessing significance of the gene set tests, CiDD performed 1000 runs of the differential expression analyses, permuting the BRAF mutant status of samples within each run. Fifteen KEGG gene sets were associated with the BRAF V600E status (FDR adjusted P-value <= 0.05). To incorporate PETACC3-derived pathways as part of the pathway analysis, a list of the top 20 pathways based on an average ranking within the TCGA and PETACC3-derived pathway lists is provided in Table 1. Because raw gene expression data was not available for the PETACC3-derived signature, gene set tests were not performed. Instead, for the PETACC3-derived analysis, hypergeometric tests were applied to identify KEGG pathways enriched with genes from this signature. Twenty-seven KEGG pathways are enriched with genes from the PETACC3-derived signature (P-value <= 0.05). The pathway ordering in Table 1 reflects the average of the P-value ranks within each set (complete results are provided in Supplementary Results). These pathways are consistently related to CRC biology such as the top ranked pathway (“Colorectal Cancer”) and other pathways related to TGFβ signaling (“TGF Beta Signaling Pathway”), which are well known for their role in CRC. Additionally, it is known that the BRAF gene plays a role in controlling cellular proliferation and differentiation through regulation of the MAP kinase signaling pathway (29), and the “MAPK Signaling Pathway” is also represented in the top ranked pathways.
Table 1. The top 20 ranked pathways associated with BRAF V600E status based on systematic TCGA gene expression analyses presented with those derived from the independent PETACC3-based analyses.
The table is ordered by the overall rank of each pathway where the overall rank represents an average rank across both the TCGA- and PETACC-derived analyses. P-values and ranks for pathways associated for both the TCGA- and PETACC-derived analyses are shown. These pathways are consistently related to CRC biology such as the top-ranked pathway “Colorectal Cancer” and the “TGF Beta Signaling Pathway” in addition to the “MAPK Signaling Pathway” which is known to play a role in BRAF-mutant CRC.
| Pathways | TCGA P-value | PETACC3 P-value | TCGA rank | PETACC3 rank | Average rank | Overall rank |
|---|---|---|---|---|---|---|
| Colorectal Cancer | 0.021 | 0.003 | 9 | 4 | 6.5 | 1 |
| Bladder Cancer | 0 | 0.017 | 2 | 18 | 10 | 2 |
| Pathways in Cancer | 0.05 | 0.004 | 15 | 6 | 10.5 | 3 |
| Chemokine Signaling Pathway | 0.04 | 0.012 | 11 | 16 | 13.5 | 4 |
| JAK-STAT Signaling Pathway | 0.053 | 0.006 | 20 | 11 | 15.5 | 5 |
| Axon Guidance | 0.057 | 0.003 | 26 | 5 | 15.5 | 6 |
| FC Epsilon RI Signaling Pathway | 0.021 | 0.05 | 7 | 27 | 17 | 7 |
| TGF Beta Signaling Pathway | 0.066 | 0.001 | 34 | 2 | 18 | 8 |
| Dorso Ventral Axis Formation | 0.057 | 0.006 | 25 | 12 | 18.5 | 9 |
| Peroxisome | 0.066 | 0.006 | 33 | 10 | 21.5 | 10 |
| MAPK Signaling Pathway | 0.057 | 0.032 | 24 | 23 | 23.5 | 11 |
| ABC Transporters | 0.068 | 0.018 | 37 | 19 | 28 | 12 |
| ERBB Signaling Pathway | 0.069 | 0.008 | 46 | 14 | 30 | 13 |
| FC Gamma R Mediated Phagocytosis | 0.062 | 0.069 | 30 | 31 | 30.5 | 14 |
| Tryptophan Metabolism | 0.037 | 0.16 | 10 | 52 | 31 | 15 |
| B Cell Receptor Signaling Pathway | 0.083 | 0 | 61 | 1 | 31 | 16 |
| Prion Diseases | 0.04 | 0.144 | 14 | 49 | 31.5 | 17 |
| Epithelial Cell Signaling in Helicobacter Pylori Infection | 0.068 | 0.039 | 39 | 24 | 31.5 | 18 |
| T Cell Receptor Signaling Pathway | 0.06 | 0.081 | 28 | 37 | 32.5 | 19 |
| Neuroactive Ligand Receptor Interaction | 0.021 | 0.234 | 3 | 67 | 35 | 20 |
Finally, we used CiDD to identify an 11-pair k-TSP classifier for predicting the BRAF V600E status of independent samples using the TCGA data set. The classifier gene pairs are listed in Supplementary Table S1.
