Abstract
The nucleotide sequences of the pncA genes within 55 multidrug-resistant pyrazinamide-resistant Mycobacterium tuberculosis clinical isolates were determined. Fifty-three out of the 55 isolates were pyrazinamidase (PZase) negative. Four strains contained a wild-type pncA gene, and PZase activity was undetectable in two of these strains. Seven of the 18 identified pncA mutations found have not been described in previous studies.
Portugal remains the country with the highest rate of notified cases of Mycobacterium tuberculosis in the European Union. In 2002, the Portuguese Health Authorities reported a tuberculosis (TB) incidence of 39.5 cases per 100,000 people nationwide and that 2.3% of primary TB cases involved multidrug resistance (MDR-TB cases) (Programa Nacional de Controlo da Tuberculose, Ponto da situação epidemiológica e de desempenho em 2002, available at the Direcção Geral de Saúde website [http://www.dgsaude.pt]). According to the National Laboratory Surveillance System for Resistance TB, 4,170 isolates from notified cases were tested for drug susceptibility between 2000 and 2001. Of these isolates, only 56.5% were tested for pyrazinamide (PZA) resistance (3). One hundred sixty-two isolates (6.9%) were resistant to PZA, including 1.3% that were monoresistant.
Recently, the M. tuberculosis pyrazinamidase (PZase) gene (pncA) was identified (14). Mutations in the pncA gene are considered the major mechanism of PZA resistance in M. tuberculosis (14), but resistant strains containing the wild-type gene have been described, suggesting additional resistance mechanisms besides a lack of PZase activity (16).
In an attempt to define the molecular basis of PZA resistance and to expand the profile of pncA mutations worldwide, we determined the nucleotide sequences of the pncA genes of 55 clinical isolates of M. tuberculosis initially found to be resistant to PZA and compared the PZase activities of these strains. These strains, isolated in the years 2000 and 2001, were collected in several hospital units in the Lisbon, Portugal, area. Fifty-five MDR-TB isolates and eight susceptible M. tuberculosis isolates were tested for susceptibility to PZA by the BACTEC MGIT 960 method with BACTEC MGIT 960 PZA test medium at a reduced pH of 5.9 and a 100-μg/ml concentration of PZA (Becton Dickinson) according to the manufacturer's manual. Almost all (53 of 55) PZA-resistant isolates were also PZase negative, and there was production of PZase in all PZA-susceptible isolates.
The 55 isolates had previously been subjected to IS6110 restriction fragment length polymorphism (RFLP-IS6110) analysis with restriction enzyme PvuII and a 245-bp IS6110 probe according to the international standard recommendations (17) and analyzed with Bionumerics software (Applied Maths, Inc., Kortrijk, Belgium).
Qualitative PZase activity analysis was performed with Dubos broth agar containing 100 μg of PZA/ml and 2 mg of sodium pyruvate/ml as described by Wayne (18).
Mutations in the sequence of the pncA gene were identified by comparison with the wild-type M. tuberculosis H37Rv pncA gene sequence (14) by using PCR sequencing. The entire pncA open reading frame, as well as 124 bp of the upstream sequence and 59 bp of the downstream sequence, was amplified by PCR by using the primers and conditions described by Morlock et al. (11). A 744-bp PCR product was generated with primers pncA-11 and pncA-8. Amplifications were carried out with a GeneAmp PCR system 9600 thermocycler (Perkin-Elmer Corp., Foster City, Calif.). The PCR-purified PCR product was subjected to a sequencing reaction by using a BigDye Terminator cycle sequencing kit with AmpliTaq DNA polymerase. The pncA amplicons were sequenced by using four internal primers according to Morlock et al. (11): pncA-10 and pncA-2R to sequence nucleotide residues 20 to 406 of the 744-bp amplicon, and pncA-6 and pncA-9 to sequence residues 318 to 720. Sequences were analyzed with BioEdit software (version 5.0.9.1; T. A. Hall Software). Every time a strain had no mutation in the pncA gene or in the upstream and downstream sequences, the sequence analysis was repeated.
