Abstract
Rhesus and human cytomegalovirus (RhCMV and HCMV, respectively) exhibit comparable inhibition by benzimidazole nucleosides, including 2,5,6-trichloro-(1-β-d-ribofuranosyl)benzimidazole (TCRB), and pyrrolo[2,3-d]pyrimidines. The two HCMV protein targets of TCRB, UL89 and UL56, are highly conserved with their RhCMV homologues. These data indicate that infection of rhesus macaques with RhCMV represents a useful model to test novel anti-HCMV drugs.
Human cytomegalovirus (HCMV) is a betaherpesvirus that establishes a lifelong, persistent, but asymptomatic, infection in healthy individuals. However, it can cause substantial morbidity and mortality in immunocompromised patients (1), including immunosuppressed transplant recipients and those with AIDS. HCMV is also a leading cause of birth defects after congenital infection of fetuses.
Antiviral drugs approved for therapy of HCMV infection include ganciclovir (GCV), valganciclovir (vGCV), cidofovir (CDV), foscarnet (phosphonoformic acid [PFA]), and fomivirsen. GCV, vGCV, CDV, and PFA all inhibit virus replication through interaction with the viral DNA polymerase, encoded by the HCMV UL54 gene. Each of these drugs has low oral bioavailability and/or dose-related toxicities that have limited their clinical usefulness (16, 31). GCV and vGCV cause substantial neutropenia and myelosuppression, and both CDV and PFA cause renal toxicity. Drug resistance is another problem in therapy with GCV, vGCV, or PFA and has been shown to occur in cell culture systems with CDV. HCMV resistance to GCV occurs predominantly from mutations in the virally encoded phosphotransferase (UL97) (9, 26) but can also arise from mutations in the DNA polymerase gene (UL54) (36). Resistance to PFA or CDV arises from mutations in UL54 (10, 11). Some of the mutations in UL54 confer cross-resistance to GCV and CDV or to GCV and PFA (10, 11, 34). Fomivirsen is an antisense phosphorthioate oligonucleotide that is limited to intravitreal treatment of HCMV retinitis only, whereas the other approved drugs are used systemically. There are clear clinical needs for safer anti-HCMV drugs with more favorable pharmacological properties and a need to develop new drugs that inhibit HCMV targets other than DNA polymerase.
One promising class of anti-CMV compounds is the benzimidazole nucleosides (39). Unlike other nucleoside analogs, some members of this group that are active against HCMV are not phosphorylated (21) and are not substrates for DNA polymerase (22, 40). Two potent members of this class, 2,5,6-trichloro-1-(β-d-ribofuranosyl)benzimidazole (TCRB) and its 2-bromo derivative (BDCRB), exert their activity by blocking the cleavage of concatemeric CMV DNA into genome-length pieces for packaging (40). Resistance to TCRB or BDCRB is conferred by mutations in HCMV genes UL89 and UL56 (22, 40).
An attractive animal model to help in the development of anti-CMV drugs is infection of rhesus macaques (Macaca mulatta) with rhesus CMV (RhCMV). This model is a strong recapitulation of the natural history of HCMV. RhCMV is endemic in populations of macaques, and immunocompetent monkeys do not display clinical signs of disease after natural infection or experimental inoculation (23, 41). RhCMV infection results in a lifelong asymptomatic persistence, characterized by periodic shedding of virus (2, 14), similar to HCMV infection of immunocompetent humans (1). However, like HCMV, RhCMV causes significant morbidity in macaques without a fully functional immune system. Fetal macaques experimentally infected with RhCMV (6, 25, 38) or monkeys coinfected with RhCMV and immunodeficiency-inducing retroviruses (simian immunodeficiency virus [4, 17-19, 35] or type D simian retrovirus [30]) exhibit clinical sequelae almost identical to those observed in HCMV-infected humans in comparable clinical settings. Moreover, allograft recipients can develop fulminant RhCMV infections during immunosuppressive treatment (D. M. Bouley and P. A. Barry, unpublished data). In addition to these similarities in pathogenicity, the genomes of RhCMV and HCMV are essentially colinear, and viral proteins representing identified or potential antiviral targets have a high degree of amino acid identity (13). As would be expected with strong conservation of target proteins, RhCMV has been shown to be similar to HCMV in susceptibility to GCV or PFA (37). The potential utility of the nonhuman primate model is further strengthened by the strong evolutionary relationship of the natural hosts of RhCMV and HCMV in terms of development, physiology, and immunology.
