Abstract
New findings lead to a revised understanding of the substrates’ binding order, the role of the substrate as an activator, and the observed lag phase in the FDTS catalyzed reaction.
Thymidylate synthases catalyze the reductive methylation of 2´-deoxyuridine-5´-monophosphate (dUMP) to 2´-deoxythymidine-5´-monophosphate (dTMP), one of the DNA building blocks. While the mechanism of the classical thymidylate synthase (TS) is well established,1–3 the mechanism of the newly discovered flavin dependent thymidylate synthase (FDTS)4–6 is not well understood.7 Several experimental observations reported in recent years led to two intriguing suggestions regarding the mechanism of FDTS. The first study,8 examined the oxidation of reduced nicotinamide-adenine dinucleotide 2’-phosphate (NADPH) under aerobic conditions, by following the decrease in 340 nm absorption in the presence or absence of different reactants. This study concluded that the substrate dUMP serves as an activator in the reductive half reaction (involving reduction of oxidized flavin adenine dinucleotide: FAD→FADH2). This conclusion was of substantial interest as dUMP only reacts during the oxidative half reaction (FADH2→FAD). A second study,9 examined the oxidized flavin reduction (following 454 nm absorption) and the findings agreed with the conclusions of ref 8. Additionally, ref 9 suggested that the reductive half reaction presents a lag phase whose duration depends on dUMP concentration. This was quite intriguing as such long lag phases are rare in most chemical and enzymatic reaction and could present a unique kinetic phenomenon. In the current communication, new findings indicate a revised binding order of ligands to the enzyme. Apparently, dUMP is not involved in the reductive half reaction but it does activate the oxidative half reaction even in the absence of the methylene donor (5,10-methylene-5,6,7,8-tetrahydrofolate, CH2H4folate). The new data also clarify the origin of the dUMP-dependent lag phase and reinforce the reported activation constant of dUMP (Kf). In contrast to previous interpretations,8,9 dUMP actually serves as an oxidation activator, rather than a reduction activator.
