Abstract
A significant contemporary question in enzymology involves the role of protein dynamics and hydrogen tunneling in enhancing enzyme catalyzed reactions. Here, we report a correlation between the donor-acceptor distance (DAD) distribution and intrinsic kinetic isotope effects (KIEs) for the dihydrofolate reductase (DHFR) catalyzed reaction. This study compares the nature of the hydride transfer step for a series of active-site mutants, where the size of a side chain that modulates the DAD (I14 in E. coli DHFR) is systematically reduced (I14V, I14A, and I14G). The contributions of the DAD and its dynamics to the hydride transfer step were examined by the temperature dependence of intrinsic KIEs, hydride transfer rates, activation parameters, and classical molecular dynamics (MD) simulations. Results are interpreted within the framework of the Marcus-like model where the increase in the temperature dependence of KIEs arises as a direct consequence of the deviation of the DAD from its distribution in the wild type enzyme. Classical MD simulations suggest new populations with larger average DADs, as well as broader distributions, and a reduction in the population of the reactive conformers correlated with the decrease in the size of the hydrophobic residue. The more flexible active site in the mutants required more substantial thermally activated motions for effective H-tunneling, consistent with the hypothesis that the role of the hydrophobic side chain of I14 is to restrict the distribution and dynamics of the DAD and thus assist the hydride transfer. These studies establish relationships between the distribution of DADs, the hydride-transfer rates, and the DAD’s rearrangement toward tunneling-ready states. This structure-function correlation shall assist in the interpretation of the temperature dependence of KIEs caused by mutants far from the active site in this and other enzymes, and may apply generally to C-H→C transfer reactions.
Keywords: Enzyme Dynamics, Hydrogen transfer, Tunneling, Isotope Effect, Dihydrofolate Reductase, Marcus-like Model
Introduction
Despite intense studies during the past century, many questions regarding enzyme catalysis still remain unanswered. Specifically, the physical features of enzyme catalyzed reactions continue to be part of a vigorous debate. The transition state (TS) approach introduced by Pauling,1 which suggested that enzymes work by preferentially binding the activated form of the substrate(s) at the TS, still remains the generally accepted description. This static view of enzyme catalysis is likely to be correct; however it fails to address the mechanism by which physical features that contribute to that TS stabilization (e.g., electrostatics and steric effects) arise along the reaction coordinate. An approach that provides a more dynamic view of the catalytic process suggests that the thermal motions of both enzyme and substrates are essential for the chemical transformations catalyzed by an enzyme. A large amount of experimental and theoretical evidence supports such a view for certain enzymes, however the relationship between enzyme dynamics2 and bond activation needs further description. Analogous issues are of equal importance in solution reactions, and here we take advantage of the ability to control the reactants’ relative orientation in an enzyme’s active site to examine the nature of C-H→C transfer reactions in general. Several experimental and theoretical approaches provide an examination of hydrogen tunneling and its environmental coupling.3–14 Here, we focus on the temperature dependence of the intrinsic kinetic isotope effect (KIE), and on single turnover rates, as a means to probe the nature of the hydride transfer.3,12,15,16
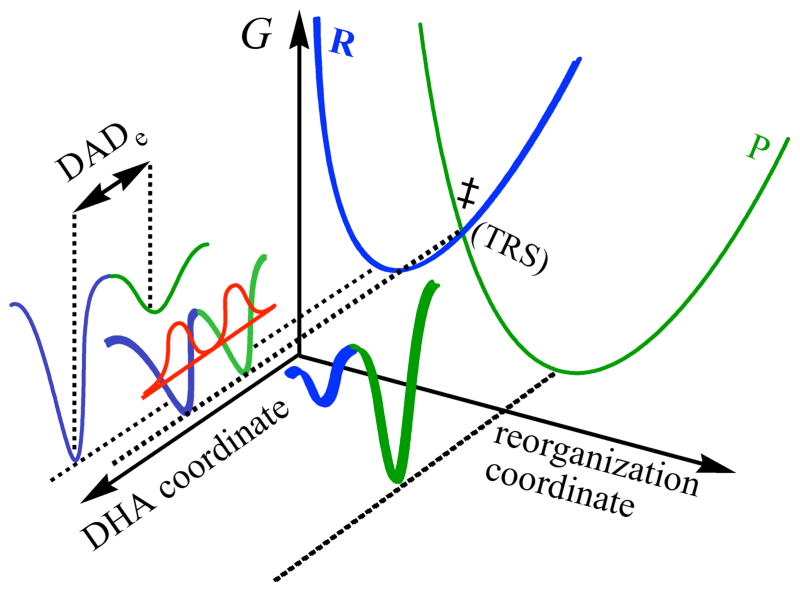
Experimental data collected by this approach led to the development of several phenomenological working models, which are extensions of the Marcus theory for electron transfer.7,17,18 They are referred to as Marcus-like models (also addressed in literature as environmentally coupled tunneling,19 rate-promoting vibrations,20 vibrationally enhanced tunneling,21 and more. In these models, illustrated in Figure 1, heavy-atom motions along the reorganization coordinate bring the system to a point where donor and acceptor wells are degenerate and where tunneling of the hydride is possible (i.e., a tunneling ready state, TRS). Thus, environmental reorganization occurs before hydride transfer takes place.3,4,6,7,12,18,22–24 At that point, the tunneling probability will depend on the overlap between the donor and acceptor wavefunctions (the first exponential inside the integral of eq. 1, referred to as the Franck-Condon term), which is sensitive to the mass of the tunneling particle, donor-acceptor distance (DADe)25 and its fluctuation along the DHA coordinate (Figure 1). Eq. 1 summarizes, in general terms, the form of the rate equation of various Marcus-like models,
Figure 1.
Illustration of a Marcus-like model. The reorganization coordinate represents the heavy-atom motions that carry the system to the TRS(‡). The blue and green correspond to the reactant and product states, respectively. The DHA coordinate (donor-hydrogen-acceptor) represents the fluctuations of the DADe. The red curve represents the wave functions of the hydrogen nucleus.
| (1) |
where C(T) represents the fraction of reactive complexes. The first exponential term is referred to as the Marcus-term and gives the probability of reaching a TRS. This term is a function of λ, the work associated with reorganization of the heavy-atoms, the reaction’s exoergicity (ΔG°), and the absolute temperature T. Importantly, while C and the Marcus-term are mostly isotopically insensitive they generate much of the temperature dependence in the reaction’s rate. The second exponent represents the Franck-Condon term (FC), an overlap integral that determines the tunneling probability for each isotope as a function of donor-acceptor distance. The last exponent has been referred to as the “gating” term, and represents the Boltzman distribution of DADes at a given temperature. This term is temperature sensitive, and together with the FC term is the source of the temperature dependency of the KIEs. The last two exponents are integrated over all the DADes sampled by the system.