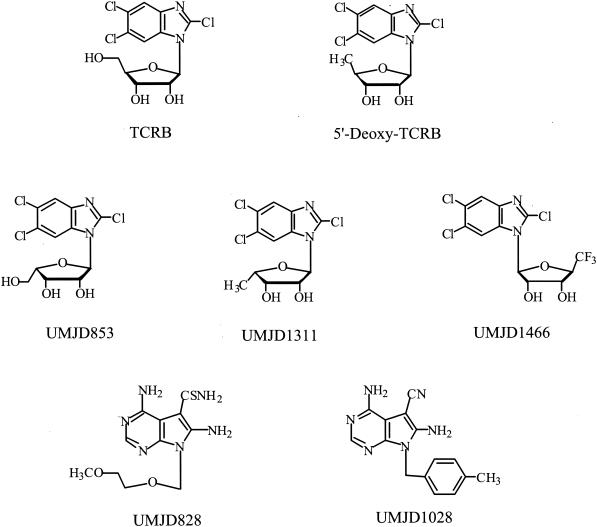
In the work reported here, we have examined the susceptibilities of RhCMV to TCRB, selected other benzimidazole nucleosides, and two pyrrolo[2,3-d]pyrimidine nucleoside analogs that are also highly active against HCMV (15, 33). The compounds used in this study are shown in Fig. 1. The syntheses and antiviral properties of UMJD076 (TCRB), UMJD697 (5′-deoxy-TCRB), UMJD853, UMJD1311, and the pyrrolo[2,3-d]pyrimidine nucleoside analogs UMJD828 and UMJD1028 have been published previously (27, 32, 33, 39). The compound UMJD1466 was synthesized by a method similar to that described for TCRB (39); antiviral activity data for this compound have not been published previously.
FIG. 1.
Structures of benzimidazole nucleosides and pyrrolo[2,3-d]pyrimidine analogs (UMJD828 and UMJD1028) active against human and rhesus CMV.
The 68-1 strain of RhCMV (2) and the Towne strain of HCMV (28) were used for these studies. RhCMV was propagated in primary dermal fibroblasts (RhDF) from rhesus macaques, and virus stocks were prepared as previously described (5). HCMV was propagated in human embryonic lung fibroblasts (HELF) (kindly provided by J. Weideman, University of California, Davis). Cultures were maintained in Dulbecco's modified Eagle medium (DMEM), supplemented with fetal bovine serum (FBS) (20% for RhDF and 10% for HELF), 100 U of penicillin per ml, and 100 μg of streptomycin per ml at 37°C in a humidified atmosphere of 5% CO2.
