Abstract
The enterocin CRL35 biosynthetic gene cluster was cloned and sequenced. The sequence was revealed to be highly identical to that of the mundticin KS gene cluster (S. Kawamoto, J. Shima, R. Sato, T. Eguchi, S. Ohmomo, J. Shibato, N. Horikoshi, K. Takeshita, and T. Sameshima, Appl. Environ. Microbiol. 68:3830-3840, 2002). Short synthetic peptides were designed based on the bacteriocin sequence and were evaluated in antimicrobial competitive assays. The peptide KYYGNGVSCNKKGCS produced an enhancement of enterocin CRL35 antimicrobial activity in a buffer system.
Antibiotic resistance has been a great concern in recent years due to the extensive use of classical antibiotics. The development of new classes of antimicrobial agents has become increasingly important. One plausible alternative could be the application of cationic peptides as antimicrobial substances (6). Among them, bacteriocins produced by lactic acid bacteria have attracted increasing attention because they are not only active in a nanomolar range but also have no toxicity to hosts (4). On the other hand, they are ribosomally synthesized peptides, which creates the possibility of improving the characteristics of each peptide in order to enhance either their activities or spectra of action. Thus, an essential preliminary step should be to study the relationship between primary structure of the antimicrobial peptides and cell specificity (20). For instance, Fimland et al. (9) described the inhibition of several pediocin-like bacteriocins by a 15-mer fragment which spans from the center toward the C terminus of pediocin PA-1 (9). This finding suggests that this segment would interact with the putative cell target of these peptides.
Enterocin CRL35 is an antilisterial pediocin-like bacteriocin produced by Enterococcus mundtii CRL35 (previously known as E. faecium), a strain isolated from an Argentinean artisanal cheese (11). This compound also displays an important activity against herpes simplex virus types I and II (18, 19).
Although the mechanism of action of enterocin CRL35 was recently described (15), only part of the NH2-terminal sequence has been revealed to date (8). In the present work we report the complete sequence of the enterocin CRL35 gene cluster and analyze the putative function of the enterocin domains.
Genetic characterization.
In order to determine the nucleotide sequence of enterocin CRL35, several PCRs were carried out with known enterocin primers. A PCR product with the expected size was obtained with mundticin KS primers (12). Total DNA from E. mundtii AT06 (kindly provided by Marjon Bennik [3]) was used as a positive control. No results were found with ent A, ent B, ent P, ent L50 AB, ent AS-48, and bac 31 primers (7).
The enterocin CRL35 biosynthetic cluster was amplified with the following primers: mun1F (5′ GCAAACCGATAAGAATGTGGGAT 3′) and mun7R (5′ TATACATTGTCCCCACAACC 3′). The resulting 3,128-bp PCR product was purified (GFX PCR DNA and Gel Band Purification kit; Amersham Biosciences) and cloned with a Zero Blunt TOPO PCR Cloning kit (Invitrogen). The recombinant plasmid pE35 was transformed in Escherichia coli DH10B (Stratagene, La Jolla, Calif.), generating an E. coli strain able to produce enterocin CRL35 (data not shown). DNA sequencing of the enterocin CRL35 cluster and the mundticin AT06 structural gene were performed by the automated DNA sequencing service from Universidad de San Martín (ATCG-Servicio de Secuenciación de ADN, Buenos Aires, Argentina). Nucleotide and amino acid sequence homology searches were performed with the BLAST programs at the National Center of Biotechnology Information (1).