Validation of the TCGA-derived gene-pair classifier for predicting BRAF V600E status
In order to validate the TCGA-derived gene expression analyses, we compared the performance of a previously reported BRAF V600E gene expression classifier derived from the PETACC3 clinical trial (15) against the gene expression classifier that we identified from the TCGA data set.
The PETACC3-derived gene expression signature consists of 193 up-regulated and 92 down-regulated probes. These probes correspond to 224 unique genes. The research group also developed a 64-gene TSP classifier (these genes are defined in Supplementary Table S2) based on Affymetrix probe IDs for predicting the BRAF V600E status of CRCs. We translated these probe IDs to Entrez gene IDs so the classifier could be applied to RNA sequencing and Agilent test data sets. To assess the robustness of their gene expression results, we applied the gene-based PETACC3-derived classifier to TCGA samples that were retrieved and annotated with BRAF mutation statuses by CiDD. When applied to TCGA RNA sequencing data, the PETACC3-derived classifier resulted in 93.3% sensitivity and 83.5% specificity for detecting BRAF V600E samples.
To assess the quality of the systematic TCGA-derived classifier generated by CiDD, we compared the performance of the TCGA- and PETACC3-derived classifiers on 3 independent data sets (see Table 2) – two have been previously published and are available in the Gene Expression Omnibus (30,31) and the third is the CCLE data set. The sensitivity and specificity of both classifiers are comparable on the GSE35896 and GSE42284 data sets with the PETACC3-derived classifier exhibiting small improvements in specificity. The PETACC3-derived classifier achieved 100% sensitivity but only 30% specificity for BRAF status prediction on the CCLE large intestine data set. The TCGA-derived classifier had lower sensitivity (71%) but achieved better specificity (62%). These results suggest that the systematically obtained BRAF V600E classifier from CiDD is comparable to the published PETACC3-derived signature and that the TCGA-derived classifier may even have improved specificity for distinguishing BRAF wild-type cell lines from the BRAF mutant cell lines.
Table 2. Performance of the TCGA- and PETACC3-derived BRAF V600E CRC classifiers when applied to independent gene expression data sets.
The sensitivity and specificity of both classifiers are comparable with the PETACC3-derived classifier exhibiting small improvements in specificity on the GSE35896 and GSE42284 data sets. The TCGA-derived classifier had lower sensitivity (71%) but achieved better specificity (62%) on the CCLE data set. These results suggest that the systematically obtained BRAF V600E classifier from CiDD is comparable to the published PETACC3-derived signature and that the TCGA-derived classifier may even have improved specificity for distinguishing BRAF wild-type cell lines from the BRAF mutant cell lines.
| Data Set | TCGA-derived classifier | PETACC3-derived classifier | ||
|---|---|---|---|---|
| sensitivity | specificity | sensitivity | specificity | |
| GSE35896 (n = 62) (Affymetrix U133 Plus 2.0 Array) | 4/6 (0.67) | 39/56 (0.70) | 4/6 (0.67) | 45/56 (0.80) |
| GSE42284 (n = 178) (Agilent Homo sapiens 37K DiscoverPrint_19742) | 33/36 (0.92) | 91/142 (0.64) | 33/36 (0.92) | 107/142 (0.75) |
| CCLE LARGE_INTESTINE (n = 57) (Affymetrix U133 Plus 2.0 Array) | 5/7 (0.71) | 31/50 (0.62) | 7/7 (1.00) | 15/50 (0.30) |
Candidate drug therapies for BRAF V600E CRC
Using both TCGA and PETACC3-derived gene expression signatures, CiDD identified candidate drugs to treat BRAF V600E CRCs. Drugs with a negative enrichment score and a permutation P-value less than 0.1 using the TCGA and PETACC3-derived gene expression signatures are listed in Table 3 and Supplementary Table S3 respectively. Three compounds, Gefitinib, MG-262 and Trapidil, were identified in both lists. Independent research groups have recently shown that EGFR inhibitors such as Gefitinib and proteosome inhibitors such as MG-262 are effective drugs for treatment of colorectal tumors with BRAF mutations (4,32). Trapidil is a novel candidate drug that inhibits phosphodiesterase and TXA2. The full candidate drug reports are provided in Supplementary Results.
