Abstract
Embryos allocate cells to the three germ layers in a spatially ordered sequence. Human embryonic stem cells (hESCs) can generate the three germ layers in culture, however, differentiation is typically heterogeneous and spatially disordered. Here we show that geometric confinement is sufficient to trigger self-organized patterning in hESCs. In response to BMP4, these colonies reproducibly differentiate to an outer trophectoderm-like ring, an inner ectodermal circle and a ring of mesendoderm expressing primitive-streak markers in between. Fates are defined relative to the boundary with a fixed length scale: small colonies correspond to the outer layers of larger ones. Inhibitory signals limit the range of BMP4 signaling to the colony edge and induce a gradient of Activin/Nodal signaling that patterns mesendodermal fates. These results demonstrate that the intrinsic tendency of stem cells to make patterns can be harnessed by controlling colony geometries, and provide a quantitative assay for studying paracrine signaling.
Introduction
During gastrulation, the cells of the embryo are allocated into three germ layers in an ordered spatial sequence1. In mammalian embryos, epiblast cells located on the interior of the embryo migrate to form the definitive endoderm on the outside of the embryo proper and the mesoderm between the endoderm and epiblast. Cells that remain in the epiblast differentiate to ectoderm. Despite the existence of numerous protocols to differentiate hESCs towards cells of these three germ layers2–5, it is unclear to what degree this spatial order can be recapitulated in vitro. Achieving this would allow the molecular dissection in the human system of the intercellular communication that is responsible for embryonic patterning and would also be an important step towards generating spatially ordered tissues for clinical purposes.
Studies in fish, frog, and mouse embryos have established that spatial patterning during gastrulation is under the control of the Activin/Nodal, BMP, and Wnt pathways. In both mouse embryos and embryoid bodies derived from mouse embryonic stem cells (mESCs), these three pathways form a positive feedback loop that establishes polarity6,7. These same three pathways can be manipulated to differentiate hESCs to any of the three germ layers2–5, however, because these protocols have been optimized to yield pure populations, it is unclear whether stem cells are capable of faithfully generating early embryonic patterns. Simple application of growth factors tends to produce multiple fates without inducing consistent spatial order8,9. Previous studies using control of colony geometries noted a shift in the proportion of cells adopting different fates as the colony size was changed but did not observe spatial organization10–12.
Here, we show that cells confined to circular micropatterns and differentiated with BMP4 produce an ordered array of germ layers along the radial axis of the colony. This order results from self-organized signaling which confines response to the BMP4 to the colony border while inducing a broader gradient of Activin/Nodal signaling to pattern mesendodermal fates. Control of fates is established from the border of the colony so that as colony size is reduced the central fates are lost. Thus, given minimal geometric and signaling cues, hESCs will self-organize to generate embryonic patterns.
Results
Prepatterning in pluripotent hESC colonies
We investigated whether hESCs could give rise to spatially ordered germ layers. We focused on differentiating cells with BMP4 ligand because it represents an early step in the embryonic signaling cascade that initiates gastrulation1,6. hESCs rapidly differentiate in response to BMP4, in contrast to Activin/Nodal and Wnt signaling both of which also play a role in the pluripotent state9,13–15. The results of BMP4 treatment on hESCs have been controversial with some groups reporting differentiation to trophectoderm16–19 while others have reported a mixture of embryonic and extra-embryonic mesoderm20.
Under standard culture conditions, hESCs grow in colonies that exhibit a wide range of sizes and shapes (Fig. 1A). Differentiation of embryonic stem cells with BMP4 led to spatial patterns of differentiation that differed drastically between neighboring colonies (Supplementary Fig. 1). We hypothesized that the heterogeneity in colony geometries could affect cell-cell signaling and result in a loss of reproducible spatial disorder upon differentiation. We thus evaluated the effects of using micropatterned technology to grow cells in colonies of precisely controlled size and geometry (Fig. 1B).