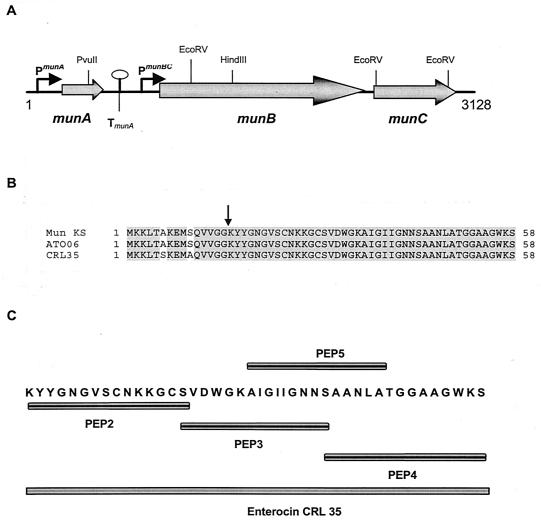
Nucleotide sequence analysis of the enterocin CRL35 gene cluster revealed an identical gene organization (Fig. 1A) and a high sequence identity (DNA:DNA) with the mundticin KS cluster (12). The deduced amino acid sequence of enterocin CRL35 (munA) exhibited two amino acid substitutions at positions 6 (A to S) and 10 (S to A) in the leader peptide (Fig. 1B). As with the mundticin KS cluster, two genes were found in the enterocin CRL35 cluster downstream of the structural munA gene: munB and munC, which code for the maturation transport protein and the immunity protein, showing 96 and 98% amino acid identity, respectively, with corresponding sequences from mundticin KS. Sequence analysis of the mundticin AT06 structural gene showed an enterocin prepeptide 100% identical to mundticin KS (Fig. 1B; accession number P80925), indicating that the last two-amino-acid inversion (SK) described in the originally published mature peptide sequence was probably due to an artifact of the NH2-terminal sequencing process (3, 12).
FIG. 1.
(A) Genetic organization of the enterocin CRL35 biosynthetic cluster. ORFs are represented by horizontal arrows. Putative promoters (P) and transcriptional terminators (T) with a stem-loop structure are shown. Relevant restriction enzyme sites are also shown. (B) Protein sequence alignment of mundticin KS (mun KS), mundticin AT06 (ATO06), and enterocin CRL35 (CRL35) prepeptides. The arrow indicates the double-glycine (GG) processing site of the prepeptides. (C) Schematic representation of enterocin CRL35 and the four truncated derivatives.
The high similarity between enterocin CRL35 and mundticin KS clusters could be the result of an evolutionarily recent horizontal gene transfer, considering that both clusters are carried by plasmids (12). Southern and dot blot analyses revealed that enterocin CRL35 genes are carried in a 50-kb plasmid (data not shown). Moreover, the partial sequencing of the enterocin CRL35 plasmid showed some genes involved in plasmid mobilization (data not shown).
Effect of enterocin CRL35 in combination with short synthetic peptides on Listeria innocua 7 viability.
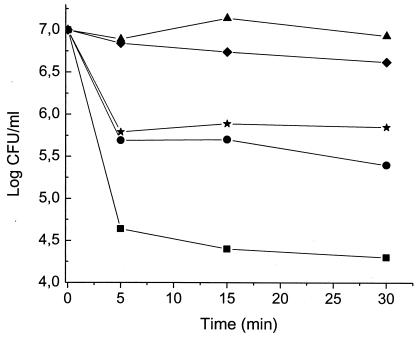
On the basis of the deduced amino acid sequence, the bacteriocin and four fragments spanning different regions of active enterocin CRL35 were synthesized (Fig. 1C) (Bio-Synthesis, Lewisville, Tex.). The purity of the synthetic peptides (greater than 80%) was verified by mass spectrometry (matrix-assisted laser desorption ionization-time of flight) and high-performance liquid chromatography. L. innocua 7 was provided by Unité de Recherches Laitiéres et Génetique Appliqueé, Institut National de la Recherche Agronomique (Jouy-en-Josas, France). The viability assays were carried out as described by McAuliffe et al. (13). As is shown in Fig. 2, the presence of either peptide 3 (SVDWGKAIGIIGNNS) or peptide 5 (KAIGIIGNNSAANLA) inhibited the activity of enterocin CRL35. These results are in agreement with those previously published by Fimland et al. (9), suggesting the implication of this region in interaction with a cellular receptor. On the other hand, the addition of peptide 2 (KYYGNGVSCNKKGCS) enhanced enterocin CRL35 activity. Indeed, the amount of remaining viable cells after treatment with the combination of enterocin and peptide 2 was 10-fold lower than that after treatment of the control with enterocin alone. The presence of peptide 4 (SAANLATGGAAGWKS) produced a slight inhibition of the bacteriocin action. None of the truncated derivatives alone showed any activity, even at concentrations as high as 100 μM.