Table 3. Candidate drug compounds identified systematically by CiDD for BRAF V600E CRC based on the TCGA-derived gene expression signature.
Nine drugs were identified having both a negative enrichment score and a maximum permutation P-value of 0.1. Three of these drugs (*) were also identified using the PETACC3-derived gene expression signature.
| Compound | Enrichment score | Permutation p-value | Specificity |
|---|---|---|---|
| gefitinib* | −0.995 | 0.016 | 0.000 |
| 2-deoxy-D-glucose | −0.977 | 0.051 | 0.022 |
| 5286656 | −0.967 | 0.075 | 0.038 |
| yohimbic acid | −0.901 | 0.003 | 0.000 |
| amrinone | −0.884 | 0.001 | 0.003 |
| trapidil* | −0.852 | 0.004 | 0.016 |
| mycophenolic acid | −0.735 | 0.024 | 0.048 |
| withaferin A | −0.679 | 0.026 | 0.054 |
| MG-262* | −0.656 | 0.073 | 0.141 |
Cancer cell lines that most resemble BRAF V600E CRC
In order to identify candidate cell lines for in vitro testing, CiDD analyzed data from the CCLE. From 947 cell lines in the CCLE, CiDD identified 48 large intestine samples that we consider to be representative of colorectal tumors. Then CiDD reduced this number to 7, representing those large intestine cell lines that have BRAF V600E mutations. Using the 11 gene-pair k-TSP classifier generated by CiDD, 5 of these cell lines were predicted to be BRAF V600E on the basis of having similar gene expression profiles to the TCGA BRAF V600E mutated CRCs. The five identified cell lines include RKO, SNUC5, CL34, COLO205 and HT29. OUMS23 and SW1417 are the two BRAF V600E large intestine cell lines that are predicted to be BRAF wild-type by the TCGA-derived classifier.
Discussion
As genomic technologies have ushered in the potential for targeted drug development, large-scale public genomic databases have matured in size, scope and information content to complement this effort. It is thus advantageous, and indeed possibly necessary, to apply computational genomics to inform the drug discovery process. While subgroup classification for prognostic assessment and therapeutic planning has been applied clinically for decades, especially among hematologic malignancies and in some solid tumors such as breast cancers, other tumor types such as CRCs appear phenotypically homogenous and are thus clinically indistinguishable. In order to reveal subclasses for these tumors and to generalize their genome-based classification, the use of genetic and transcriptomic analyses may prove essential. Systems biology tools such as CMap, and we believe CiDD, help fill this need of identifying candidate interventions that target specific pathways deregulated in these tumor subclasses. In this regard, CMap provided the original approach to guide drug development based on transcriptomic data. CiDD is taking this approach further by extending CMap with the clinical and molecular data of TCGA along with the high-throughput experiments of the CCLE for the purposes of systematic cancer drug discovery. While current public resources such as that of TCGA are impressive, they are likely just a beginning. The basic logic of CiDD naturally extends to utilization of forthcoming, larger-scale databases from drug perturbation experiments and genetic and transcriptomic sequencing of tumors of a wider array of sizes and associated clinical outcomes.
We believe CiDD is the first framework that supports systematic drug discovery based on user-specified TCGA clinical phenotypes and molecular characteristics. CiDD allows researchers to perform the following: (1) assess whether or not a mutation or clinical phenotype is associated with a gene expression signature, (2) identify candidate drugs to target this gene expression signature, and (3) identify cell lines for subsequent in vitro drug experimentation. We have illustrated the power of such an approach in a meaningful application to CRCs with somatic mutations in BRAF. CiDD also offers utility to researchers simply wishing to interrogate and organize TCGA data, as it can be applied to create an inventory of available TCGA data with particular clinical or genomic features, such as available data sets or patients with particular mutations, independently of its drug identification capabilities.