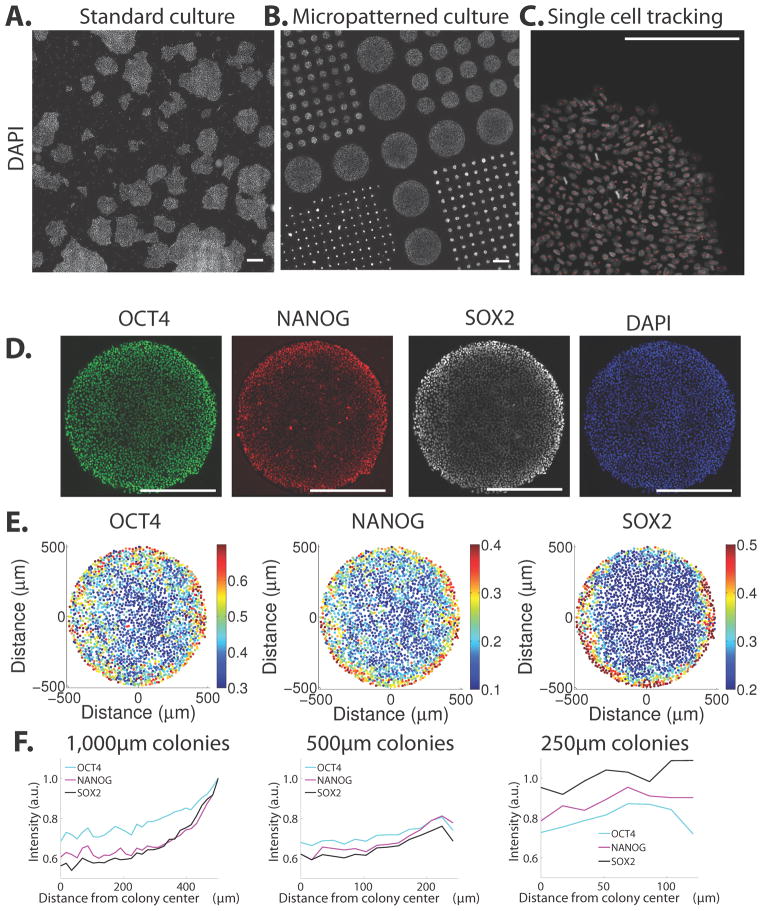
Figure 1. Stem cells grown in the pluripotent state show prepatterning in micropatterned culture.
(A–B) Tiled scans of RUES2 hESCs grown under standard (A) and micropatterned (B) conditions show heterogeneous and standardized colony geometries, respectively. (C) A single image from the tiled scan with all cells identified computationally. (D) Immunofluorescence analysis shows cells in the micropatterned colonies maintain expression of pluripotency markers (E) Quantification of expression of markers of the colony shown in (D). Each dot represents a single cell and the color represents the intensity of the indicated marker normalized to the intensity of the DAPI stain. (F) Quantification of average nuclear intensity from immunofluorescence data shows that markers are elevated at the colony edges. In all cases, nuclei were identified using the DAPI nuclear counterstain and the intensity of the indicated markers was normalized to the DAPI intensity to prevent imaging artifacts. All scale bars are 500 μm.
hESCs grown on micropatterns for 24 hours in pluripotency conditions maintained pluripotent morphology and expression of markers OCT4, SOX2, and NANOG (Fig. 1D). We first examined whether signaling and pluripotency markers are uniformly expressed throughout the colony. We used computational methods to identify all cells in a large number of colonies and to quantify the expression of different markers with single-cell resolution (Fig. 1C). In the largest colonies, pluripotency markers are uniformly expressed over the center of the colony but rise towards the edges over a range of approximately 150μm (Fig. 1E–F).
Signaling pathway activity for the BMP, Activin/Nodal, and Wnt pathways as measured by the activity of the SMAD1/5/8, SMAD2/3 and β-CATENIN signal transducers were all elevated towards the edges of the colonies as well (Supplementary Fig. 2). In intermediate sized colonies (500μm diameter), the size of the region of elevated expression was approximately the same. Colonies 250μm and smaller uniformly expressed the pluripotency markers at the levels of the cells at the edges of larger colonies (Fig. 1F). These trends were observed in nearly every colony (Supplementary Fig. 3). These results reveal a previously unappreciated spatial ordering in pluripotent hESC colonies: the edges of colonies differ from their centers and control of this difference is exerted from the edge inwards. As a result, small colonies are equivalent to the edges of large colonies.