As in other studies (2, 6, 9-11, 14-16), the frequency of pncA mutations in the Portuguese PZA-resistant isolates analyzed was very high, 94%. The results obtained are presented in Table 1. Seven of the 18 identified pncA mutations have not been described in previous studies (2, 5, 7, 8, 10, 11, 14-16). There was a thymine-to-cytosine point mutation at position 359, resulting in proline instead of leucine, in 26% of the isolates, making this the most common type of pncA mutation in this study. These 13 isolates represented at least two different clones defined by RFLP-IS6110 analysis (Fig. 1), indicating that they are actually different strains which happened to acquire the same type of mutation. Since pncA mutations occur randomly along the whole gene, apparently the same mutations of the pncA gene would rarely be present in unrelated isolates. Indeed, the two RFLP patterns belong to cluster A (13) now known as cluster Lisboa, a cluster of highly related strains that are responsible for the majority of MDR-TB in Portugal. The fact that this most common mutation has not been described in other studies is in agreement with the observation that cluster Lisboa is almost limited to Portugal, also being found in some neighboring countries (RFLP-IS6110 patterns of these strains were compared with those in the international MDR-TB database established at the Rijksintituut voor Volksgezondheid en Milieu, Bilthoven, The Netherlands).
TABLE 1.
pncA nucleotide and amino acid changes in PZA-resistant M. tuberculosis clinical isolates from Portugal
| Mutation site | Nucleotide change | Amino acid changea | No. of isolates |
|---|---|---|---|
| −11 | A→G | Mutation in promoter | 2 |
| 2 | T2C | Met1→Thr* | 1 |
| 137 | C137T | Ala46→Val | 1 |
| 152 | A152C | His51→Pro | 1 |
| 193 | A insertion | Frameshift | 1 |
| 212 | A212G | His71→Arg | 3 |
| 250 | C insertion | Frameshift* | 4 |
| 286 | A286G | Lys96→Glu | 3 |
| 290 | T insertion | Frameshift* | 1 |
| 359 | T359C | Leu120→Pro* | 13 |
| 374 | T374G | Val125→Gly* | 8 |
| 391 | GG insertion | Frameshift | 1 |
| 395 | G395A | Gly132→Asp | 1 |
| 406 | G406C | Asp136→His | 1 |
| 421 | C421T | Gln141Stop | 1 |
| 439 | CG insertion | Frameshift* | 3 |
| 476 | T476C | Leu159→Pro* | 5 |
| 511 | G511A | Ala171→Thr | 1 |
FIG. 1.
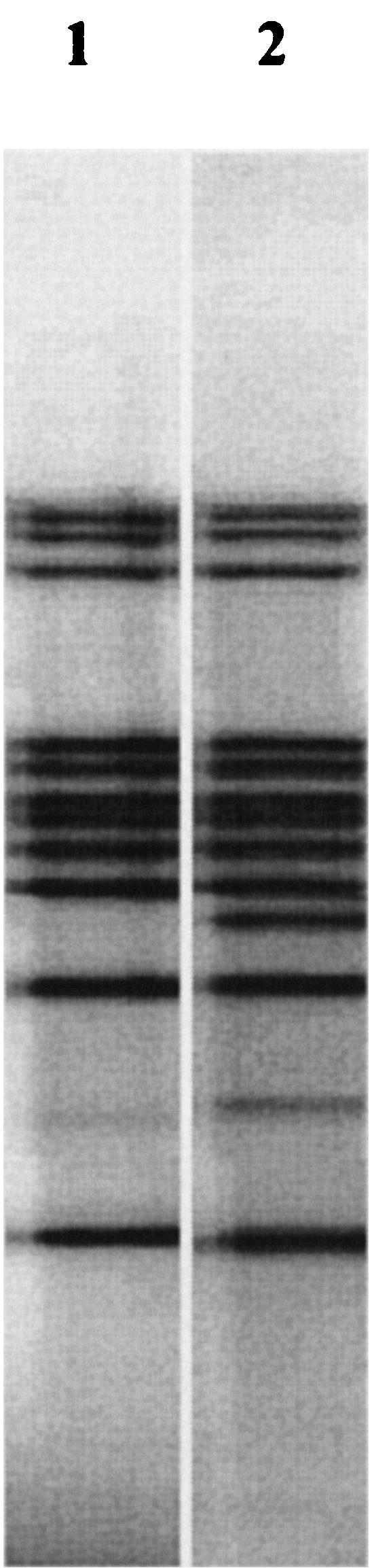
RFLP-IS6110 patterns of strains from cluster Lisboa. Lane 1, Lisboa3; lane 2, Lisboa2.