One of the most crucial steps in the BRAF V600E analysis was identifying a gene expression signature associated with the BRAF V600E mutation and generating a classifier for predicting mutation status. In both of these cases, we showed that the signature and classifier of the CiDD framework are comparable to those identified from the published PETACC3-derived analyses (15). Similarly to the PETACC3-derived signature and classifier, the CiDD-generated signature was composed of genes representative of known pathways associated with the BRAF V600E mutation, most notably the “MAPK Signaling Pathway”, and the performance of the classifier on independent data sets generated from orthogonal gene expression technologies showed robustness. The advantage of CiDD analyses is that they are systematic studies of generally available datasets. We did not have to generate any of our own experimental data, and the gene expression analyses can be relatively easily replicated and repeated for other mutation or clinical phenotypes.
Once we validated the gene expression signature, we used CiDD to identify candidate compounds for tumors harboring the well-known BRAF V600E mutation. Since the initial communication of the presence of mutations in the kinase BRAF in cancer (33), activating mutations have been described in several malignancies with different frequencies such as hairy cell leukemia (100%), melanoma (50–60%), thyroid carcinoma (30–50%) and CRC (10%) (34). The most frequently identified mutation is a valine-to-glutamic acid substitution at codon 600 (V600E) that activates the signaling cascade downstream of MEK and ERK (33). Other mutations have been found at the same codon and are considered equivalent in terms of oncogenic activation (34). Therefore, substantial efforts were invested on developing ATP-competitive RAF inhibitors such as Vemurafenib and Dabrafenib to specifically target the MAPK pathway. Yet, the clinical success of BRAF inhibition has been variable and highly dependent on the tumor context. In this regard, Vemurafenib has demonstrated improvement in survival in patients diagnosed with stage IV melanomas harboring the BRAF V600E mutation (35). However, this degree of clinical benefit has not been observed in the same molecular context in CRCs and papillary thyroid cancers (36). This is probably secondary to the intrinsic mechanisms of resistance to BRAF inhibition that are specific to the tumor context (34). BRAF mutations in the context of metastatic CRCs have been associated with poor prognosis and an aggressive disease course contrasting with cases in early stages. In addition, they have a characteristic clinical phenotype consistent with older age at diagnosis, female gender, right-sided location and the presence of high levels of microsatellite instability (37,38).
Two strategies have been suggested to overcome the primary resistance to BRAF inhibition in CRC biology. One strategy that has been supported independently by two different groups is the inhibition of the EGFR pathway by using monoclonal antibodies against EGFR (such as Cetuximab) or kinase inhibitors (such as Gefinitib and Erlotinib) in combination with BRAF inhibitors. EGFR is activated by feedback mechanisms upon BRAF inhibition, thus reactivating ERK via RAS and CRAF, therefore combinations of EGFR and BRAF inhibition will synergize in terms of activity (4,39–41). The second strategy is based on targeting the proteasome pathway. This has demonstrated specific activity against BRAF V600E mutant CRC cell lines and tumor xenografts. This set of experiments was performed using classical (Bortezomib) and novel (Carfilzomib) proteasome inhibitors and demonstrated similar activity. However, as opposed to EGFR feedback, proteasome inhibition seems to function independently of BRAF inhibition (32). CiDD has been able to identify both types of compounds (EGFR and proteasome inhibitors) as candidate drugs through an agnostic approach, thus providing a biological validation of the value of CiDD as a screening tool to identify novel drugs to be tested and further developed in specific tumor subtypes.