FIG. 2.
Effect of enterocin CRL35 (Ent) alone and in combination with truncated derivatives on the viability of L. innocua 7 cells. Cell viability, expressed as CFU/milliliter, was determined at the indicated times. Results represent the averages of three independent experiments. •, 0.2 nM enterocin CRL35; ▪, 500 nM enterocin CRL35 plus peptide 2; ▴, 500 nM enterocin CRL35 plus peptide 3; ★, 500 nM enterocin CRL35 plus peptide 4; ♦, 500 nM enterocin CRL35 plus peptide 5.
Effect of enterocin CRL35 combined with short synthetic peptides on the transmembrane potential of L. innocua 7.
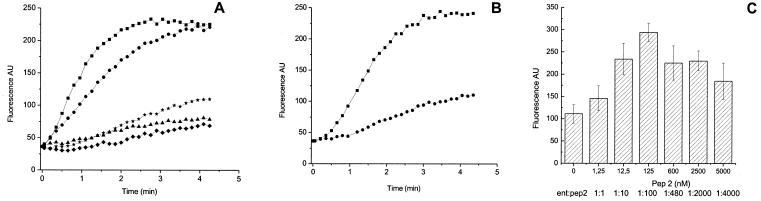
As is the case with other bacteriocins, enterocin CRL35 dissipates both components of the proton motive force (Δp) of sensitive cells (15). Therefore, the modification of the transmembrane electrical potential (ΔΨ) using the potential-sensitive dye 3,3′-dipropylthiacarbocyanine iodide DiSC3 (2, 5) (Molecular Probes, Eugene, Oreg.) was studied upon the addition of enterocin CRL35 alone or in combination with the peptide fragments. This experiment was performed with a Shimatzu RF-5301 PC spectrofluorometer at 30°C, with excitation and emission wavelengths of 622 and 674 nm, respectively. Complete dissipation was obtained in the presence of 1 μM valinomycin. The ΔΨ depolarization induced by the bacteriocin was greatly inhibited by peptides 3 and 5, although a slight dissipation could be observed (Fig. 3A). On the contrary, combination of enterocin CRL35 with peptide 2 produced a faster depolarization of the electrical potential (Fig. 3A). Surprisingly, even though the bactericidal activity of enterocin CRL35 was slightly affected by peptide 4, ΔΨ dissipation was inhibited to some extent. This finding could indicate that ΔΨ depolarization, which is a process generally associated with the bacteriocin effect, would not be the key step in the mechanism of action of this bacteriocin. Indeed, it was recently found that sublethal concentrations of enterocin CRL35 could dissipate ΔΨ without altering the viability of Listeria cells (14). Therefore, it must be stressed that although ΔΨ determination is a useful and valuable tool, it needs to be associated with viability experiments in order to determine the real effect of these kinds of peptides.
FIG. 3.
(A) Dissipation of ΔΨ induced by enterocin CRL35 alone and in combination with truncated derivatives on L. innocua 7 cells. The results are the means of five independent experiments. •, 0.5 nM enterocin CRL35; ▪, 500 nM enterocin CRL35 plus peptide 2; ▴, 500 nM enterocin CRL35 plus peptide 3; ★, 500 nM enterocin CRL35 plus peptide 4; ♦, 500 nM enterocin CRL35 plus peptide 5. (B) Dissipation of ΔΨ induced by 0.2 nM enterocin CRL35 alone (•) and the combination of pep2 plus enterocin CRL35 (ratio 100:1) (▪) on L. innocua 7 cells. (C) Dissipation of ΔΨ induced by enterocin CRL35 alone and in combination of different concentrations of peptide 2 on L. innocua 7 cells. AU, arbitrary units.