Focus reduction assays were used for drug susceptibility assays in RhDF cells for RhCMV and HELF for HCMV. Cells were seeded into 24-well tissue culture plates at a density of 5 × 104 cells per well in 1 ml of culture medium and incubated overnight. Prior to virus infections, culture medium was removed, cells were washed three times with phosphate-buffered saline (PBS), and cells were preincubated for 45 min in DMEM containing 2% FBS and drugs at the concentrations to be tested. After this preincubation, medium was removed and cultures were infected with approximately 50 focus-forming units of virus in 0.5 ml of DMEM with 2% FBS and drugs at appropriate concentrations. After adsorption for 2 h at 37°C, the inoculum was removed, and cells were overlaid with 1.5 ml of DMEM containing 2% FBS, 0.8% agarose (Sea-Plaque), and drugs at appropriate concentrations. After 7 to 10 days, the overlay was removed, and cells were fixed with acetone-methanol (1:1) at 4°C for 20 min, washed three times with PBS, and stored at −20°C until immunocytochemical staining of foci. Immediately prior to staining, fixed cells were washed three times with PBS and then incubated for 2 h at 25°C with either rabbit anti-RhCMV IE1 antibody (1:3,200 dilution in PBS containing 0.1% Tween 20) or a monoclonal antibody to HCMV IE1 (clone MAB810; Chemicon, Temecula, Calif.) (23). Plates were subsequently washed three times with PBS and then incubated with a 1:800 dilution of biotinylated goat anti-rabbit antibody (Vector Laboratories) for 1 h. Plates were washed three times with PBS and then incubated with horseradish peroxidase-conjugated avidin-biotin complex (avidin-biotin complex from Vector Laboratories) for 60 min. Foci of infected cells were visualized by staining with the peroxidase substrate 3,3′-diaminobenzidine and counted with a 10× objective on an Olympus CK40 microscope. Foci were considered positive if there were, at least, four clustered cells that had IE1 nuclear staining. Data were plotted as a percentage of control foci (no drug) versus inhibitor concentrations. The concentrations required to inhibit focus formation by 50% (EC50) were obtained directly from the linear portions of those plots.
The susceptibilities of RhCMV and HCMV to TCRB were compared to determine whether the two primate CMVs exhibited comparable sensitivities to a compound that is directed against viral enzymes involved in the cleavage of DNA for packaging. RhCMV was inhibited by TCRB with an EC50 slightly lower (threefold) than that of HCMV (Table 1). The EC50 we observed for TCRB against HCMV in these studies (2.5 μM) was comparable to that (2.9 μM) of previous studies (39). We also compared susceptibilities of RhCMV and HCMV to other benzimidazole nucleosides that previously had been shown (21, 27) to inhibit HCMV replication in vitro (Fig. 1). The 5′-deoxy analog of TCRB (5′-dTCRB) was more active against HCMV (0.37 μM) than was TCRB (Table 1). The EC50 we observed for 5′-dTCRB was comparable to that (0.58 μM) of previous studies (21). 5′-dTCRB was similarly more active than TCRB against RhCMV (0.04 versus 0.9 μM) (Table 1). RhCMV was inhibited by the three other benzimidazole nucleosides (UMJD853, UMJD1311, and UMJD1466) with EC50s slightly higher (1.6- to 3-fold) than the EC50s for HCMV.
TABLE 1.
EC50s for drugs against HCMV and RhCMV
| Drug | EC50a
|
|
|---|---|---|
| HCMV | RhCMV | |
| Benzimidazole nucleosides | ||
| TCRB | 2.5 | 0.90 (0.06) |
| 5′-dTCRB (UMJD697) | 0.37 | 0.04 (0.01) |
| UMJD853 | 3.2 | 5.0b |
| UMJD1311 | 0.24 | 0.76 (0.32) |
| UMJD1466 | 0.45 | 0.71 (0.14) |
| Pyrrolo[2,3-d]pyrimidines | ||
| UMJD828 | 0.62 | 1.4 (0.14) |
| UMJD1028 | 0.35 | 1.6 (0.23) |
EC50s are the means of three experiments (standard deviations shown in parentheses) for RhCMV and the averages of two experiments for HCMV.
Drug UMJD853 was analyzed in two experiments for RhCMV.
Because two of these compounds, UMJD1311 and UMJD1466, act early in the virus replication cycle by a mechanism different from TCRB (J. C. Drach and L. B. Townsend, unpublished observations), it was of interest to determine whether two unrelated nonnucleoside antiviral agents that also act early in the replication cycle of HCMV by an unknown mechanism (15) were active against RhCMV. The pyrrolo[2,3-d]pyrimidines UMJD828 and UMJD1028 (15, 32, 33) were active against RhCMV (Table 1), although RhCMV was 2.2- and 4.6-fold less sensitive, respectively, to these compounds than HCMV.