CiDD also addresses the important issue of identifying appropriate publicly available cell lines as pre-clinical models for cancer researchers. Systematic comparisons between cancer cell lines and tumor samples from human tissues have documented substantial differences between the two, emphasizing the importance of making genomically informed choices when identifying cell lines as pre-clinical models of a tumor subtype (42). The CCLE provides mutation and gene expression data that allow CiDD to make these molecularly informed decisions in selecting cell lines. In our BRAF V600E analysis, CiDD identified 7 large intestine cell lines harboring the BRAF V600E mutation. However, only 5 of the 7 were predicted to be BRAF V600E based on CiDD’s gene expression classifier, suggesting heterogeneity among the BRAF V600E mutated cell lines. CiDD prioritized those cell lines into 2 groups for in vitro testing, proposing that 5 of the 7 BRAF V600E mutated large intestine cell lines more closely resemble the TCGA CRC BRAF V600E tumors at a gene expression level. We note however, that there may be a more ideal strategy for obtaining cell lines for in vitro testing for researchers wishing to deviate from the use of publicly available cell lines. The use of isogenic cell lines in drug experiments has been shown to be very effective, thus allowing for direct association of the sensitivity of a drug with a specific mutation (43). As an example, in our BRAF mutant application, a researcher could obtain a colon cancer cell line that is wildtype for BRAF, then create a second identical cell line from this cell line except that it has a mutation in BRAF.
CiDD has some limitations that could restrict its application in specific situations. Primarily, CiDD is dependent on identifying a gene expression signature representative of a phenotype of interest. In some cases, there may be no gene expression signature associated with a clinical phenotype or mutation. In other clinical contexts, such as for rare mutations and infrequent clinical phenotypes, CiDD may not have the power to identify the true underlying gene expression signature associated with the phenotype, because CiDD is limited by the number of samples available in TCGA with that specific phenotype. In these rare-phenotype analyses, CiDD may fail to identify a statistically significant gene expression signature representative of the phenotype of interest. Researchers interested in rare clinical or molecular subgroups will need to consider alternative strategies for increasing their sample sizes. These strategies may include aggregating TCGA tumor types or grouping mutations or clinical phenotypes in biologically meaningful ways, such as aggregating rare mutations at a gene or pathway level to increase the sample size. The CiDD command that generates gene expression signatures based on defined mutations provides support for aggregating mutations by listing amino acid substitutions explicitly, by specifying types of mutations (such as nonsense mutations) or by defining sets of mutations based on gene and gene set membership. Additionally, the CiDD framework does not support the identification of candidate drug combinations to target tumor subtypes. CMap provides drug-perturbed data that were generated by applying compounds to cell lines one compound at a time. If future drug-perturbed data sets provide gene expression data of multiple compounds being applied to cell lines, incorporation of this data into CiDD should be relatively straightforward. As an alternative, the computational identification of multiple interacting candidate drugs based on current data sets is a potential area for future CiDD development.
Of course, these limitations apply more generally for these difficult scenarios and are not unique to CiDD. In fact, CiDD helps address these limitations by being easy to run and repeat to test multiple hypotheses quickly. Further, CiDD is a framework rather than a specific method per se. As public databases evolve and expand, and as robust statistical methodologies mature for cross-platform expression-based signature identification, CiDD can be adapted to incorporate these improved components. In this sense, what we have demonstrated here is a “lower bound” of sorts, and we expect more powerful findings to emerge from such efficient systems-based computation. Finally, the field of gene expression analysis, particularly for identifying signatures of cancer subtypes, has been criticized for failing to adhere to standards of repeatability (44). Our software facilitates repeatability and even enables replication of findings with external data sets. In all of these aspects, we expect the community of cancer genomic researchers to benefit from, and further contribute to, this framework.
Supplementary Material
Acknowledgments
Financial Information: This work was supported by the Schissler Foundation (F.A. San Lucas), the Conquer Cancer Foundation of the American Society of Clinical Oncology, Young Investigator Award (E. Vilar), by the National Institutes of Health grants 1R03CA176788-01A1 (E. Vilar), U24 CA143883 (P.A. Scheet), U01 GM 92666 (P.A. Scheet), R01HG005859 (F.A. San Lucas, J. Fowler, P.A. Scheet) and 1R01CA172670-01 (E.S. Kopetz), and by The University of Texas MD Anderson Cancer Center Core Support Grant.
The authors thank The Cancer Genome Atlas (TCGA) research network for publicly sharing their data. The software and results published here are in large part based upon data generated by the TCGA project, which was established by the NCI and NHGRI. Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at http://cancergenome.nih.gov.