The positive effect obtained with peptide 2 is more pronounced at low bacteriocin concentrations (Fig. 3B). The enhancement of enterocin CRL35 effect on ΔΨ supports the results found in the viability assays. To elucidate the optimum ratio between the bacteriocin and peptide 2, different concentrations of the latter compound were added to keep the concentration of enterocin CRL35 constant at 0.2 nM. As shown in Fig. 3C, a ratio of 100:1 (peptide 2:bacteriocin) was found to be optimal.
Although the modifications to bactericidal activity produced by all the peptides were detected in the experiments mentioned above, no significant change in the enterocin CRL35 MIC was observed. The lack of concordance between the former and latter findings is not unexpected. Galvin et al. observed that lacticin 3147 displayed antimicrobial activity against some pathogenic bacteria in buffer but did not do so on agar plates, pointing out that time kill curves would be a more accurate method to determine the true potential of these kinds of microbicidal peptides (10). In addition, from a pharmacodynamic point of view the MICs provide only limited information of the kinetics of the antimicrobial action. Furthermore, the MIC does not provide information on the rate of bactericidal activity (16).
This report constitutes the first evidence of a fragment, derived from the NH2-terminal region of a pediocin-like bacteriocin, which enhances bacteriocin activity. The main effect of peptide 2 would be the increase in bacteriocin killing rate. Because this domain would be involved in the initial unspecific binding with the membrane and would not be related to pore formation (5, 17), a plausible explanation might be that the first interaction of peptide 2 with the membrane would facilitate later binding and correct insertion of enterocin CRL35. These findings could open a new field of research into the development of short peptides that exert strong enhancement on the antimicrobial activity of class II bacteriocins. The next step would be the design of peptide 2 variants with improved characteristics. Although further studies involving pharmacokinetics as well as immunogenicity and toxicity tests must be carried out, this interesting positive association might be successfully used in both food preservation and medical treatments.
Nucleotide sequence accession number.
The DNA sequence of the enterocin CRL35 gene cluster has been deposited in GenBank under accession number AY398693.
Acknowledgments
We thank Néstor Katz for his collaboration on the fluorescence experiments.
Financial support was provided by ANPCyT (Buenos Aires, Argentina) with grant PICT09-04632 and by CONICET (Buenos Aires, Argentina).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennik, M., A. Verheul, T. Abee, G. Naaktgeboren-Stoffels, L. Gorris, and E. Smid. 1997. Interactions of nisin and pediocin PA-1 with closely related lactic acid bacteria that manifest over 100-fold differences in bacteriocin sensitivity. Appl. Environ. Microbiol. 63:3628-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennik, M. H., B. Vanloo, R. Brasseur, L. G. Gorris, and E. J. Smid. 1998. A novel bacteriocin with a YGNGV motif from vegetable-associated Enterococcus mundtii: full characterization and interaction with target organisms. Biochem. Biophys. Acta 1373:47-58. [DOI] [PubMed] [Google Scholar]
- 4.Cleveland, J., T. J. Montville, I. F. Nes, and M. L. Chikindas. 2001. Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71:1-20. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y., R. D. Ludescher, and T. J. Montville. 1997. Electrostatic interactions, but not the YGNGV consensus motif, govern the binding of pediocin PA-1 and its fragments to phospholipid vesicles. Appl. Environ. Microbiol. 63:4770-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diep, D., and I. F. Nes. 2002. Ribosomally antibacterial peptides in gram-positive bacteria. Curr. Drug Targets 3:107-122. [DOI] [PubMed] [Google Scholar]