Classical thymidylate synthase (TS, EC 2.1.1.45) catalyses the reductive methylation of dUMP and CH2H4folate, yielding dTMP and 7,8-dihydrofolate (H2folate). This enzyme is a target of several cytostatic drugs (chemotherapeutics and antibiotics).7 The alternative TS is a flavin dependent thymidylate synthase (FDTS, EC 2.1.1.148). The gene encoding for FDTS (ThyX) is present in several severe human pathogens and thus FDTS represents a new antibiotic target.4,7,8,10,11 FDTS is a homotetramer that contains one tightly bound FAD cofactor per subunit.5,9,12–16 In contrast to TS, the CH2H4folate in the FDTS reaction is not the reducing agent. During the reductive half reaction, oxidized FAD (yellow, due to an absorbance band at 454 nm) is reduced to colorless FADH2 by nicotinamides, dithionite, or other reducing agents. It remains largely unclear how the methylene group is transferred from CH2H4folate to dUMP, but this enigma is beyond the scope of this communication. Instead, we focus on the relationship between the two half reactions and between the two competing reactants: the natural cofactor, CH2H4folate, which leads to production of dTMP, and oxygen (O2), which leads to production of hydrogen peroxide (H2O2). The second reaction, denoted oxidase activity, while common to most flavin containing enzymes,17 is likely to be a side reaction because several studies, including the current one, examined FDTS from the anaerobic hyperthermophile Thermotoga maritima.8,9,16
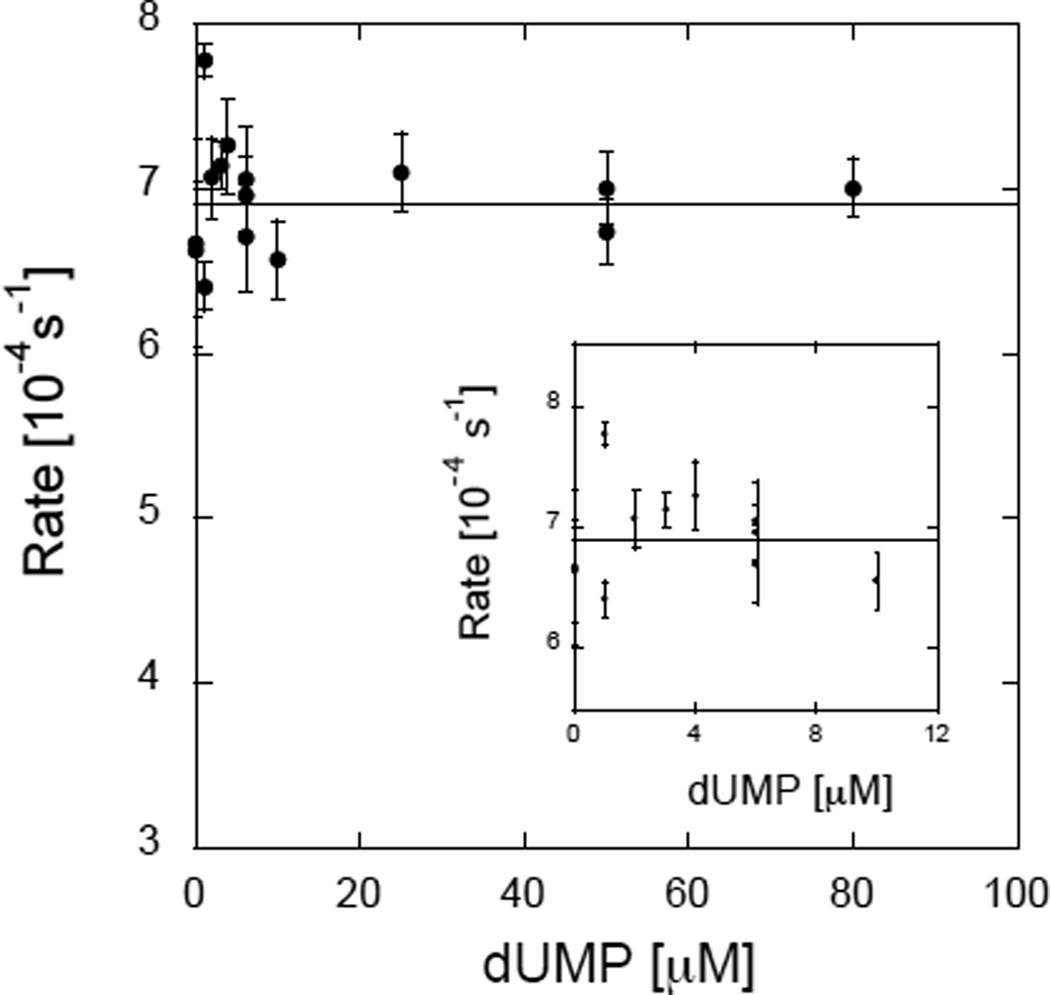
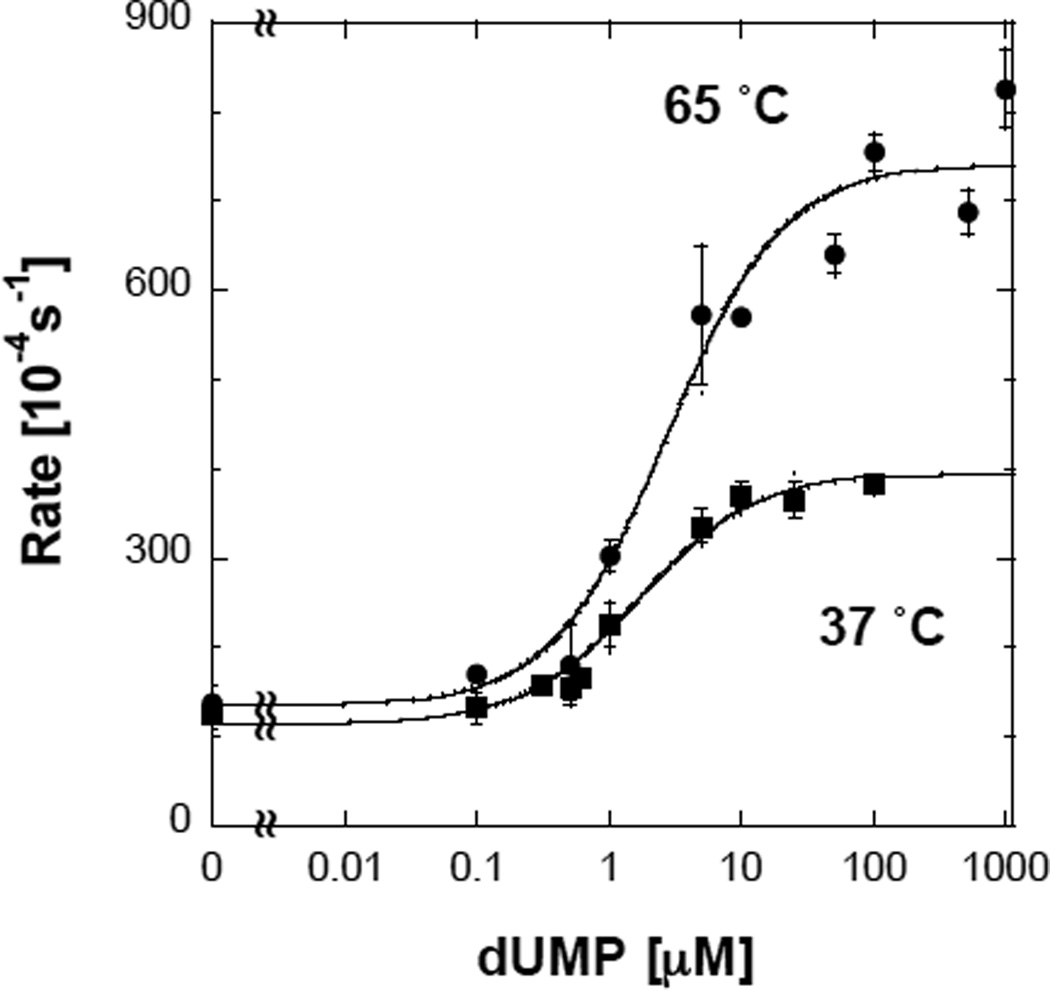
Recently, we measured the single turnover reduction of FDTS from T. maritima by NADPH under oxygen depleted conditions18 in the absence of CH2H4folate and with various concentrations of dUMP. The reduction of enzyme-bound FAD by NADPH was measured (454 nm absorbance) over time and a sigmoidal lag phase was observed.9 The duration of the lag-phase decreased with increasing concentration of dUMP leading to the determination of effective binding constant (Kf), at 37 and 80 °C (the