Several researchers employed these models to rationalize their experimental results,9,12,13,16,23,26,27 where temperature-independent KIEs indicate a system wherein the reactants’ relative orientation and electronic states are optimized for H-tunneling and thermally-activated fluctuations of the DADe do not affect the KIE. On the other hand, temperature-dependent KIEs indicate a system wherein the reactants’ orientation and relative dynamics are not optimal and thermally activated gating motions have a different effect on the transfer of H than that of D or T. These models have been instrumental in qualitatively rationalizing trends in both the temperature and pressure dependence of kinetic data. In recent years, there have been modest attempts to quantitatively link the experimental data to the potential energy surface describing the gating coordinate (to link it to Equation 1.).13,28–30 However, due to the complexity of the DADe’s potential surface, and the fact that the DAD’s fluctuations do not constitute a normal mode, there are certain limitations, in e.g. determining a precise DADe and its fluctuations.31 MD simulations may be necessary to visualize the atomic level structural changes that affect the DADe in these reactions.
Here, we assess the modulation of the DADe by examining the relationship between the H-transfer rates and temperature dependence of KIEs, and the distribution and fluctuations of the DADc25 via MD simulations, through the targeted mutagenesis of an active site residue. Several other recent studies examined the effects of active site mutations, especially residues that are in direct contact with the substrates, on H-transfer reactions. In the study of a ketoglutarate-dependent dioxygenase, residue F159, which is in a position where the size of its side chain might affect the DADe, was systematically mutated to L, V and A, leading to reduced rates for substrate binding and C-H bond cleavage.32 For this system where a hydrogen radical is transferred to Fe(IV)=O (probably through a proton coupled electron transfer), elevated KIEs were measured at two different temperatures for the F159L and the F159V mutants which was in accordance with a longer DADe as analyzed in terms of a Marcus-like model. However, fluctuations in the DADe were not directly examined. In another relevant study on horse liver alcohol dehydrogenase, V203 (a residue analogous to I14 in DHFR) was mutated to alanine, to lead to a longer DADc.33,34 The 2° KIEs suggested reduced tunneling effects in the mutant, and the crystal structures of mutants implied an increase in the ground-state DADcs, but no temperature effects or fluctuations/distributions of the DAD were examined. Most recently, a study on a thermophilic alcohol dehydrogenase employed a series of active site mutants which led to enormously elevated Arrhenius prefactors below the physiological temperature range.14 In this case, no clear trend was observed between the size of the active site residue and the size of the Arrhenius prefactors. Moreover, mutations had little effect on activation parameters at physiologically optimal temperatures. Recent studies on morphinone reductase allowed for the examination of DAD modulation by pressure and temperature dependence through the two active-site residues that are in direct contact with the NADH cofactor, N189 and W106.13,35 For both residues only the alanine mutant was examined.
In the present work, we build upon these previous studies through examination of intrinsic KIEs, and we also expand the study executed by Scrutton and co-workers13 by systematically examining the correlation between changes in kinetics and the population distribution along the DAD coordinate through the gradual reduction of the size of a residue that directly modulates DADe. Careful selection of the residue of interest allowed us to minimize any electrostatic changes in the active site and focus on changes to the DAD. In addition to measuring KIEs, we used MD simulations to assess the fraction of reactive complexes and distribution of DADcs. The model system in this work is dihydrofolate reductase (DHFR) from E. coli, an enzyme that has been the subject of extensive experimental and theoretical studies over the years, and is well suited for the study reported here.36,37 DHFR is a small, flexible, monomeric protein (18 kDa), that catalyzes the conversion of 7,8-dihydrofolate (H2F) to 5,6,7,8-tetrahydrofolate (H4F) with the stereospecific transfer of a hydride from the pro-R C4 position of the nicotinamide ring to the si face of C6 of the dihydropterin ring. The complex kinetic mechanism of DHFR has been determined from equilibrium binding, steady-state, and presteady-state kinetic studies.36,37
Previous measurements of intrinsic KIEs and their temperature dependence implied quantum mechanical tunneling of the hydride in the wild-type DHFR (wtDHFR) catalyzed reaction.38 Kinetic studies of several distal mutants of DHFR indicated an important role of remote residues in modulating the H-tunneling process.27,39,40 The latter studies supported the hypothesis that a set of equilibrated conformational changes are coupled to the chemical transformation (C-H→C transfer), as predicted by hybrid quantum/classical MD simulations.41,42 Nonetheless, the long distance and complex relations between these remote residues and the active site prohibited a microscopic explanation of the effect of the mutants on KIEs and on the DAD. Reproducing similar observations via active site mutations, which have more straightforward effects on DADe and DADc, is critical in rationalizing analogous effects caused by remote mutants or altered conditions.
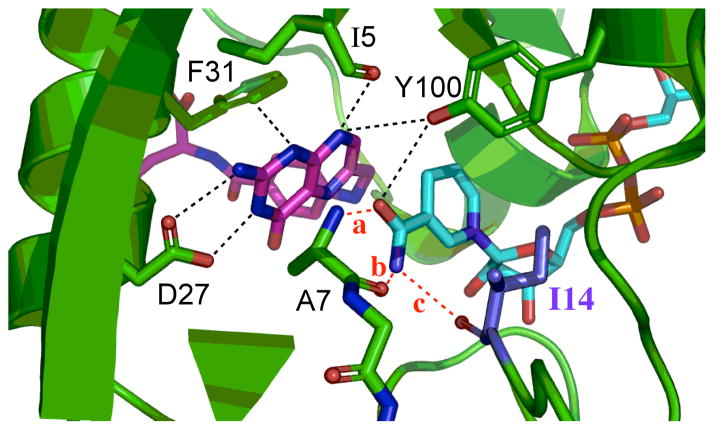
The focus of the present study is an active site residue, Ile 14, a hydrophobic residue situated behind the nicotinamide ring of the cofactor (NADPH) that keeps the nicotinamide ring, i.e., the H-donor, in close proximity to the pterin ring of the substrate, i.e., the H-acceptor (Figure 2). Importantly, this hydrophobic side-chain is typical to many nicotinamide dependent enzymes, which further broadens the application of the current study. Two-dimensional heteronuclear (1H-15N) magnetic relaxation studies of DHFR demonstrated that this residue exists as two rotamers, about the χ1 dihedral angle (χ1 :N-Cα-Cβ-Cγ1), in solution; the more populated +gauche rotamer and smaller but significantly populated trans rotamer.43 However, in both open and occluded crystal structures of wtDHFR, I14 residue is observed as only the +gauche rotamer. Modeling suggested that if this residue occupied the trans rotameric state, it would clash with the nicotinamide ring. In order for residue 14 to exist as a trans rotamer, the nicotinamide ring would be displaced towards the pterin ring. Brook and co-workers44 computed the free energy surfaces corresponding to the χ1 dihedral angle of residue 14, along the reaction coordinate, as part of the conformational change of the Met20 loop. They found that the trans rotamer population is observed only in the open state and in high-energy conformations leading to the occluded state of that loop. Only the +gauche and small amount of −gauche populations are present in the closed state; the state in which chemistry takes place. Moreover, they observed that the free energy barriers separating the functional important conformational states of the Met20 loop of DHFR are small and can be populated via thermal fluctuations.44 These two rotameric populations are of importance in analyzing our MD simulation (see Results and Discussion).