Abbreviations
- CCLE
Cancer Cell Line Encyclopedia
- CiDD
Cancer in silico Drug Discovery
- CMap
Connectivity Map
- CRC
colorectal cancer
- MSigDB
Molecular Signatures Database
- PETACC
Pan-European Trial Adjuvant Colon Cancer
- TCGA
The Cancer Genome Atlas
- TSP
top scoring pairs
Footnotes
Conflicts of interest: The authors disclose no potential conflicts of interest.
References
- 1.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600 mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–14. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lievre A, Bachet J-B, Boige V, Cayre A, Le Corre D, Buc E, et al. KRAS Mutations As an Independent Prognostic Factor in Patients With Advanced Colorectal Cancer Treated With Cetuximab. J Clin Oncol. 2008 Jan 20;26:374–9. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, et al. KRAS Mutations and Primary Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012 Jan 26;483:100–3. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 5.McDermott U, Settleman J. Personalized Cancer Therapy With Selective Kinase Inhibitors: An Emerging Paradigm in Medical Oncology. J Clin Oncol. 2009 Oct 26;27:5650–9. doi: 10.1200/JCO.2009.22.9054. [DOI] [PubMed] [Google Scholar]
- 6.Rehm HL. Disease-targeted sequencing: a cornerstone in the clinic. Nat Rev Genet. 2013;14:295–300. doi: 10.1038/nrg3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen AR, Bedard PL. Clinical application of high-throughput genomic technologies for treatment selection in breast cancer. Breast Cancer Res. 2013;10:11. doi: 10.1186/bcr3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim T-M. Clinical applications of next-generation sequencing in colorectal cancers. World J Gastroenterol. 2013;19:6784. doi: 10.3748/wjg.v19.i40.6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter H, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, et al. Cancer-Specific High-Throughput Annotation of Somatic Mutations: Computational Prediction of Driver Missense Mutations. Cancer Res. 2009 Aug 4;69:6660–7. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010 Apr;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng PC. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003 Jul 1;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102:13550–5. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011 May 5;27:1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamb J. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science. 2006 Sep 29;313:1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 15.Popovici V, Budinska E, Tejpar S, Weinrich S, Estrella H, Hodgson G, et al. Identification of a Poor-Prognosis BRAF-Mutant-Like Population of Patients With Colon Cancer. J Clin Oncol. 2012 Mar 5;30:1288–95. doi: 10.1200/JCO.2011.39.5814. [DOI] [PubMed] [Google Scholar]
- 16.Muzny DM, Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012 Jul 18;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012 Mar 28;483:603–307. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandmann T, Kummerfeld SK, Gentleman R, Bourgon R. gCMAP: user-friendly connectivity mapping with R. Bioinformatics. 2013 Oct 15;30:127–8. doi: 10.1093/bioinformatics/btt592. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X-Z, Yin A-H, Lin D-J, Zhu X-Y, Ding Q, Wang C-H, et al. Analyzing Gene Expression Profile in K562 Cells Exposed to Sodium Valproate Using Microarray Combined with the Connectivity Map Database. J Biomed Biotechnol. 2012;2012:1–8. doi: 10.1155/2012/654291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinonen H, Nieminen A, Saarela M, Kallioniemi A, Klefstrom J, Hautaniemi S, et al. Deciphering downstream gene targets of PI3K/mTOR/p70S6K pathway in breast cancer. BMC Genomics. 2008;9:348. doi: 10.1186/1471-2164-9-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McArt DG, Dunne PD, Blayney JK, Salto-Tellez M, Van Schaeybroeck S, Hamilton PW, et al. Connectivity Mapping for Candidate Therapeutics Identification Using Next Generation Sequencing RNA-Seq Data. In: Stieger K, editor. PLoS ONE. Vol. 8. 2013. Jun 26, p. e66902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, et al. DrugBank 3.0: a comprehensive resource for “Omics” research on drugs. Nucleic Acids Res. 2010 Nov 8;39:D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunther S, Kuhn M, Dunkel M, Campillos M, Senger C, Petsalaki E, et al. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res. 2007 Dec 23;36:D919–D922. doi: 10.1093/nar/gkm862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2013 Nov 7;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smyth GK. Bioinformatics and computational biology solutions using R and Bioconductor. Springer; 2005. Limma: linear models for microarray data; pp. 397–420. [Google Scholar]
- 26.Varemo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013 Feb 26;41:4378–91. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan AC, Naiman DQ, Xu L, Winslow RL, Geman D. Simple decision rules for classifying human cancers from gene expression profiles. Bioinformatics. 2005 Aug 16;21:3896–904. doi: 10.1093/bioinformatics/bti631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Damond J. ktspair. 2013 Aug 29; Available from: http://www.inside-r.org/packages/cran/ktspair/docs/ordertsp.