Spatial ordering in differentiated micropatterned colonies
Treatment of micropatterned hESC colonies with 50 ng/ml BMP4 led to morphological differentiation within 24 hours of treatment with a dense ring of cells forming at a reproducible radius within the colony with larger, more spread cells radially to the inside and outside (Fig. 2A). We next examined whether the reproducible cell geometries lead to organized germ layer differentiation.
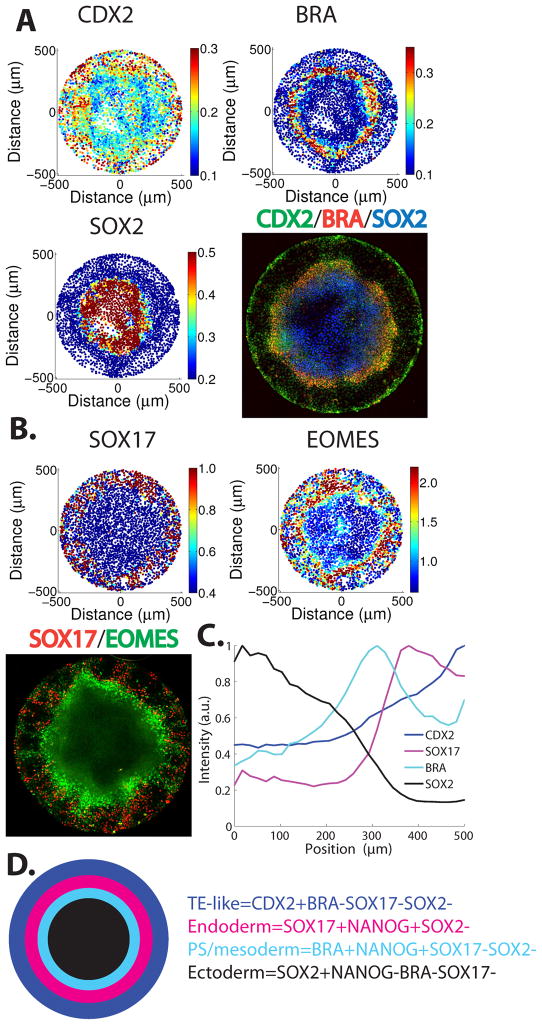
Figure 2. Stem cells differentiated on micropatterns form self-organized spatial patterns.
(A–B) Immunofluoresence for fate markers shows patterns along the radial axis of the colonies. Cells were seeded on micropatterned coverslips, grown overnight, and then treated with BMP4 for 42 hours. Each panel corresponds to a single colony while each dot corresponds to a single cell. (C) Quantification of immunofluorescence data showing that germ layer markers are induced at particular radii. (D) Schematic of the results of 42hours of BMP4 treatment in micropatterned culture.
Transcription factors that are essential for pluripotency are reused during differentiation in order to both activate and repress lineage-specific factors21–24. Following the spatial dynamics of pluripotency markers can thus aide in the identification of germ layers. SOX2 expression is specifically maintained during ectodermal differentiation but down-regulated during mesendoermal differentiation, while OCT4 and NANOG follow the opposite pattern. After 42 hours of micropatterned differentiation, SOX2 is only expressed at the center of the 1000μm colonies (Fig. 2A). This pattern results from a dramatic rise in SOX2 protein levels while OCT4 and NANOG are down-regulated in this region (Supplementary Fig. 4). The elevated SOX2 expression and the exclusion of NANOG, BRACHYURY (BRA), EOMESODERMIN (EOMES), SOX17 and Nodal signaling (see below) identifies the center with prospective ectoderm22. Markers that would distinguish subpopulations of the ectoderm such as KERATIN14 for epidermal cells and PAX6 or SOX1 for neural cells arise only later in development5,25.