Figure 2.
The active-site of DHFR from E. coli (PDB ID 1RX2) emphasizing the role of Ile14 (metallic blue) as a support of the nicotinamide ring of NADP+. The nicotinamide ring is highlighted in light blue and the folate in magenta. Several other residues that form hydrogen bonding with the amide of NADPH are highlighted as well as I14 and A7. Three distinct hydrogen bonds, labeled in red, are: (a): NADPH(O-amide)-Ala-7(H); (b): NADPH(H72)-Ala-7(O); and (c): NADPH(H71)-Ile-14(O). The pterin ring is also immobilized in the active site via tight van der Waals interactions with F31, and strong hydrogen bonds to D27 and I5.
In earlier kinetic studies of residue 14 we found an order of magnitude reduction in the rate of the hydride transfer for I14A relative to the wtDHFR.45 In a more recent communication we reported the temperature dependence of the intrinsic KIEs of this mutant.38,46 Here, we extend these studies to a rigorous and systematic reduction of the side chain volume of I14 without changing its electrostatic properties. We found that the mutation leads to an increase in the magnitude of the KIEs as well as in their temperature dependence, and is also associated with reduction in hydride-transfer rate. In addition, classical MD simulations suggest new populations of DADcs, with larger average DADc with broader distributions, as well as a smaller population of the active conformers with the similar DADcs to wtDHFR. The analysis of all findings establishes a benchmark for the interpretation of KIEs and rates at the molecular level, and has significant applications to DHFRs and other enzymes.
Materials and Methods
Detailed experimental and molecular modeling methods are available in the Supplementary Information (SI Text). A brief description is provided here.
Expression, purification and kinetic experiments
WT DHFR and its mutant I14A have been expressed and purified as described elsewhere.45 Constructs of the I14V and I14G DHFR mutants were generated similarly to other DHFR mutants, and this is detailed in the SI Text. Synthesis, purification and storage of the radioactively labeled materials used in KIE studies are described in detail elsewhere and the SI Text.47,48 Briefly, [Ad-14C]-NADPH and (R)-[4-3H]NADPH were synthesized in order to measure 1° H/T KIEs, and (R)-[Ad-14C,4-2H]-NADPH and (R)-[4-3H]NADPH were used to determine 1° D/T KIEs. KIEs were measured competitively, where the two isotopically labeled cofactors were consumed by an excess of dihydrofolate in the presence of the enzyme. The reactions were quenched at different time points, and samples were analyzed as described in the SI Text. Intrinsic KIEs were calculated from the observed values as described in the SI Text and elsewhere.49 The isotope effects on the activation parameters for the intrinsic KIEs were calculated by a nonlinear fit to the Arrhenius equation for KIEs:
| (2) |
where kl and kh are the rates for light and heavy isotopes, respectively, Al/Ah is the isotope effect on the Arrhenius preexponential factor, ΔEa h-l is the difference in energy of activation between the two isotopes, R is the gas constant and T is the absolute temperature. Pre-steady-state kinetics were measured at 25°C, pH 7, using a stopped-flow apparatus (Applied Photophysics Ltd., Leatherhead, U.K.). Each value is reported with standard deviation, and represents at least 2 independent measurements.
MD simulations
MD simulations were performed using the Amber9 package.50 Details regarding the system setup, ligand parameterization, MD runs, data fitting and analysis are presented in the SI Text. Briefly, all simulations were run starting from the crystal structure of wtDHFR in complex with folate and NADP+ (PDB entry 1RX2).50,51 The folate and NADP+ ligands were replaced by N5 protonated 7,8-dihydrofolate (H2F) and NADPH respectively. The protonation state for all ionizable residues was set corresponding to pH 7, and the final protein structure was solvated with a previously equilibrated truncated octahedron box of TIP3P water molecules. The size of the box ensures that all protein atoms are at least 9 Å away from the edges of the box. Subsequent mutations were performed in silico by changing the corresponding amino acid in the original structure. During the simulations AMBER99 force field52 parameters were used for all residues. The NADP+ and NADPH parameters were taken from the Amber parameter database.53 Gaff force field54 parameters together with RESP55 charges calculated at the HF/6-31G* level were used to generate the parameter files for H2F. All simulations were run with the PMEMD module of the AMBER9 package.50,51 An equilibration protocol was applied that consists of performing an initial energy minimization, followed by a slow heating to the desired temperature using a linear temperature ramp from 100 to 300 K during 80 ps at constant volume and a subsequent pressure stabilization run at 300 K and 1 bar over 100 ps. Position frames, which were used for analyzing trajectories, were collected at 2 ps intervals. The ptraj module of Amber9 was used to analyze the data extracted from the MD simulations. Data from the DADc distribution was fitted with a combination of Gaussian and Split Gaussian functions using the open source software for non linear curve fitting, Fityk.56 For the equations and procedures used for the fitting please refer to the SI Text.
Results and Discussion
Competitive KIEs and Their Temperature Dependence
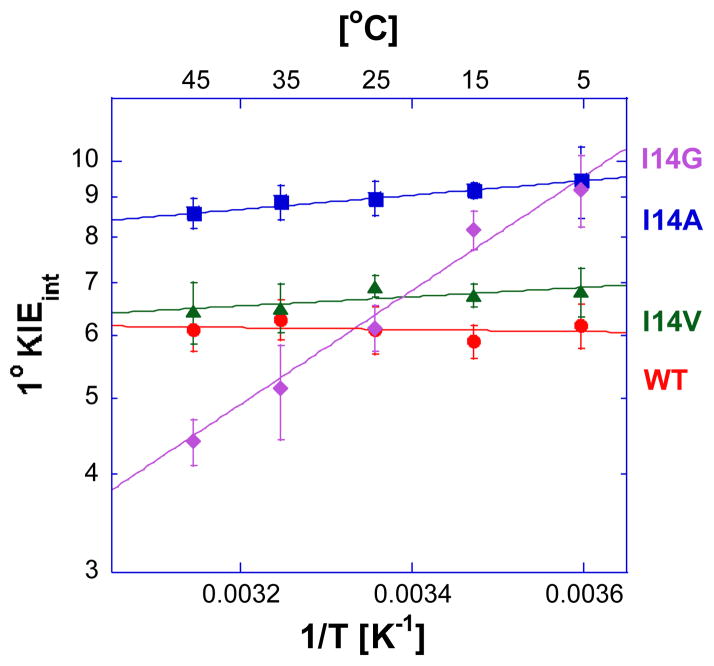
We determined the temperature dependence of the intrinsic KIEs and hydride transfer rates for a series of DHFR isoforms for which the side-chain that holds the hydride donor (nicotinamide) close to the acceptor (dihydropterin) are systematically reduced from isoleucine to glycine. Figure 3 shows Arrhenius plots of the intrinsic H/T 1° KIEs for all four enzymes. The same trend was observed for H/D and D/T KIEs as presented in the SI Text. Table 1 summarizes the volume of these side chains, their hydride transfer rates, the isotope effect on the Arrhenius preexponential factors (Al/Ah, where l denotes light and h heavy isotope), and the difference in the activation energy between light and heavy isotope (ΔEah-l=Eah−Eal).