optimal growth temperature of T. maritima). Since the observed lag-phase in FAD reduction may result from residual oxygen in the reaction mixture,19–21 we repeated the same experiments, in the presence of an in situ oxygen-consuming system (10 mM glucose, 60 units of glucose oxidase under purified Ar). Under these strictly anaerobic conditions, no lag phase was detected and FAD reduction was independent of dUMP concentration (Figure 1). Contrary to previous suggestions,8,9 dUMP does not appear to be involved in the FAD reduction and there is no indication that it binds before NADPH. However, the rate of the oxidase activity (O2→H2O2) was clearly dependent on the concentration of dUMP (Figure 2). We determined the functional constants for dUMP binding (Kf) by measuring the decrease in 340 nm absorbance as probe for NADPH oxidation under aerobic conditions using 0.1 mM NADPH, and various concentration of dUMP. The initial velocities were fitted to single activator kinetics with finite rates at low and high concentrations of dUMP (lines in Figure 2):
| [1] |
where V0 and V∞ are the reaction rates with no dUMP and at saturating dUMP, respectively. The values of Kf at 37 and 65 °C were 1.6 ±0.3 M and 2.7 ±0.6 µM, respectively. These Kfs are within experimental errors from those obtained in ref 9 using the duration of the apparent lag phase (1.4 ±0.1 µM and 2.1 ±0.1 µM at 37 °C and 80 °C, respectively). The similarities in the values of Kf determined from the duration of the lag phase and from the oxidase activity are consistent with a common origin. The fact that the lag phase was not observed under strictly anaerobic conditions indicates that this phenomenon primarily stems from the kinetics of the oxidase reaction, rather than from other factors, like conformational rearrangements of the enzyme9 or reorientation of the substrates,16 as was previously proposed.
Figure 1.
Single turnover FDTS-bound FAD’s reduction rate vs. dUMP concentration under anaerobic conditions at 37 °C. Insert, the 0–12 µM range, where the dUMP effect on steady state rates is pronounced (see Figure 2 and ref 9).
Figure 2.