Moving outward from the center region, there is a ring of BRA expression (Fig. 2A). This ring also expresses NANOG and OCT4 but not SOX2 which represses mesendodermal fates23,24 (Supplementary Fig. 5). Much of this region also expresses the mesendodermal marker EOMES and GATA6 but these markers extend further away from the colony center along the radial axis (Fig. 2B and Supplementary Fig. 5). Many of the cells in this region also express CDX2, which is expressed in a broad domain that extends to the colony border. At the level of individual cells, CDX2, EOMES, and NANOG were all coexpressed with BRA (Supplementary Figs. 6A–C, 7A–B), while SOX2 was mutually exclusive with all of these markers (Supplementary Fig. 6D–E). This set of observations is consistent with the identification of this region as primitive streak and nascent mesodermal cells (see further below).
Beyond the ring of BRA expression, we find cells expressing the definitive endoderm marker SOX17 (Fig. 2B). Its expression overlaps with the outer ridge of the cells that are positive for EOMES or GATA6 staining but extends further along the radial axis of the colony (Fig. 2B and Supplementary Figure 6G). Many of the cells expressing SOX17 also express high levels of NANOG, consistent with a role of NANOG in endodermal differentiation23 (Supplementary Fig. 7C–D). No expression of SOX2 was detected in this population (Supplementary Fig. 6F).
Finally we observe a broad ring of CDX2 expression that peaks at the very edge of the colony but extends through the mesendodermal region. The coexpression of CDX2 and BRA (Supplementary Fig. 6A) is consistent with expression of CDX2 in the mesoderm26, however, the outermost cells of the colony expressed CDX2 without expression of mesendodermal markers, BRA, SOX17, and GATA6, or pluripotency markers OCT4, NANOG and SOX2 (Supplementary Fig. 5–6). These outer cells are positioned corresponding to the extraembryonic tissue in the embryo that surrounds the epiblast, and they also displays high BMP signaling (see below) similar to the trophoectoderm in the embryo1. These cells express EOMES but at lower levels then in the mesendoderm (Supplementary Fig. 6C). The cells at the colony edge thus share many characteristics with extraembryonic trophoblast cells but the true identity of trophoblast-like cells induced by BMP4 remains the subject of debate in the literature17–20. Thus, cells confined to circular geometries and differentiated with BMP4 differentiated to all three germ layers and a trophoblast-like population in an ordered sequence along the radial axis (Fig. 2C–D).
The radial structure of the patterns is extremely reproducible (Supplementary Figs. 5 and 8). There remain small angular inhomogeneities (e.g. Fig. 2A, Supplementary Fig. 7C–D) that correlate with cell density and are impossible to control during cell seeding. We found very similar patterns for two additional hESC lines RUES1 and H1 (Supplementary Fig. 9A, B). We also found similar patterns if cells were grown on recombinant Laminin 521 instead of Matrigel or grown in mTeSR1 medium instead of conditioned medium (Supplementary Fig. 9C, D). Thus, forcing cells to grow in a confined geometry triggers spontaneous emergence of embryonic patterning in several hESC lines and under all conditions examined.