Figure 3.
Arrhenius plot of intrinsic H/T KIEs (on a log scale) for wild-type (red)38, I14V DHFR mutant (green), I14A DHFR (blue)46 and I14G DHFR (purple). The lines represent the nonlinear regression to an exponential equation. More details are presented in the SI Text.
Table 1.
Comparative kinetic parameters of the DHFR I14 mutants.
The isotope effects on the preexponential Arrhenius factors (AH/AT, AH/AD, and AD/AT) for the wtDHFR, I14V DHFR and I14A DHFR are larger than unity, and above the semiclassical limit, whereas the values for the I14G DHFR are smaller than unity and below the semiclassical limit. The traditional, semi-classical approach with the Bell correction59 would suggest that hydride transfer for the first three isozymes involves “extensive tunneling” for both heavy and light isotopes, whereas the hydride transfer for I14G DHFR would be characterized as a case with “moderate tunneling”.60,61 According to the Marcus-like model, AH/AT is affected by both the average DADe and its distribution, and reveals the difference in the nature of the hydride transfer between these mutants. A longer average DADe leads to an increase in the Al/Ah, whereas a broader distribution of DADe-s increases the temperature dependence of the KIEs resulting in a decrease in the Al/Ah. Because of this ambiguity, a better indicator for the nature of the hydride-transfer is the slope of the Arrhenius plot, (Figure 3, i.e., the ΔEa h-l, where h and l are the heavy and light isotopes, respectively) that increases as the distribution of DADes broadens. ΔEa h-l close to zero indicates that the KIEs of wtDHFR exhibits no temperature dependence (within the experimental error), and the larger ΔEa h-l for the mutants indicates that temperature dependence of the KIEs increases as the size of the hydrophobic residue behind the nicotinamide ring decreases. In terms of the Marcus-like model this means that in the case of the wtDHFR the average DADe is optimal for the hydride transfer and has a narrow distribution around the average. The I14V mutant is similar to wtDHFR, but has a slight (but statistically significant) temperature dependence on the intrinsic KIEs. The I14A mutant has the largest intrinsic KIE at 25°C, but its KIEs’ temperature dependence (ΔEa h-l) falls between the I14V and I14G DHFR. This interesting observation supports the earlier examination of distal mutants of DHFR, which concluded that in order to assess the nature of the hydride transfer it is necessary not only to measure the KIE at a single temperature but for the whole temperature range.27
According to the Marcus-like model, the inflated temperature dependence of the KIEs in the mutants reflects a broader distribution of DADe sampling that is not present in the WT. The average DADe in the mutants is too long for efficient tunneling at low temperatures, but at higher temperatures more conformations with a shorter DADe are sampled. This affects the rate for transfer by heavy isotopes more than that of protium-transfer because these can only tunnel from the shorter distances, thus leading to temperature dependent KIEs. These observations are in accordance with some of the earlier studies, e.g. a study on the soybean lipoxygenase-1 (SBL-1), which showed that a decrease in the bulk of the active site residue (residue 553, not analogous to I14, not in direct contact with the substrate) was correlated with an increase in the temperature dependence of the 1° KIE.62 Numerical modeling using a form of equation 1 provided more evidence that these remote mutations led to longer DADes. However, in other studies, in particular the one on morphine reductase, Pudney et al.13 found an increase in temperature dependence of KIEs with an increase in the size of the active-site residue. In that particular study, the residue of interest was V108, which is not in direct contact with the cofactor NADH; thus the effect of the size of the residue on the DADe was hard to assess. The other residue of interest in the same study was W106, which is in direct contact with the cofactor; however, only the alanine mutant was examined and therefore no trends were available. In the pressure-dependence studies on the same enzyme the authors suggested that an increase in the force constant of the compressive mode can lead to an increase in the magnitude of the 1° KIE even as the DAD decreases,31,63 which became a source of the extensive debate.64 Our findings are in accordance with the majority of cases where numerical modeling of the temperature dependence of the 1° KIEs using equation 1 results in decreasing temperature dependence with decreasing average DADe. From these independent studies it is evident that the relationship between the size of the side chain and the DADe might be quite complex, and might go beyond the notion that the smaller size of the residue leads to longer average DADe. These results were the driving force in the experimental approach presented here, where we determined the effect of the DADs’ distribution on the nature of the catalyzed H-transfer.
In order to asses the effect of the mutations on the hydride transfer, we measured the single turnover rates at 25 °C, pH=7.0, for all mutants and for wtDHFR.36 Fluorescence Energy Transfer (FRET) decay traces were fitted to single exponential functions with slope. These rates indicate that hydride transfer for the I14V, I14A, and I14G mutants are about 7, 40, and 1000 times slower than wtDHFR, respectively (Table 1). There is a clear correlation between the decrease in size of the hydrophobic residue, the rate and the nature of the hydride transfer. We would like to point out that the reduction of the size of this residue is likely to affect the mobility of the M20 loop and consequently affect the fraction of reactive complexes, the pKa of H2F,65 the fraction of different M20 loop conformations, and other factors that affect the multi-steps reaction’s rate, but not the hydride transfer step under study. All these affects are included in the prefactor C(T) in rate Eq. 1, but do not affect the terms in the integral, including the DAD, its dynamics and distribution. The intrinsic KIEs, on the other hand, probe the hydride transfer step per sé, and not the steps preceding it (such as protonation of N5 position of H2F, and other isotopically insensitive factors).