Four strains had no mutation in the pncA gene, even though they were resistant to PZA. Further sequence analysis of the pncA upstream region, which contains the putative pncA promoter, also failed to reveal any mutations for these strains. Two of these strains were positive for PZase activity, considered a rare event because we have found only one report of a case of a PZA-resistant strain without a pncA mutation (2). This finding indicates a possible alternative mechanism of PZA resistance that does not affect PZase activity or expression but is not very important in PZA resistance. These two strains present similar RFLP patterns and belong to a two-strain cluster. For the other two strains without pncA mutations, PZase activity was undetectable. We retested these four strains without pncA mutations for PZA susceptibility by using the BACTEC MGIT 960 PZA with 100-, 300-, and 900-μg/ml concentrations of PZA. PZase activity was also retested. The two PZase-positive strains had relatively low PZA MICs (100 to 300 μg/ml). One of the initial resistant strains confirmed to be PZase negative by the assay was found to be resistant to a 100-μg/ml concentration of PZA but susceptible when 300 μg/ml was used. This result may be an indication that a markedly low MIC (100 μg/ml) should be used for determining that these strains are truly phenotypically resistant. These findings support some previous reports (2, 4) that suggested the use of 300 instead of 100 μg of PZA per ml in the single-concentration qualitative test. It is also possible that this strain remained negative for PZase activity due to the poor sensitivity of the method used, as the relationship between PZA susceptibility and positive PZase activity is well established (19). The other PZase-negative strain had an MIC of over 900 μg/ml. PZA-resistant strains that were PZase negative were also found by others (1, 2, 8, 11, 12). Cheng et al. (2) suggested that PZA resistance is due to a pncA-regulatory gene and that mutation of this gene can affect the expression of pncA. Nevertheless, it can be noted that there was a strong correlation between loss of PZase activity and PZA resistance, making the determination of PZase activity an indirect measure of PZA susceptibility.
In this study, we have shown that most of the PZA-resistant M. tuberculosis strains analyzed, isolated in Portugal, carry mutations in the pncA gene, despite the fact that the number of strains in which no mutations were found still makes phenotypic tests necessary for the detection of PZA resistance. Although the high diversity of pncA mutations already described could be useful as a marker in tracing the outbreak or transmission of PZA-resistant M. tuberculosis isolates, one should be cautious in interpreting pncA mutation results for epidemiologic purposes.
Acknowledgments
This work was supported by Fundação Calouste Gulbenkian (project FCG 001013) and by ADEIM (Associação para o Desenvolvimento do Ensino e Investigação da Microbiologia).