The dependence of FDTS’s oxidase activity on dUMP concentration at 37 °C (squares) and 65 °C (circles).
Additional experiments resulted in several other important findings: (i) The possibility that dUMP serves as a substrate in an oxygenase reaction was eliminated by conducting the FDTS catalyzed reaction with [2-14C]dUMP, NADPH, and oxygen (no CH2H4folate). The products were analyzed using HPLC and no new radioactive peak developed during the reaction even after all the oxygen and NADPH were consumed; (ii) The formation of H2O2 was confirmed using the Amplex Red/horseradish peroxidase assay.22 Unfortunately, this assay could not be used quantitatively because NADPH is also oxidized by H2O2 under the assay conditions; (iii) The apparent KM for O2 was determined to be 9.0 ±0.5 µM by measuring initial velocities of NADPH oxidation in the absence of CH2H4folate, with varying concentrations of O2, and saturating dUMP (100 µM); and (iv) Previous findings8,9 indicated that dUMP binding to FDTS in the absence of CH2H4folate increases the rate of NADPH oxidation by a factor of about ten.
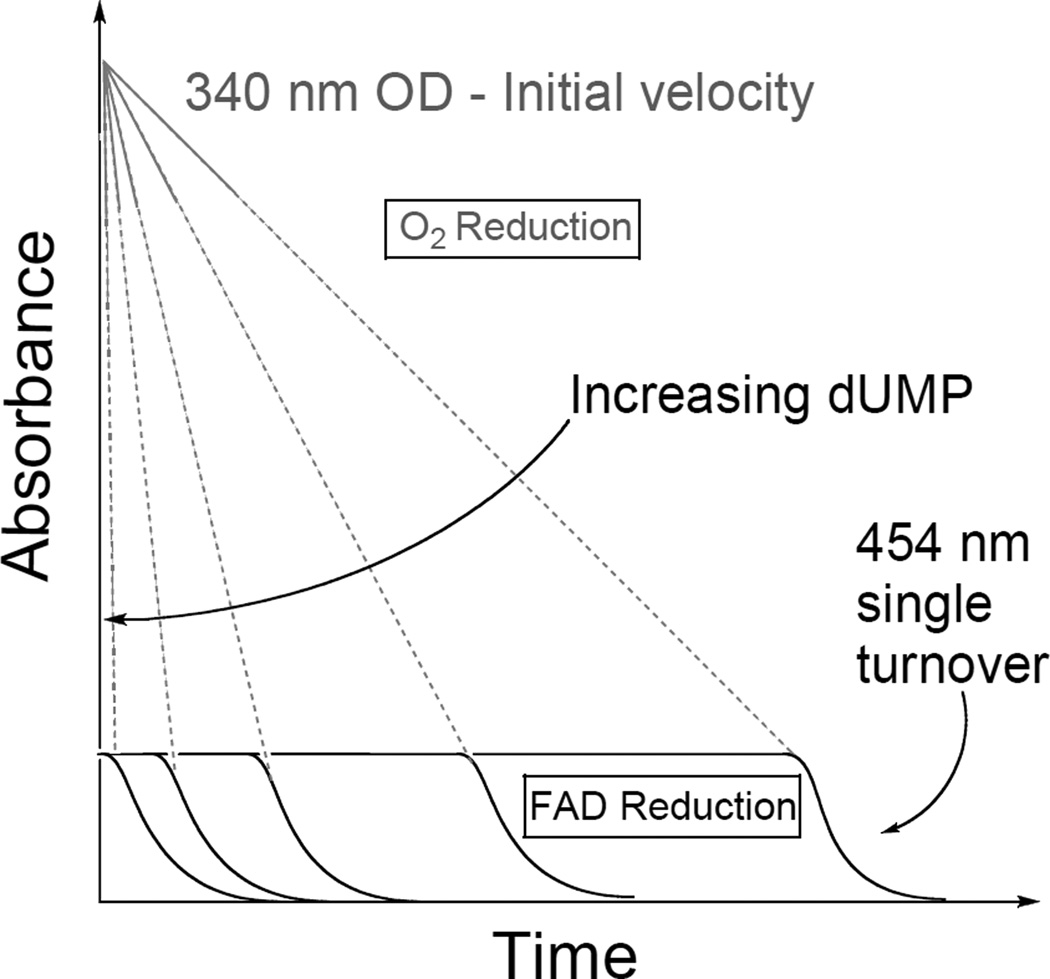
All the observations discussed above can be understood within the model illustrated in Figure 3. At the top, the blue lines represent the dUMP dependent O2 reduction to H2O2 by NADPH (detected by following 340 nm absorbance). At the bottom, the red lines represent the oxidation state of the enzyme bound flavin (detected by following 454 nm absorbance). Since the FAD reduction is rate limiting, the 454 nm absorbance does not decrease significantly in the presence of O2. The FAD is reduced (at a dUMP independent rate) only after the O2 concentration drops below its KM, leading to apparent lag-phase (red lines). This finding suggests that the reductive half reaction does not require dUMP, and a new examination of the substrates binding order is presented below.