MD Simulations on mutants of DHFR
In order to provide a detailed microscopic description of the active site structural change upon mutation of residue 14, we performed MD simulations. One 15 ns simulation was performed for wtDHFR and each mutant (see SI Text and Figures S6–S8), as was previously done for wtDHFR and I14A DHFR.46 In order to better sample the conformational space for each mutant, we also performed ten independent runs of 3 ns, differing only in initial velocities for each one of them (see SI Text and Figures S9–S11).
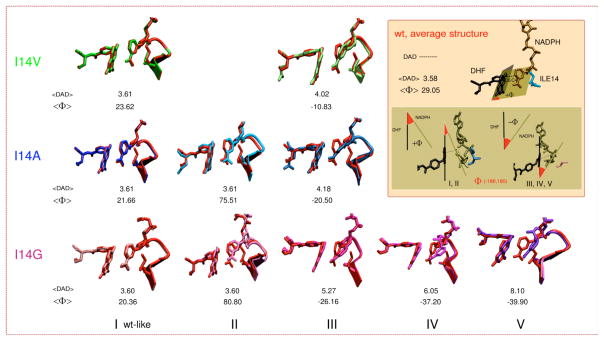
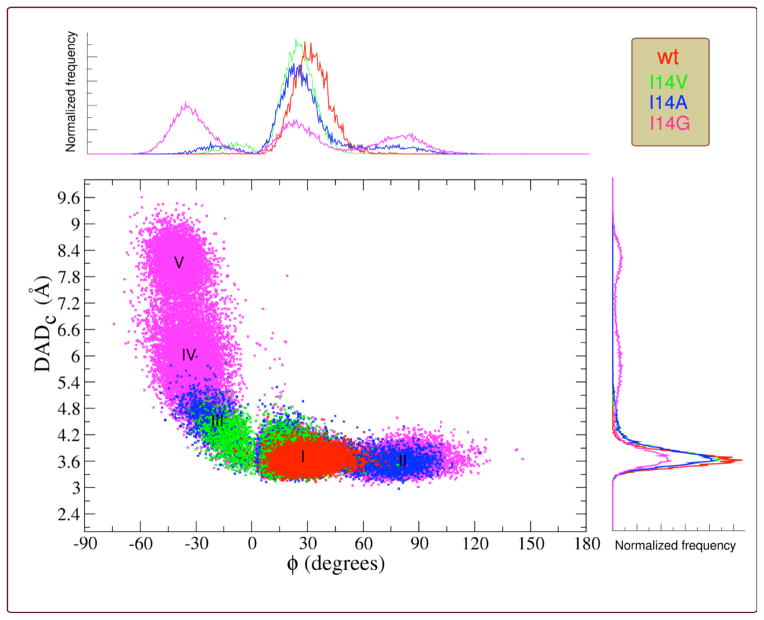
A comparison of the resulting conformations is presented in Figure 4, which also describes two parameters of interest, DADc and Φ. DADc is defined as the distance between C4 of NADPH and C6 of H2F (see Figure 4 inset). The relative donor acceptor orientation was measured as the angle (Φ) between the planes that fit the nicotinamide moiety of NADPH and the pterin ring of H2F (see Figure 4 inset). The distribution of DADcs and the orientation of donor and acceptor (Φ angle) are displayed in Figure 5 (see Figures S14–S16 for each individual mutant). The DADcs’ and Φs’ distributions (Figure 5, 1D plots on y and x-axis) were fitted to one split Gaussian or a combination of Gaussian functions and split Gaussian functions (Table 2 and Figures S17–S20). The correlation is presented as a 2-D plot in Figure 5.
Figure 4.
Structures comparing WT (red) to the different conformations found for the three mutants (see color code of each mutant on the left) together with the corresponding DADc and Φ average values (DAD in the figure refers to DADc). (I) A wt-like conformation; (II) A conformation with almost the same DADc average distance as the conformation I, but with the nicotinamide almost perpendicular to the H2F pterin ring; (III) A conformation where the nicotinamide is partially twisted to the right towards residue 14; (IV) A conformation similar to III but with a broader distribution of distances; and (V) A conformation where the nicotinamide is completely twisted towards residue 14. Inset: definition for DADc distance, Φ angle, and average values found for wtDHFR. Structures I and II display positive values for Φ, whereas structures III, IV and V (nicotinamide twisted to the right, towards residue 14) display negative values.
Figure 5.
Correlation plot between the DADc (angstroms) and the relative orientation of donor and acceptor (Φ, degrees) for wtDHFR (red), I14V (green), I14A (blue), I14G (magenta). I, II, III, IV and V indicate the different populations identified for each DADc and Φ values. Overlaid wt and mutants DADc and Φ distributions are shown on the y and x axis respectively.
Table 2.
DADc distributions for the different conformations found along MD simulation obtained from Gaussian analysis (see text).
| DHFR | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Active site conformation | <DAD> (Å) | σ (Å)c | <Φ>d (degrees) | Fraction | R | |
|
| ||||||
| wt | Ia | 3.58 | 0.16 | 29.0 | 1 | 0.9979 |
|
| ||||||
| I14V | Ia | 3.61 | 0.17 | 23.6 | 0.86 | |
| IIIb | 4.02 | 0.31 | −10.8 | 0.14 | 0.9984 | |
|
| ||||||
| I14A | Ia | 3.61 | 0.17 | 21.7 | 0.68 | |
| IIa | 3.61 | 0.17 | 75.5 | 0.13 | 0.9980 | |
| IIIb | 4.18 | 0.49 | −20.5 | 0.19 | ||
| Ia | 3.60 | 0.19 | 20.4 | 0.33 | ||
|
| ||||||
| I14G | IIa | 3.60 | 0.19 | 80.8 | 0.26 | 0.9943 |
| IIIb | 5.27 | 0.82 | −26.2 | 0.09 | ||
| IVb | 6.05 | 0.67 | −37.2 | 0.22 | ||
| Vb | 8.10 | 0.36 | 39.9 | 0.1 | ||
Fitted to a split-Gaussian,
Fitted to a Gaussian.
For split-Gaussian σ is the average of both parts.
Φ separately from the DADc. See SI for the procedure and parameters (Table S3).
The results indicate that reduction in the size of the hydrophobic residue behind the nicotinamide ring leads to an increase in the flexibility of the nicotinamide ring. As a result, the nicotinamide ring can not only explore more orientations at the same DADcs, but also this increase in the flexibility leads to an increase in the average DADc, a broader DADc distributions, as well as new DADc populations (Table 2 and Figures 4–5). The different populations are labeled I, II, III, IV and V; the structures that represent these different populations are presented in Figure 4 for each mutant, and are overlaid with the average wt-DHFR structure.