REFERENCES
- 1.Bishop, K. S., L. Blumberg, A. P. Trollip, A. N. Smith, L. Roux, D. F. York, and P. Kiepiela. 2001. Characterisation of the pncA gene in Mycobacterium tuberculosis isolates from Gauteng, South Africa. Int. J. Tuberc. Lung Dis. 5:952-957. [PubMed] [Google Scholar]
- 2.Cheng, S. J., L. Thibert, T. Sanchez, L. Heifects, and Y. Zhang. 2000. pncA mutations as a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis: spread of a monoresistant strain in Quebec, Canada. Antimicrob. Agents Chemother. 44:528-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furtado, C., and L. Brum. 2003. Vigilāncia laboratorial da resistência aos antibacilares em Portugal em 2000-2001. Rev. Port. Pneumol. IX:279-291. [DOI] [PubMed] [Google Scholar]
- 4.Heifets, L. B. 1999. Pyrazinamide, p. 668-676. In V. L. Yu, T. C. Merigan, and S. L. Barriere (ed.), Antimicrobial therapy and vaccines. Williams and Wilkins, Baltimore, Md.
- 5.Hirano, K., M. Takahashi, Y. Kazumi, Y. Fukasawa, and C. Abe. 1997. Mutation in pncA is a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis. Tuber. Lung Dis. 78:117-122. [DOI] [PubMed] [Google Scholar]
- 6.Lee, K. W., J. Lee, and K. Jung. 2001. Characterization of pncA mutations of pyrazinamide-resistant Mycobacterium tuberculosis in Korea. J. Korean Med. Sci. 16:537-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemaitre, N., W. Sougakoff, C. Truffot-Pernot, and V. Jarlier. 1999. Characterization of new mutations in pyrazinamide-resistant strains of Mycobacterium tuberculosis and identification of conserved regions important for the catalytic activity of the pyrazinamidase pncA. Antimicrob. Agents Chemother. 43:1761-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marttila, H. J., M. Marjamaki, E. Vyshnevskaya, B. I. Vyshnevskiy, T. F. Otten, A. V. Vasilyef, and M. K. Viljanen. 1999. pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis isolates from northwestern Russia. 1999. Antimicrob. Agents Chemother. 43:1764-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClatchy, J. K., A. Y. Tsang, and M. S. Cernich. 1981. Use of pyrazinamidase activity in Mycobacterium tuberculosis as a rapid method for determination of pyrazinamide susceptibility. Antimicrob. Agents Chemother. 20:556-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mestdagh, M., P. A. Fonteyne, L. Realini, R. Rossau, G. Jannes, W. Mijs, K. A. L. De Smet, F. Portaels, and E. Van den Eeckhout. 1999. Relationship between pyrazinamide resistance, loss of pyrazinamidase activity, and mutations in the pncA locus in multidrug-resistant isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 43:2317-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morlock, G. P., J. T. Crawford, W. R. Butler, S. E. Brim, D. Sikes, G. H. Mazurek, C. L. Woodley, and R. C. Cooksey. 2000. Phenotipic characterization of pncA mutants of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park, S. K., J. Y. Lee, C. L. Chang, M. K. Lee, H. C. Son, C. M. Kim, H. J. Jang, H. K. Park, and S. H. Jeong. 2001. pncA mutations in clinical Mycobacterium tuberculosis isolates from Korea. BMC Infect. Dis. 1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Portugal, I., M. J. Covas, L. Brum, M. Viveiros, P. Ferrinho, J. Moniz Pereira, and H. David. 1999. Outbreak of multiple-drug-resistant tuberculosis in Lisbon: detection by restriction length polymorphism analysis. Int. J. Tuberc. Lung Dis. 3:207-313. [PubMed] [Google Scholar]
- 14.Scorpio, A., and Y. Zhang. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2:662-667. [DOI] [PubMed] [Google Scholar]
- 15.Scorpio, A., P. Lindholm-Levy, L. Heifets, R. Gilman, S. Siddiqi, M. Cynamon, and Y. Zhang. 1997. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 41:540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sreevatsan, S., X. Pan, Y. Zhang, B. N. Kreiswirth, and J. M. Musser. 1997. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob. Agents Chemother. 41:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wayne, L. G. 1974. Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am. Rev. Respir. Dis. 109:147-151. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, Y., and D. Mitchison. 2003. The curious characteristics of pyrazinamide: a review. Int. J. Tuberc. Lung Dis. 7:6-21. [PubMed] [Google Scholar]