Figure 3.
Illustration of the relationship between rate of NADPH oxidation (340 nm absorbance, in grey) and the lag phase observed in ref 9 (454 nm absorbance, in black). While consuming the oxygen the 340 nm absorbance decreases (grey lines), but the 454 nm absorbance is hardly effected indicating that the FAD is still oxidized, and in accordance with the reductive half reaction being rate limiting.9 Only after the oxygen is mostly consumed, is FAD reduced. The oxidative half reaction (grey) is dUMP dependent, but the reductive half reaction (black) appears independent of dUMP concentration.
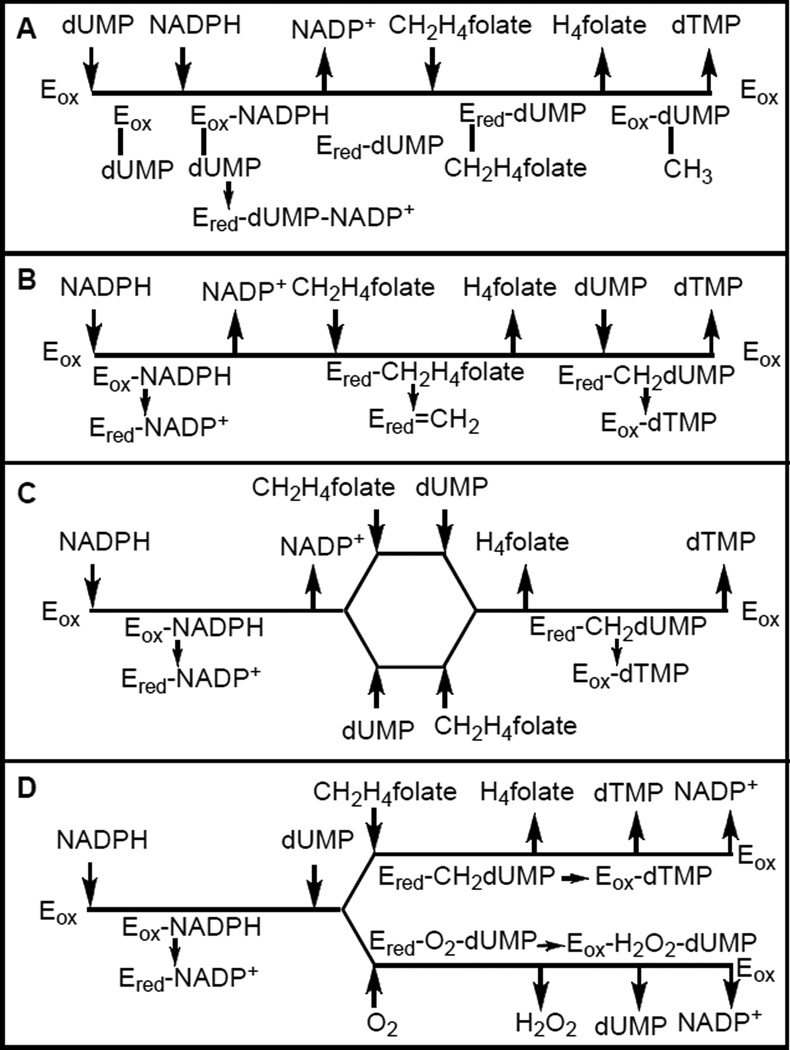
The fact that dUMP appeared to enhance NADPH consumption led Myllykallio, Liebl, and their co-workers to propose that dUMP binds to the free, oxidized, enzyme and enhances the reductive half reaction.8 They proposed an ordered binding mechanism that is illustrated in Scheme 1A. McClarty and co-workers, on the other hand, conducted steady state studies by following tritium release from [5-3H]dUMP, and reported that their kinetic examination of the two natural substrates, dUMP and CH2H4folate, resulted in parallel pattern in a double reciprocal plot analysis (Lineweaver Burke).10 That pattern suggested that CH2H4folate binds to the reduced (FADH2) enzyme, transfers its methylene to the enzyme, and that the product H4folate is released before dUMP binds. These observations lead to the kinetic scheme illustrated in Scheme 1B.10
Scheme 1.