- 7.Du Toit, M., C. M. A. P. Franz, L. M. T. Dicks, and W. H. Holzapfel. 2000. Preliminary characterization of bacteriocins produced by Enterococcus faecium and Enterococcus faecalis isolated from pig faeces. J. Appl. Microbiol. 88:482-494. [DOI] [PubMed] [Google Scholar]
- 8.Farías, M. E., R. N. Farías, A. P. de Ruiz Holgado, and F. Sesma. 1996. Purification and N-terminal amino acid sequence of enterocin CRL 35, a ‘pediocin-like’ bacteriocin produced by Enterococcus faecium CRL 35. Lett. Appl. Microbiol. 22:417-419. [DOI] [PubMed] [Google Scholar]
- 9.Fimland, G., R. Jack, G. Jung, I. F. Nes, and J. Nissen-Meyer. 1998. The bactericidal activity of pediocin PA-1 is specifically inhibited by a 15-mer fragment that spans the bacteriocin from the center toward the C terminus. Appl. Environ. Microbiol. 64:5057-5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galvin, M., C. Hill, and R. P. Ross. 1999. Lacticin 3147 displays activity in buffer against gram-positive bacterial pathogens which appear insensitive in standard plate assays. Lett. Appl. Microbiol. 28:355-358. [DOI] [PubMed] [Google Scholar]
- 11.Giori, G. S., G. F. Valdez, A. P. Ruíz Holgado, and G. Oliver. 1983. Microflora of Tafi cheese: changes during manufacture and maturation. J. Food Prot. 46:518-521. [DOI] [PubMed] [Google Scholar]
- 12.Kawamoto, S., J. Shima, R. Sato, T. Eguchi, S. Ohmomo, J. Shibato, N. Horikoshi, K. Takeshita, and T. Sameshima. 2002. Biochemical and genetic characterization of mundticin KS, an antilisterial peptide produced by Enterococcus mundtii NFRI 7393. Appl. Environ. Microbiol. 68:3830-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McAuliffe, O., M. P. Ryan, R. P. Ross, C. Hill, P. Breeuwer, and T. Abee. 1998. Lacticin 3147, a broad-spectrum bacteriocin which selectively dissipates the membrane potential. Appl. Environ. Microbiol. 64:439-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minahk, C. J., F. Dupuy, and R. D. Morero. 2004. Enhancement of antibiotic activity by sub-lethal concentrations of enterocin CRL35. J. Antimicrobiol. Chemother. 53:240-246. [DOI] [PubMed] [Google Scholar]
- 15.Minahk, C., and R. Morero. 2003. Inhibition of enterocin CRL35 antibiotic activity by mono and divalent ions. Lett. Appl. Microbiol. 37:374-379. [DOI] [PubMed] [Google Scholar]
- 16.Mueller, M., A. de la Peña, and H. Derendorf. 2004. Issues in pharmacokinetics and pharmacodynamics of anti-infective agents: kill curves versus MIC. Antimicrob. Agents Chemother. 48:369-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quadri, L., L. Z. Yan, M. E. Stiles, and J. C. Vederas. 1997. Effect of amino acid substitutions on the activity of carnobacteriocin B2. J. Biol. Chem. 272:3384-3388. [DOI] [PubMed] [Google Scholar]
- 18.Wachsman, M. B., V. Castilla, A. P. de Ruiz Holgado, R. A. de Torres, F. Sesma, and C. E. Coto. 2003. Enterocin CRL35 inhibits late stages of HSV-1 and HSV-2 replication in vitro. Antivir. Res. 58:17-24. [DOI] [PubMed] [Google Scholar]
- 19.Wachsman, M. B., M. E. Farias, E. Takeda, F. Sesma, A. P de Ruiz Holgado, R. A. de Torres, and C. E. Coto. 1999. Antiviral activity of enterocin CRL35 against herpesviruses. Int. J. Antimicrob. Agents 12:293-299. [DOI] [PubMed] [Google Scholar]
- 20.Yang, L. Z., A. C. Gibbs, M. E. Stiles, D. S. Wishart, and J. C. Vederas. 2000. Analogues of bacteriocin: antimicrobial specificity and interaction of leucocin A with its enantiomer, carnobacteriocin B2, and truncated derivatives. J. Med. Chem. 43:4579-4581. [DOI] [PubMed] [Google Scholar]