The RhCMV genes for the two protein homologues of HCMV that are targets of TCRB, UL89 and UL56, were cloned and sequenced, and the predicted open reading frames (ORFs) were determined. The UL56 ORF was contained within a subgenomic, molecular clone that had been previously characterized for the UL55 (glycoprotein B) ORF (20). Almost all of the UL89b ORF was contained within a restriction fragment cloned from the RhCMV genome and identified by sequence comparison to the HCMV genome (8; also data not shown). The cDNA transcript was mapped for exon junctions and the initiation or termination sites of transcription by rapid amplification of cDNA ends (RACE) (12) using the SMART-RACE cDNA kit from Clontech (Palo Alto, Calif.). For 5′ RACE, reverse transcription was initiated using primer 539 (5′ GATACAGACCCACCGCGGCAATCC 3′), and RNA purified from RhCMV-infected RhDF was used as a template, similar to published protocols (5, 24). The 5′ and 3′ RACE cDNAs were amplified with primers 537 (5′ CCCGTACCAGAAGCCTTGCGGTTG 3′) and 536 (5′ CGACAAACGCCTAGCCGTTGAACAG 3′), respectively, cloned into the TOPO-TA vector (Invitrogen), and sequenced (Davis Sequencing, Davis, Calif.). All sequence analyses were performed with SeqWeb (version 2; Accelrys, San Diego, Calif.).
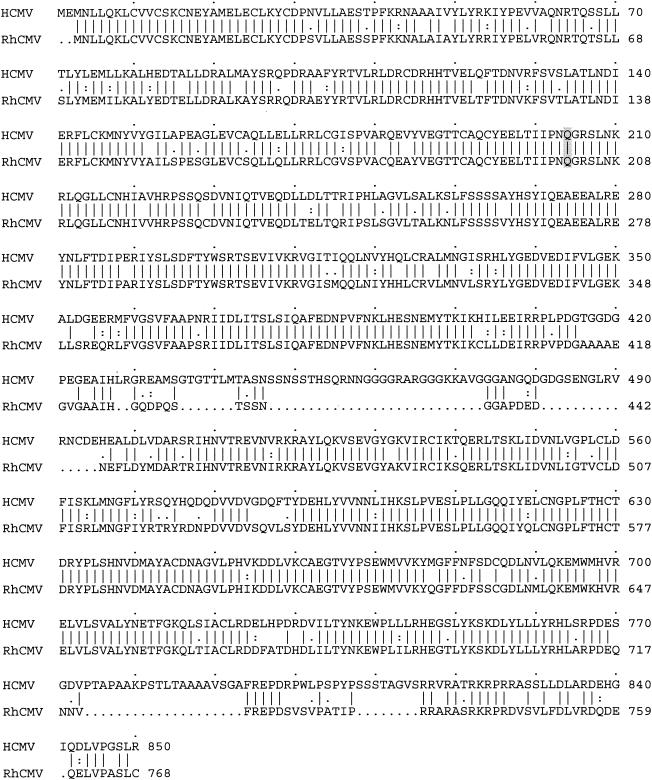
There was an extremely high conservation of amino acid sequence for both UL89 and UL56 (87 and 82% identity, respectively) of these two viruses (Fig. 2 and 3). There was 100% identity in the regions of both genes contiguous to known mutations in HCMV that confer resistance to TCRB and BDCRB, amino acid 204 in UL56 (21) and amino acids 344 and 355 in UL89 of the Towne strain of HCMV (40). The levels of identity for these two proteins were some of the highest that have been observed between the RhCMV and HCMV ORFs that have been analyzed (3, 7, 20, 24, 37; P. A. Barry, unpublished data). By comparison, these two viruses have 59 and 54% amino acid sequence identity in the DNA polymerase (UL54) and phosphotransferase (UL97) proteins, respectively (37). Differences between the UL89 ORF of primate CMV were evenly distributed throughout the predicted protein (Fig. 2). RNA analysis of the RhCMV-infected cells confirmed that the UL89 transcript consisted of a singly spliced message with an intron of approximately 3.5 kbp (data not shown). The DNA sequence of the intron termini contained the consensus 5′ GT…AG 3′ bases. The terminal base of a consensus polyadenylation sequence (5′ AATAAA 3′) was located 16 bases upstream of the site of polyadenylation. The position of the exon-exon junction was identical for HCMV and RhCMV (Fig. 2). For UL56, there was strong conservation throughout the protein except for two areas of discordance characterized by the presence of two gapped regions (amino acids 435 to 495 and 774 to 815 [RhCMV coordinates]) (Fig. 3).