The DADc distribution for wtDHFR was fitted to one split Gaussian function and the residue I14 was only observed as the +gauche rotamer during the time scale of the simulation. This result agrees well with Brooks’s work,44 where this rotamer represents the major population observed for the closed conformation of wtDHFR. The DADc distribution for the I14V mutant was fitted to a combination of one split and one normal Gaussian functions suggesting that there are two populations present during the simulation time. The average structure for the major population is very similar to the wtDHFR (structure I in Figure 4), displaying similar DADc and Φ distributions, but with a slight increase in the average DADc as well as a deviation of the average Φ value found in the wtDHFR. The smaller population (structure III in Figures 4 and 5) has a larger average DADc with wider distribution, where new Φ values are explored, manifesting themselves as a partial twisting of the nicotinamide ring. In the case of the I14A mutant, the correlation plot (Figure 4) shows that the conformational space explored is bigger compared to the I14V mutant. The simulations in this case suggest three populations: (i) the wt-like population; (ii) a new population with similar DADc to I, but a different Φ distribution (Structure II); and (iii) the Structure III, equivalent to the one found for the I14V mutant, but with a slightly larger average DADc and Φ, which also exhibits broader distributions, indicating more flexibility in the location of the nicotinamide ring (see Figure 4 and 5). Population III is not expected to contribute to a reactive conformation (average DAD too large), therefore leading to a smaller constant C(T), from the equation 1, which is in accordance with a reduction in rate of the hydride transfer (Table 1). On the other hand the new population (II) exhibits the same average DADc as the population I, however it is expected to be by far less reactive due to the substantial deviation of the Φ angle from 30°. As visible in figure 4, the nicotinamide ring of NADPH is almost perpendicular to the pterin ring of H2F, making the HOMO-LUMO orbital overlap very poor, which has a deleterious effect on the rate.66 Consequently, population II is not likely to be reactive, but its exact reactivity (if any) is hard to determine without high-level QM/MM calculations that are beyond the scope of this work. Lastly, the hydrogen-bonds (two to residue A7 and one to the backbone of I14) that hold the NADPH amide moiety (see Figure 2), show a broader distribution, that is in accord with the elevated flexibility of nicotinamide ring in the I14A mutant relative to I14V and wtDHFR.
MD simulations for the mutant with the smallest side chain, I14G DHFR, suggested all five significant populations. In this case, the three H-bond interactions mentioned before, ((a), (b) and (c) in Figure 2) are longer and weaker than in the wt and I14V/ I14A mutants. The last population, V, has one of the H-bonds (c), completely broken, while the other two (a) and (b), are only weakly established (see Figures S12–S13 in the SI Text). Once this H-bond is broken the structure remains in conformation V and doesn’t return to wt-like or other conformations during the time scale of the simulation (the simulation was extended to 30 ns). The breakage of the H-bonds, particularly the (c) H-bond, results from the diminishment in the side chain of the residue 14, and contributes to the increase in the flexibility over other mutants and the ability to explore a broader conformational space that leads to a non-reactive population (DADcs > 6 Å). Finally, Table 2 lists the fraction of each population found, which appear to correlate well with the reduced rate of H-transfer. In all the cases, the wt-like population is the predominant one, although its relative contribution decreases in proportion to the size of the side chain, leading to decrease in the CT term in Eq. 1. A similar effect on rates was obtained in a recent study where modeling of the conformational sampling that lead to elevated Arrhenius prefactors (below the physiologically optimal temperatures) for the thermophilic alcohol dehydrogenase showed an increase in the fractional occupation of catalytically inactive regions of conformational space.14 More importantly in this work, a correlation was observed between the temperature dependence of the intrinsic KIEs (ΔEaT-H, Table 1) and the DADc, its σ values, and the Φ angle (Table 2) for the reactive population (I). As the ΔEaT-H increases (Table 1), the average DADc of population I increases and its distribution widens, while the average Φ angle decreases (Table 2). This important correlation is also visualized in Figure 5, where the reactive population (short DADc and 0° > Φ > 60°) is larger for the WT (red), smaller and with broader distribution toward longer DADs for I14V (green) and more so for I14A (blue). The population I for I14G is too small to be clearly observed in Figure 5 (for a better view see Figure S16), but has the largest DADcs distribution (σ) and smallest Φ (Table 2).
The simulations provide molecular insight and support to the interpretation of the kinetic findings via the Marcus-like model: the smaller side chain acts to broaden the sampling of configurations within the reactive ternary complex along the reaction coordinate. This leads to a steeper temperature dependence of intrinsic KIEs and reduced hydride transfer rates. The MD simulations also revealed the importance of the donor-acceptor orientation (DADc and Φ), and its relative distribution, which affects the overlap between the HOMO and LUMO.25
Conclusions
The relationship between the DAD and its distribution and dynamics to the rate of hydride-transfer and the temperature dependence of intrinsic KIEs has been examined. The model system was DHFR, which like many nicotinamide dependent enzymes has a hydrophobic amino acid behind this cofactor that positions the nicotinamide ring close to its counter substrate. Here we examined the effect of systematic active site mutations that decrease the size of the side chain of this amino acid (I14 to V, A, and G) on the enzyme catalyzed hydride transfer step. The hydride-transfer rates and the temperature dependence of intrinsic KIEs were measured, and the ensemble of ternary complex structures was assessed through MD simulations. The findings suggested that residue I14 participates in the restrictive active-site motions that modulate the DAD. The findings confirm the relations predicted by the Marcus –like model, where longer DADs and broader distributions of DADs lead to a steeper temperature dependence of intrinsic KIEs (ΔEa h-l, determined mostly by the integral in Eq. 1) and the smaller fraction of reactive conformations (C(T) in Eq. 1) contributes to the reduction of the hydride transfer rate. The correlation that was found between DAD distributions and the temperature dependence of the intrinsic KIEs provides a valuable benchmark for understanding the active site dynamics and H-tunneling in DHFR catalyzed reactions, including remote mutations or altered protein scaffolds, for which it has not been trivial to rationalize the kinetic findings at a molecular level. It is also likely to be relevant to C-H→C reactions in other enzymes, as well as non-enzymatic reactions in general, as the correlation found here is likely to hold whether the DADes are imposed by proteins’ scaffold or by solvent molecules.
The current findings suggest that this series of DHFR mutants is suitable for more in-depth examination by a broad variety of researchers applying a variety of methods. Future studies of this system may include high-level hybrid classical-quantum mechanical MD simulations,22,67 structural studies,68 examination of the systems dynamics at millisecond to nanosecond time scales using NMR relaxation experiments,10 and femtosecond to picosecond time-scale motions using two-dimensional infrared spectroscopy.16 Those experiments will allow more in depth examination of possible correlations between structure, dynamics, and the nature of H-transfer in a well-defined environment imposed by the enzyme, and should reflect on C-H→C reactions in general.