Proposed kinetic mechanisms for FDTS A. Graziani et al. (2006)16, B. Griffin et al. (2005)10, C. Agrawal et al. (2004)14. D. This work (see text).
We conducted similar steady state experiments while following [2-14C]dUMP conversion to [2-14C]dTMP and an intersecting double reciprocal plot of dUMP vs. CH2H4folate suggested that these two substrates bind sequentially as illustrated in Scheme 1C.14 Similar analysis of the relationship between NADPH and dUMP or CH2H4folate also indicated that the product of the reductive half reaction (NADP+) is not released from the enzyme until both dUMP and CH2H4folate bind, and presumably until the end of the oxidative half reaction. Scheme 1D illustrates this mechanism, where the bottom path (oxidase activity) is faster than the top path (TS activity).8 We suggest that O2 and CH2H4folate compete for the reduced enzyme (FDTS bound FADH2) after dUMP binding. dUMP enhances the rate of the oxidative half reaction and while the oxidase reaction is faster than the reaction with CH2H4folate, this cofactor slows down the consumption of NADPH in the presence of O2,8 in accordance with substrate competition.
In summary, two new sets of experiments were conducted in order to examine the role of dUMP as an activator, rather than a substrate of FDTS. In contrast to previous reports, the substrate of the oxidative half reaction (dUMP) does not affect the reductive half reaction (Figure 1). In the absence of the second substrate (CH2H4folate), dUMP enhances the reaction of molecular oxygen with the reduced enzyme (Figure 2) with an “effector” binding constant (Kf) close to 2 µM. This constant presented a weak temperature dependency (e.g., about 1 µM per 30 °C). This relatively tight binding with a weak temperature dependency is typical of entropy driven processes. A plausible molecular explanation may involve a dUMP induced protein rigidity prior to O2 reduction. The lower oxidase activity in the absence of dUMP may result from multiple conformations with only a small fraction of reactive conformations. The binding of dUMP may induce conversion of the protein into the more reactive ensemble of conformations.
Future efforts will examine whether dUMP also activates its reaction with CH2H4folate. Allosteric substrate (dUMP) activation will be pursued under anaerobic conditions with CH2H4folate as the oxidizing agent (Hill constant values larger than unity may indicate such substrate activation). Additionally, the nature of the competition between CH2H4folate and O2 for the reduced enzyme will be examined via kinetic analysis of their mutual inhibition. While the oxidase activity might not be natural to FDTS from the anaerobic T. maritima, it may serve as a probe for hidden features such as the tight binding of dUMP to the reduced enzyme, a much stronger interaction than predicted from KM values of dUMP.16
Acknowledgments
This work was supported by NIH R01 GM65368-65301 and NSF CHE- 0133117 to AK, and NIH U0133154 GM0074898 to SAL.