FIG. 2.
GAP analysis of the HCMV and RhCMV UL89 proteins. The GAP program of SeqWeb, which is based on the algorithm of Needleman and Wunsch (29), was used to align the UL89 proteins of RhCMV (GenBank accession number AY536265) and HCMV (AD169; SwissProt number P16732). The two predicted ORFs are 87% identical and 91% similar, with three single amino acid gaps (.). Amino acids that are identical (|), highly conserved (:), and less conserved (.) in the two ORFs are indicated between the two sequences. Exon-exon junctions (arrows) and mutations in HCMV UL89 that cause resistance to TCRB and BDCRB (shading) are shown. The GenBank accession number for RhCMV UL89 is AY536265.
FIG. 3.
GAP analysis of the HCMV and RhCMV UL56 ORFs. The two predicted ORFs are 82% identical and 86% similar, with seven gaps (.) in the sequences. Amino acids that are identical (|), highly conserved (:), and less conserved (.) in the two ORFs are indicated between the two sequences. The single mutation in HCMV UL56 that causes resistance to TCRB and BDCRB is indicated by shading. The RhCMV UL56 protein sequence from the molecularly cloned fragment was identical to that found in the subsequently published sequence of the RhCMV genome (13) (GenBank accession number AY186194). The HCMV UL56 sequence is from the Towne strain (SwissProt accession number AAC40818) (22).
We conclude from these data that RhCMV and HCMV are comparably inhibited by benzimidazole nucleosides, such as TCRB, that act against DNA processing late in the viral replication cycle, and pyrrolo[2,3-d]pyrimidines that act at an unknown early stage. The utility of the RhCMV model for future studies is enhanced by the observations that the EC50s for inhibition of murine, rat, or guinea pig CMV by BDCRB are more than 50-fold higher than for HCMV or RhCMV (42). The exceedingly high homologies within the UL56 and UL89 proteins of primate CMVs further suggest that in vivo analyses of these compounds against RhCMV infection will be especially relevant to a better understanding of bioactivity and mechanisms of drug resistance.
Expanding the available options of effective chemotherapeutic agents would dramatically enhance treatment of HCMV disease. Further utilization of the rhesus macaque model should facilitate this expansion. As described above, the spectrum of diseases associated with HCMV have all been reported for experimental studies of RhCMV, enabling testing of antivirals in clinical settings that accurately reflect the human condition. The RhCMV genome has been sequenced recently (13), and the RhCMV genome is closer in sequence to HCMV than any other CMV used in an animal model. Sensitive assays have been developed to quantify viral loads either by infection in culture (5) or real-time PCR detection of RhCMV genomes in plasma or mucosal fluids (14, 35). In sum, a sound foundation is in place to test new drugs, such as the benzimidazole ribonucleosides and pyrrolo[2,3-d]pyrimidines, in a primate host.
Acknowledgments
We thank Shan Shan Zhou for excellent technical assistance.
This work was supported in part by grants from the National Institutes of Health to P.A.B. (RO1 HL36859 and R03 AI053208) and to the California National Primate Research Center (RR000169).
REFERENCES
- 1.Alford, C. A., and W. J. Britt. 1993. Cytomegalovirus, p. 227-255. In B. Roizman, R. J. Whitley, and C. Lopez (ed.), The human herpesviruses. Raven Press, Ltd., New York, N.Y.