- 29.Cantwell-Dorris ER, O’Leary JJ, Sheils OM. BRAFV600E: Implications for Carcinogenesis and Molecular Therapy. Mol Cancer Ther. 2011 Mar 8;10:385–94. doi: 10.1158/1535-7163.MCT-10-0799. [DOI] [PubMed] [Google Scholar]
- 30.Roepman P, Schlicker A, Tabernero J, Majewski I, Tian S, Moreno V, et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition: Molecular subtypes in colorectal cancer. Int J Cancer. 2014 Feb 1;134:552–62. doi: 10.1002/ijc.28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlicker A, Beran G, Chresta CM, McWalter G, Pritchard A, Weston S, et al. Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines. BMC Med Genomics. 2012;5:66. doi: 10.1186/1755-8794-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zecchin D, Boscaro V, Medico E, Barault L, Martini M, Arena S, et al. BRAF V600E is a determinant of sensitivity to proteasome inhibitors. Mol Cancer Ther. 2013 Oct 9;12:2950–61. doi: 10.1158/1535-7163.MCT-13-0243. [DOI] [PubMed] [Google Scholar]
- 33.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 34.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nat Med. 2013 Nov 7;19:1401–9. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- 35.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopetz S, Desai J, Chan E, Hecht JR, O’Dwyer J, Lee RG, et al. PLX4032 in metastatic colorectal cancer patients with mutant BRAF tumors. J Clin Oncol. 2010;28:3534. [Google Scholar]
- 37.Tanaka H, Deng G, Matsuzaki K, Kakar S, Kim GE, Miura S, et al. BRAF mutation, CpG island methylator phenotype and microsatellite instability occur more frequently and concordantly in mucinous than non-mucinous colorectal cancer. Int J Cancer. 2006 Jun 1;118:2765–71. doi: 10.1002/ijc.21701. [DOI] [PubMed] [Google Scholar]
- 38.Tran B, Kopetz S, Tie J, Gibbs P, Jiang Z-Q, Lieu CH, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer. 2011 Oct 15;117:4623–32. doi: 10.1002/cncr.26086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-Mediated Reactivation of MAPK Signaling Contributes to Insensitivity of BRAF-Mutant Colorectal Cancers to RAF Inhibition with Vemurafenib. Cancer Discov. 2012 Mar 1;2:227–35. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang H, Higgins B, Kolinsky K, Packman K, Bradley WD, Lee RJ, et al. Antitumor Activity of BRAF Inhibitor Vemurafenib in Preclinical Models of BRAF-Mutant Colorectal Cancer. Cancer Res. 2012 Feb 1;72:779–89. doi: 10.1158/0008-5472.CAN-11-2941. [DOI] [PubMed] [Google Scholar]
- 41.Mao M, Tian F, Mariadason JM, Tsao CC, Lemos R, Dayyani F, et al. Resistance to BRAF Inhibition in BRAF-Mutant Colon Cancer Can Be Overcome with PI3K Inhibition or Demethylating Agents. Clin Cancer Res. 2013 Feb 1;19:657–67. doi: 10.1158/1078-0432.CCR-11-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013 Jul;9:4. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samuels Y, Diaz LA, Schmidt-Kittler O, Cummins JM, DeLong L, Cheong I, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005 Jun;7:561–73. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 44.Baggerly K. Disclose all data in publications. Nature. 2010 Sep 23;467:401–401. doi: 10.1038/467401b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.