Notes and references
- 1.Carreras CW, Santi DV. Annu. Rev. Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 2.Finer-Moore JS, Santi DV, Stroud RM. Biochemistry. 2003;42:248–256. doi: 10.1021/bi020599a. [DOI] [PubMed] [Google Scholar]
- 3.Stroud RM, Finer-Moore JS. Biochemistry. 2003;42:239–247. doi: 10.1021/bi020598i. and many cited therein. [DOI] [PubMed] [Google Scholar]
- 4.Myllykallio H, Lipowski G, Leduc D, Filee J, Forterre P, Liebl U. Science. 2002;297:105–107. doi: 10.1126/science.1072113. [DOI] [PubMed] [Google Scholar]
- 5.Lesley SA, Kuhn P, Godzik A, Deacon AM, Mathews I, Kreusch A, Spraggon G, Klock HE, McMullan D, Shin T, Vincent J, Robb A, Brinen LS, Miller MD, McPhillips TM, Miller MA, Scheibe D, Canaves JM, Guda C, Jaroszewski L, Selby TL, Elsliger M-A, Wooley J, Taylor SS, Hodgson KO, Wilson IA, Schultz PG, Stevens RC. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhn P, Lesley SA, Mathews II, Canaves JM, Brinen LS, Dai X, Deacon AM, Elsliger MA, Eshaghi S, Floyd R, Godzik A, Grittini C, Grzechnik SK, Guda C, Hodgson KO, Jaroszewski L, Karlak C, Klock HE, Koesema E, Kovarik JM, Kreusch AT, McMullan D, McPhillips TM, Miller MA, Miller M, Morse A, Mon K, Ouyang J, Robb A, Rodrigues K, Selby TL, Spraggon G, Stevens RC, Taylor SS, Van den Bedem H, Velasquez J, Vincent J, Wang X, West B, Wolf G, Wooley J, Wilson IA. Proteins: Structure, function, and genetics. 2002;49:142–145. doi: 10.1002/prot.10202. [DOI] [PubMed] [Google Scholar]
- 7.Chernyshev A, Fleischmann T, Kohen A. Appl. Microbiol. Biotech. 2006 doi: 10.1007/s00253-006-0763-1. In press. [DOI] [PubMed] [Google Scholar]
- 8.Graziani S, Xia Y, Gurnon JR, Van Etten JL, Leduc D, Skouloubris S, Myllykallio H, Liebl U. J. Biol. Chem. 2004;279:54340–54347. doi: 10.1074/jbc.M409121200. [DOI] [PubMed] [Google Scholar]
- 9.Mason A, Agrawal N, Washington MT, Lesley SA, Kohen A. Chem. Commun. 2006:1781–1783. doi: 10.1039/b517881a. [DOI] [PubMed] [Google Scholar]
- 10.Griffin J, Roshick C, Iliffe-Lee E, McClarty G. J. Biol. Chem. 2005;280:5456–5467. doi: 10.1074/jbc.M412415200. [DOI] [PubMed] [Google Scholar]
- 11.Mathews II, Deacon AM, Canaves JM, McMullan D, Lesley SA, Agarwalla S, Kuhn P. Structure. 2003;11:677–690. doi: 10.1016/s0969-2126(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 12.Leduc D, Graziani S, Lipowski G, Marchand C, Le Maréchal P, Liebl U, Myllykallio H. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7252–7257. doi: 10.1073/pnas.0401365101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sampathkumar P, Turley S, Ulmer JE, Rhie HG, Sibley CH, Hol WGJ. J. Mol. Biol. 2005;352:1091–1104. doi: 10.1016/j.jmb.2005.07.071. [DOI] [PubMed] [Google Scholar]
- 14.Agrawal N, Lesley SA, Kuhn P, Kohen A. Biochemistry. 2004;43:10295–10301. doi: 10.1021/bi0490439. [DOI] [PubMed] [Google Scholar]
- 15.Gattis SG, Palfey BA. J. Am. Chem. Soc. 2005;127:832–833. doi: 10.1021/ja0432214. [DOI] [PubMed] [Google Scholar]
- 16.Graziani S, Bernauer J, Skouloubris S, Graille M, Zhou CZ, Marchand C, Decottignies P, van Tilbeurgh H, Myllykallio H, Liebl U. J. Biol. Chem. 2006;281:24048–24057. doi: 10.1074/jbc.M600745200. [DOI] [PubMed] [Google Scholar]
- 17.Mattevi A. Trends Biochem. Sci. 2006;31:276–283. doi: 10.1016/j.tibs.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Ar gas was bubled through all the solutions, but the enzyme's stock solution, and the reaction was conducted under Ar atmosphere.
- 19.We thank Nigel Scrutton from the University of Manchester, UK, for indicating this option to us.
- 20.Gibson QH, Swoboda BE, Massey V. J. Biol. Chem. 1964;239:3927–3934. [PubMed] [Google Scholar]
- 21.Neujahr HY, Kjellen KG. J. Biol. Chem. 1978;253:8835–8841. [PubMed] [Google Scholar]
- 22.Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. Anal. Biochem. 1997;253:162–168. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]