- 2.Asher, D. M., J. C. J. Gibbs, D. J. Lang, D. C. Gadjusek, and R. M. Chanock. 1974. Persistent shedding of cytomegalovirus in the urine of healthy rhesus monkeys. Proc. Soc. Exp. Biol. Med. 145:794-801. [DOI] [PubMed] [Google Scholar]
- 3.Barry, P. A., D. J. Alcendor, M. D. Power, H. Kerr, and P. A. Luciw. 1996. Nucleotide sequence and molecular analysis of the rhesus cytomegalovirus immediate-early gene and the UL121-117 open reading frames. Virology 215:61-72. [DOI] [PubMed] [Google Scholar]
- 4.Baskin, G. B. 1987. Disseminated cytomegalovirus infection in immunodeficient rhesus macaques. Am. J. Pathol. 129:345-352. [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, W. L., V. Kirchoff, G. S. Pari, and P. A. Barry. 2002. Replication of rhesus cytomegalovirus in life-expanded rhesus fibroblasts expressing human telomerase. J. Virol. Methods 104:135-146. [DOI] [PubMed] [Google Scholar]
- 6.Chang, W. L., A. F. Tarantal, S. S. Zhou, A. D. Borowsky, and P. A. Barry. 2002. A recombinant rhesus cytomegalovirus expressing enhanced green fluorescent protein retains the wild-type phenotype and pathogenicity in fetal macaques. J. Virol. 76:9493-9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y.-N., K.-T. Jeang, T. Lietman, and G. S. Hayward. 1995. Structural organization of the spliced immediate-early gene complex that encodes the major acidic nuclear (IE1) and transactivator (IE2) proteins of African green monkey cytomegalovirus. J. Biomed. Sci. 2:105-130. [DOI] [PubMed] [Google Scholar]
- 8.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 9.Chou, S., A. Erice, M. C. Jordan, G. M. Vercellotti, K. R. Michels, C. L. Talarico, S. C. Stanat, and K. K. Biron. 1995. Analysis of the UL97 phosphotransferase coding sequence in clinical cytomegalovirus isolates and identification of mutations conferring ganciclovir resistance. J. Infect. Dis. 171:576-583. [DOI] [PubMed] [Google Scholar]
- 10.Chou, S., G. Marousek, S. Guentzel, S. E. Follansbee, M. E. Poscher, J. P. Lalezari, R. C. Miner, and W. L. Drew. 1997. Evolution of mutations conferring multidrug resistance during prophylaxis and therapy for cytomegalovirus disease. J. Infect. Dis. 176:786-789. [DOI] [PubMed] [Google Scholar]
- 11.Cihlar, T., M. D. Fuller, and J. M. Cherrington. 1998. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J. Virol. 72:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frohman, M. A., M. K. Dush, and G. R. Martin. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85:8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen, S. G., L. I. Strelow, D. C. Franchi, D. G. Anders, and S. W. Wong. 2003. Complete sequence and genomic analysis of rhesus cytomegalovirus. J. Virol. 77:6620-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huff, J. E., R. Eberle, J. Capitanio, S.-S. Zhou, and P. A. Barry. 2003. Differential detection of B virus and rhesus cytomegalovirus in rhesus macaques. J. Gen. Virol. 84:83-92. [DOI] [PubMed] [Google Scholar]
- 15.Jacobson, J. G., T. E. Renau, M. R. Nassiri, D. G. Sweier, J. M. Breitenbach, L. B. Townsend, and J. C. Drach. 1999. Nonnucleoside pyrrolopyrimidines with a unique mechanism of action against human cytomegalovirus. Antimicrob. Agents Chemother. 43:1888-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobson, M. A. 1994. Current management of cytomegalovirus disease in patients with AIDS. AIDS Res. Hum. Retrovir. 10:917-923. [DOI] [PubMed] [Google Scholar]