Acknowledgments
This work was supported by NSF CHE-0133117 and BSF-2007256 (for AK) and 1RO1GM092946 (for SJB). MD simulations were done using resources provided by the Open Science Grid, which is supported by the National Science Foundation and the U. S. Department of Energy’s Office of Science.
Abbreviations
- DHFR
dihydrofolate reductase
- DAD
donor-acceptor distance
- MD
molecular dynamics
- KIE
kinetic isotope effect
- TRS
tunneling ready state
- H2F
7,8-dihydrofolate
Footnotes
Supporting Information Available: Details pertaining to KIE experiments, MD simulations and stopped-flow experiments. Moreover, average H/D and D/T intrinsic KIE values with errors obtained at different temperatures are presented. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Pauling L. Nature. 1948;161:707–709. doi: 10.1038/161707a0. [DOI] [PubMed] [Google Scholar]
- 2.Here we define dynamics as any motion along the reaction coordinate and not as only non-equilibrium motions.
- 3.Kohen A. In: Isotope effects in chemistry and biology. Kohen A, Limbach HH, editors. Ch 28. Taylor & Francis, CRC Press; Boca Raton, FL: 2006. pp. 743–764. [Google Scholar]
- 4.Schwartz SD. In: Isotope effects in chemistry and biology. Kohen A, Limbach HH, editors. Ch 18. Taylor & Francis, CRC Press; Boca Raton, FL: 2006. pp. 475–498. [Google Scholar]
- 5.Truhlar DG. In: Isotope effects in chemistry and biology. Kohen A, Limbach HH, editors. Ch 22. Taylor & Francis, CRC Press; Boca Raton, FL: 2006. pp. 579–620. [Google Scholar]
- 6.Warshel A, Olsson MHM, Villa-Freixa J. In: Isotope effects in chemistry and bilogy. Kohen A, Limbach HH, editors. Taylor & Francis, CRC Press; Boca Raton, FL: 2006. pp. 621–644. [Google Scholar]
- 7.Kiefer PM, Hynes JT. In: Isotope effects in chemistry and biology. Kohen A, Limbach HH, editors. Ch 21. Taylor & Francis, CRC Press; Boca Raton, FL: 2006. pp. 549–578. [Google Scholar]
- 8.Sutcliffe MJ, Masgrau L, Roujeinikova A, Johannissen LO, Hothi P, Basran J, Ranaghan KE, Mulholland AJ, Leys D, Scrutton NS. Philos Trans R Soc B, Biol Sci. 2006;361:1375–1386. doi: 10.1098/rstb.2006.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan F, Gadda G. J Am Chem Soc. 2005;127:17954–17961. doi: 10.1021/ja0560377. [DOI] [PubMed] [Google Scholar]
- 10.Boehr DD, McElheny D, Dyson HJ, Wright PE. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 11.Hammes-Schiffer S. In: Quantum Tunnelling in Enzyme-Catalysed Reactions. Allemann RK, Scrutton NS, editors. Royal Society of Chemistry; Cambridge: 2009. pp. 79–104. [Google Scholar]
- 12.Nagel ZD, Klinman JP. Chem Rev. 2010;110:PR41–67. doi: 10.1021/cr1001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pudney CR, Johannissen LO, Sutcliffe MJ, Hay S, Scrutton NS. J Am Chem Soc. 2010;132:11329–11335. doi: 10.1021/ja1048048. [DOI] [PubMed] [Google Scholar]
- 14.Nagel ZD, Dong M, Bahnson BJ, Klinman JP. Proc Natl Acad Sci U S A. 2011;108:10520–10525. doi: 10.1073/pnas.1104989108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hay S, Sutcliffe MJ, Scrutton NS. In: Quantum Tunnelling in Enzyme-Catalysed Reactions. Scrutton NS, Allemann RK, editors. RSC Publishing; 2009. pp. 199–218. [Google Scholar]
- 16.Bandaria JN, Dutta S, Nydegger MW, Rock W, Kohen A, Cheatum CM. Proc Natl Acad Sci USA. 2010;107:17974–17979. doi: 10.1073/pnas.0912190107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcus RA, Sutin N. Biochem Biophys Acta. 1985;811:265–322. [Google Scholar]
- 18.Marcus RA. J Phys Chem B. 2007;111:6643–6654. doi: 10.1021/jp071589s. [DOI] [PubMed] [Google Scholar]
- 19.Francisco WA, Knapp MJ, Blackburn NJ, Klinman JP. J Am Chem Soc. 2002;124:8194–8195. doi: 10.1021/ja025758s. [DOI] [PubMed] [Google Scholar]
- 20.Antoniou D, Caratzoulas S, Kalyanaraman C, Mincer JS, Schwartz SD. Eur J Biochem. 2002;269:3103–3112. doi: 10.1046/j.1432-1033.2002.03021.x. [DOI] [PubMed] [Google Scholar]
- 21.Sutcliffe MJ, Scrutton NS. Eur J Biochem. 2002;13:3096–3102. doi: 10.1046/j.1432-1033.2002.03020.x. [DOI] [PubMed] [Google Scholar]
- 22.Villa J, Warshel A. J Phys Chem B. 2001;105:7887–7907. [Google Scholar]
- 23.Roston D, Kohen A. Proc Natl Acad Sci. 2010;107:9572–9577. doi: 10.1073/pnas.1000931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammes-Schiffer S. Acc Chem Res. 2006;39:93–100. doi: 10.1021/ar040199a. [DOI] [PubMed] [Google Scholar]
- 25.Two different, yet related DADs are used in this paper: The first, DADe, represents the donor-acceptor distance in the energy space, i.e., the distance between the hydrogen in the reactant state and in the product state (e.g., Figure 1, the double-well system along the DHA coordinate). DADe depends on the overlap between the electronic orbitals of the donor and acceptor, and is most relevant to the kinetic measurements presented here. The second, DADc, refers to the donor-acceptor distance between two carbons that donate, or accept the hydride. The DADc is the one addressed in the structures and MD calculations. The importance of the orbital steering in catalysis has been previously shown (e.g., Mesecar AD, Stoddard BL, Koshland DE. Science. 1997;227:202–206. doi: 10.1126/science.277.5323.202.). DADc relates to DADe, through the DHA angle and the C-H distance at each stage of the reaction coordinate. E.g., when the DHA angle is 180° and the C-H is in its ground state (~1.04 Å) for both reactant and product, than DADc = DADe + 2 × 1.04 Å. Also, when the DHA angle is fixed (on the reaction’s time scale), then both DADs’ fluctuations are coherent.