- 17.Kaur, A., N. Kassis, C. L. Hale, M. Simon, M. Elliott, A. Gomez-Yafa, J. D. Lifson, R. C. Desrosiers, F. Wang, P. Barry, M. Mach, and R. P. Johnson. 2003. Direct relationship between suppression of virus-specific immunity and emergence of cytomegalovirus disease in simian AIDS. J. Virol. 77:5749-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaur, A., M. D. Daniel, D. Hempel, D. Lee-Parritz, M. S. Hirsch, and R. P. Johnson. 1996. Cytotoxic T-lymphocyte responses to cytomegalovirus in normal and simian immunodeficiency virus-infected rhesus macaques. J. Virol. 70:7725-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaur, A., M. Rosenzweig, and R. P. Johnson. 2000. Immunological memory and acquired immunodeficiency syndrome pathogenesis. Philos. Trans. R. Soc. Lond. B 355:381-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kravitz, R. H., K. S. Sciabica, K. Cho, P. A. Luciw, and P. A. Barry. 1997. Cloning and characterization of the rhesus cytomegalovirus glycoprotein B. J. Gen. Virol. 78:2009-2013. [DOI] [PubMed] [Google Scholar]
- 21.Krosky, P. M., K. Z. Borysko, M. R. Nassiri, R. V. Devivar, R. G. Ptak, M. G. Davis, K. K. Biron, L. B. Townsend, and J. C. Drach. 2002. Phosphorylation of β-d-ribosylbenzimidazoles is not required for activity against human cytomegalovirus. Antimicrob. Agents Chemother. 46:478-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krosky, P. M., M. R. Underwood, S. R. Turk, K. W. Feng, R. K. Jain, R. G. Ptak, A. C. Westerman, K. K. Biron, L. B. Townsend, and J. C. Drach. 1998. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J. Virol. 72:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lockridge, K. M., G. Sequar, S. S. Zhou, Y. Yue, C. M. Mandell, and P. A. Barry. 1999. Pathogenesis of experimental rhesus cytomegalovirus infection. J. Virol. 73:9576-9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lockridge, K. M., S. S. Zhou, R. H. Kravitz, J. L. Johnson, E. T. Sawai, E. L. Blewett, and P. A. Barry. 2000. Primate cytomegaloviruses encode and express an IL-10-like protein. Virology 268:272-280. [DOI] [PubMed] [Google Scholar]
- 25.London, W. T., A. J. Martinez, S. A. Houff, W. C. Wallen, B. L. Curfman, R. G. Traub, and J. L. Sever. 1986. Experimental congenital disease with simian cytomegalovirus in rhesus monkeys. Teratology 33:323-331. [DOI] [PubMed] [Google Scholar]
- 26.Lurain, N. S., L. E. Spafford, and K. D. Thompson. 1994. Mutations in the UL97 open reading frame of human cytomegalovirus strains resistant to ganciclovir. J. Virol. 68:4427-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Migawa, M. T., J. L. Girardet, J. A. Walker II, G. W. Koszalka, S. D. Chamberlain, J. C. Drach, and L. B. Townsend. 1998. Design, synthesis, and antiviral activity of alpha-nucleosides: d- and l-isomers of lyxofuranosyl- and (5-deoxylyxofuranosyl)benzimidazoles. J. Med. Chem. 41:1242-1251. [DOI] [PubMed] [Google Scholar]
- 28.Mocarski, E. S., A. C. Liu, and R. R. Spaete. 1987. Structure and variability of the a sequence in the genome of human cytomegalovirus (Towne strain). J. Gen. Virol. 68:2223-2230. [DOI] [PubMed] [Google Scholar]
- 29.Needleman, S. B., and C. D. Wunsch. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443-453. [DOI] [PubMed] [Google Scholar]
- 30.Osborn, K. G., S. Prahalada, L. J. Lowenstine, M. B. Gardner, D. H. Maul, and R. V. Henrickson. 1984. The pathology of an epizootic of acquired immunodeficiency in rhesus macaques. Am. J. Pathol. 114:94-103. [PMC free article] [PubMed] [Google Scholar]