- 26.Wang Z, Kohen A. J Am Chem Soc. 2010;132:9820–9825. doi: 10.1021/ja103010b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Goodey NM, Benkovic SJ, Kohen A. Proc Nat Acad Sci USA. 2006;103:15753–15758. doi: 10.1073/pnas.0606976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antoniou D, Schwartz SD. J Phys Chem B. 2001;105:5553–5558. [Google Scholar]
- 29.Knapp MJ, Klinman JP. J Am Chem Soc. 2002;124:3865–3874. doi: 10.1021/ja012205t. [DOI] [PubMed] [Google Scholar]
- 30.Meyer MJ, Tomchick DR, Klinman JP. Proc Natl Acad Sci U S A. 2008;105:1146–1151. doi: 10.1073/pnas.0710643105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hay S, Scrutton NS. Biochemistry. 2008;47:9880–9887. doi: 10.1021/bi8005972. [DOI] [PubMed] [Google Scholar]
- 32.McCusker KP, Klinman JP. J Am Chem Soc. 2010;132:5114–5120. doi: 10.1021/ja909416z. [DOI] [PubMed] [Google Scholar]
- 33.Bahnson BJ, Colby TD, Chin JK, Goldstein BM, Klinman JP. Proc Nat Acad Sci Usa. 1997;94:12797–12802. doi: 10.1073/pnas.94.24.12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colby TD, Bahnson BJ, Chin JK, Klinman JP, Goldstein BM. Biochemistry. 1998;37:9295–9304. doi: 10.1021/bi973184b. [DOI] [PubMed] [Google Scholar]
- 35.Pudney CR, McGrory T, Lafite P, Pang J, Hay S, Sutcliffe MJ, Scrutton NS. ChemBioChem. 2009;10:1379–1384. doi: 10.1002/cbic.200900071. [DOI] [PubMed] [Google Scholar]
- 36.Fierke CA, Johnson KA, Benkovic SJ. Biochemistry. 1987;26:4085–4092. doi: 10.1021/bi00387a052. [DOI] [PubMed] [Google Scholar]
- 37.Fierke CA, Benkovic SJ. Biochemistry. 1989;28:478–486. doi: 10.1021/bi00428a011. [DOI] [PubMed] [Google Scholar]
- 38.Sikorski RS, Wang L, Markham KA, Rajagopalan PTR, Benkovic SJ, Kohen A. J Am Chem Soc. 2004;126:4778–4779. doi: 10.1021/ja031683w. [DOI] [PubMed] [Google Scholar]
- 39.Wang L, Goodey NM, Benkovic SJ, Kohen A. Phil Trans R Soc B. 2006;361:1307–1315. doi: 10.1098/rstb.2006.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L, Tharp S, Selzer T, Benkovic SJ, Kohen A. Biochemistry. 2006;45:1383–1392. doi: 10.1021/bi0518242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radkiewicz J, Brooks C. J Am Chem Soc. 2000;122:225–231. [Google Scholar]
- 42.Wong KF, Selzer T, Benkovic SJ, Hammes-Schiffer S. Proc Natl Acad Sci U S A. 2005;102:6807–6812. doi: 10.1073/pnas.0408343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schnell JR, Dyson HJ, Wright PE. Biochemistry. 2004;43:374–383. doi: 10.1021/bi035464z. [DOI] [PubMed] [Google Scholar]
- 44.Arora K, Brooks CLI. J Am Chem Soc. 2009;131:5642–5647. doi: 10.1021/ja9000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams JA, Fierke CA, Benkovic SJ. Biochemistry. 1991;30:11046–11054. doi: 10.1021/bi00110a006. [DOI] [PubMed] [Google Scholar]
- 46.Stojkovic V, Perissinoti LL, Lee J, Benkovic S, Kohen A. Chem Comm. 2010;46:8974–8976. doi: 10.1039/c0cc02988b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Markham KA, Sikorski RS, Kohen A. Anal Biochem. 2004;325:62–67. doi: 10.1016/j.ab.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 48.McCracken JA, Wang L, Kohen A. Anal Biochem. 2003;324:131–136. doi: 10.1016/j.ab.2003.09.025. [DOI] [PubMed] [Google Scholar]
- 49.Northrop DB. In: Enzyme mechanism from isotope effects. Cook PF, editor. CRC Press; Boca Raton, Fl: 1991. pp. 181–202. [Google Scholar]
- 50.Case DA, et al. University of California; San Francisco: 2006. [Google Scholar]
- 51.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, III, DeBolt S, Ferguson D, Seibel G, Kollman PA. Comput Phys Commun. 1995;91:1–41. [Google Scholar]
- 52.Cheatham TE, Cieplak P, Kollman PA. J Biomol Struct Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 53.Walker RC, De Souza MM, Mercer IP, Gould IR, Klug DR. J Phys Chem B. 2002;106:11658–11665. [Google Scholar]
- 54.Wang J, Wolf RM, Caldwell JW, Kollamn PA, Case DA. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 55.Wang J, Cieplak P, Kollman PA. J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 56.Wojdyr M. J Appl Cryst. 2010;43:1126–1128. [Google Scholar]
- 57.Creighton TE. Proteins Structure and Molecular Principles. Freeman; New York, N. Y: 1984. [Google Scholar]
- 58.Andrews J, Fierke CA, Birdsall B, Ostler G, Feeney J, Roberts GC, Benkovic SJ. Biochemistry. 1989;28:5743–5750. doi: 10.1021/bi00440a007. [DOI] [PubMed] [Google Scholar]
- 59.Bell RP. The tunnel effect in chemistry. Chapman & Hall; London & New York: 1980. [Google Scholar]
- 60.Kohen A, Klinman JP. Acc Chem Res. 1998;31:397–404. [Google Scholar]
- 61.Kohen A, Klinman JP. Chem Biol. 1999;6:R191–198. doi: 10.1016/s1074-5521(99)80058-1. [DOI] [PubMed] [Google Scholar]
- 62.Meyer MJ, Tomchick DR, Klinman JP. Proc Natl Acad Sci U S A. 2008;105:1146. doi: 10.1073/pnas.0710643105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hay S, Sutcliffe MJ, Scrutton NS. Proc Natl Acad Sci U S A. 2007;104:507–512. doi: 10.1073/pnas.0608408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kamerlin SCL, Mavri J, Warshel A. FEBS Letters. 2010;584:2759–2766. doi: 10.1016/j.febslet.2010.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khavrutskii IV, Price DJ, Lee J, Brooks CLI. Protein Science. 2007;16:1087–1100. doi: 10.1110/ps.062724307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mesecar AD, Stoddard BL, Koshland DE. Science. 1997;227:202–206. doi: 10.1126/science.277.5323.202. [DOI] [PubMed] [Google Scholar]
- 67.Garcia-Viloca M, Truhlar DG, Gao J. Biochemistry. 2003;42:13558–13575. doi: 10.1021/bi034824f. [DOI] [PubMed] [Google Scholar]
- 68.Sawaya MR, Kraut J. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]