- 31.Polis, M. A., K. M. Spooner, B. F. Baird, J. F. Manischewitz, H. S. Jaffe, P. E. Fisher, J. Falloon, R. T. Davey, Jr., J. A. Kovacs, R. E. Walker, S. M. Whitcup, R. B. Nussenblatt, H. C. Lane, and H. Masur. 1995. Anticytomegaloviral activity and safety of cidofovir in patients with human immunodeficiency virus infection and cytomegalovirus viruria. Antimicrob. Agents Chemother. 39:882-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renau, T. E., C. Kennedy, R. G. Ptak, J. M. Breitenbach, J. C. Drach, and L. B. Townsend. 1996. Synthesis of non-nucleoside analogs of toyocamycin, sangivamycin, and thiosangivamycin: the effect of certain 4- and 4,6-substituents on the antiviral activity of pyrrolo[2,3-d]pyrimidines. J. Med. Chem. 39:3470-3476. [DOI] [PubMed] [Google Scholar]
- 33.Renau, T. E., L. L. Wotring, J. C. Drach, and L. B. Townsend. 1996. Synthesis of non-nucleoside analogs of toyocamycin, sangivamycin, and thiosangivamycin: influence of various 7-substituents on antiviral activity. J. Med. Chem. 39:873-880. [DOI] [PubMed] [Google Scholar]
- 34.Sarasini, A., F. Baldanti, M. Furione, C. Percivalle, R. Brerra, M. Barbi, and G. Gerna. 1995. Double resistance to ganciclovir and foscarnet of four human cytomegalovirus strains recovered from AIDS patients. J. Med. Virol. 47:237-244. [DOI] [PubMed] [Google Scholar]
- 35.Sequar, G., W. J. Britt, F. D. Lakeman, K. M. Lockridge, R. P. Tarara, D. R. Canfield, S. S. Zhou, M. B. Gardner, and P. A. Barry. 2002. Experimental coinfection of rhesus macaques with rhesus cytomegalovirus and simian immunodeficiency virus: pathogenesis. J. Virol. 76:7661-7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan, V., K. K. Biron, C. L. Talarico, S. C. Stanat, M. Davis, L. M. Pozzi, and D. M. Coen. 1993. A point mutation in the human cytomegalovirus DNA polymerase gene confers resistance to ganciclovir and phosphonylmethoxyalkyl derivatives. Antimicrob. Agents Chemother. 37:19-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swanson, R., E. Bergquam, and S. W. Wong. 1998. Characterization of rhesus cytomegalovirus genes associated with anti-viral susceptibility. Virology 240:338-348. [DOI] [PubMed] [Google Scholar]
- 38.Tarantal, A. F., S. Salamat, W. J. Britt, P. A. Luciw, A. G. Hendrickx, and P. A. Barry. 1998. Neuropathogenesis induced by rhesus cytomegalovirus in fetal rhesus monkeys (Macaca mulatta). J. Infect. Dis. 177:446-450. [DOI] [PubMed] [Google Scholar]
- 39.Townsend, L. B., R. V. Devivar, S. R. Turk, M. R. Nassiri, and J. C. Drach. 1995. Design, synthesis, and antiviral activity of certain 2,5,6-trihalo-1-(beta-d-ribofuranosyl)benzimidazoles. J. Med. Chem. 38:4098-4105. [DOI] [PubMed] [Google Scholar]
- 40.Underwood, M. R., R. J. Harvey, S. C. Stanat, M. L. Hemphill, T. Miller, J. C. Drach, L. B. Townsend, and K. K. Biron. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel, P., B. J. Weigler, H. Kerr, A. Hendrickx, and P. A. Barry. 1994. Seroepidemiologic studies of cytomegalovirus infection in a breeding population of rhesus macaques. Lab. Anim. Sci. 44:25-30. [PubMed] [Google Scholar]
- 42.Williams, S. L., C. B. Hartline, N. L. Kushner, E. A. Harden, D. J. Bidanset, J. C. Drach, L. B. Townsend, M. R. Underwood, K. K. Biron, and E. R. Kern. 2003. In vitro activities of benzimidazole d- and l-ribonucleosides against herpesviruses. Antimicrob. Agents Chemother. 47:2186-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]