Gastrulation-like events in micropatterned differentiation
The expression of BRA and the emergence of all three germ layers in the micropatterned cultures suggested that cells might be patterned by gastrulation-like events in a region resembling the primitive steak. In the embryo, FGF signaling in the primitive streak leads to cells undergoing an epithelial to mesenchymal transition (EMT), upregulating the transcription factor SNAIL and downregulating or removing E-CADHERIN (E-CAD) from the cell surface1. In micropatterned culture, the downstream effector of FGF signaling, phospho-ERK, was primarily localized to region of BRA expression (Fig. 3A). We also observed upregulation of SNAIL in this region (Fig. 3B) and E-CAD was internalized becoming more cytoplasmic rather than exclusive localization to the cell membrane (Fig. 3C)27. This demonstrates the presence of human primitive streak cells that undergo gastrulation-like events in micropatterned culture
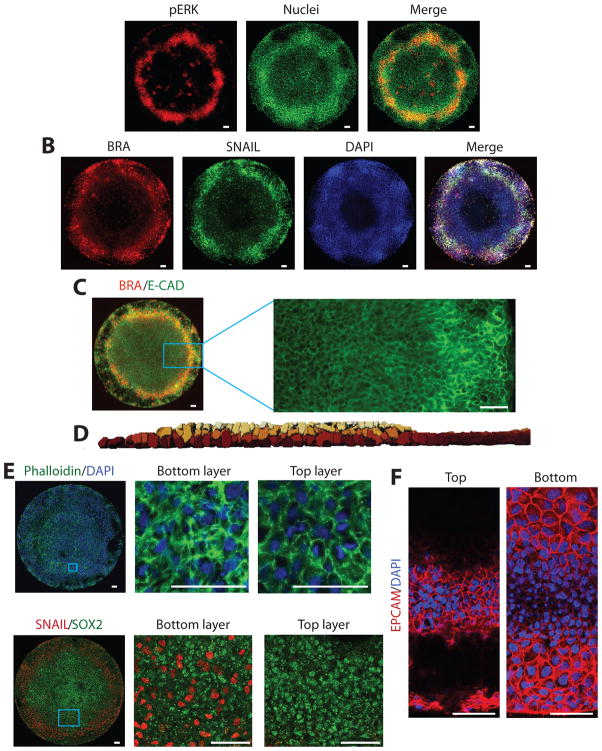
Figure 3. During differentiation cells undergo EMT in a region expressing markers of the primitive streak.
(A) Cells in the PS-like region have high levels of pERK. (B) Cells in the PS-like region express SNAIL. (C) Cells in the PS-like region internalize E-CAD. BRA expression is not shown in the blow-up for clarity (D) 3D reconstruction showing that the PS-streak like region is 2–3 cells deep. (E) Phalloidin staining reveals differences in cytoskeletal organization in the upper and lower layers of the PS-like region. The blue box in the left panel shows the region expanded in the individual confocal slices in the other two panels. (F) Immunofluorescence for EpCam shows it is only expressed in the upper epithelial layer (left) of the PS-like region and is absent from the cells below (right). Each panel is an individual confocal slice. (G) Cells in the lower region of the culture express SNAIL while those in the upper layer express the epiblast/ectoderm marker SOX2. The left panel is a maximum intensity projection of the entire confocal stack while the center and right panels show blow-ups of individual confocal slices. All scale bars are 50 μm.
To better understand these gastrulation-like processes, we examined the three-dimensional structure of the colonies. Three dimensional reconstruction of the colonies based on DAPI staining revealed that at the inside and outside of the colonies, the cells grew as a monolayer, whereas inside the primitive streak-like region, cells were piled 2–3 layers deep (Fig. 3D). We next examined the actin cytoskeleton using phalloidin and found that in the monolayered regions, as well as in the upper layer of the streak-like region, the actin was largely localized to the cell membranes, consistent with the entire top of the colony forming an epithelial layer. In the streak-like region, cells in the bottom layers displayed a more active cytoskeleton consistent with migratory cells that have undergone an EMT (Fig. 3E). Consistent with this interpretation, the entire top of the colony expressed Epithelial Cell Adhesion Molecule (EpCam)28 on the cell membranes, whereas cells in the lower layer of the streak-like region did not (Fig. 3F). Where the colonies were multiple layers thick, cells in the lower layers expressed SNAIL, whereas those in the upper layer expressed SOX2 (Fig. 3G). We also found individual SNAIL expressing cells under the epithelial layer closer to the colony center (Fig. 3G), which likely represent cells that underwent EMT at the streak-like region and then migrated under the epithelial layer of the colony. Taken together, these results suggest that cells in the BRA-expressing region undergo EMT and migrate inwards towards the culture dish and then underneath the upper epithelial layer, mimicking human gastrulation movements.
Patterns are controlled from the colony edge
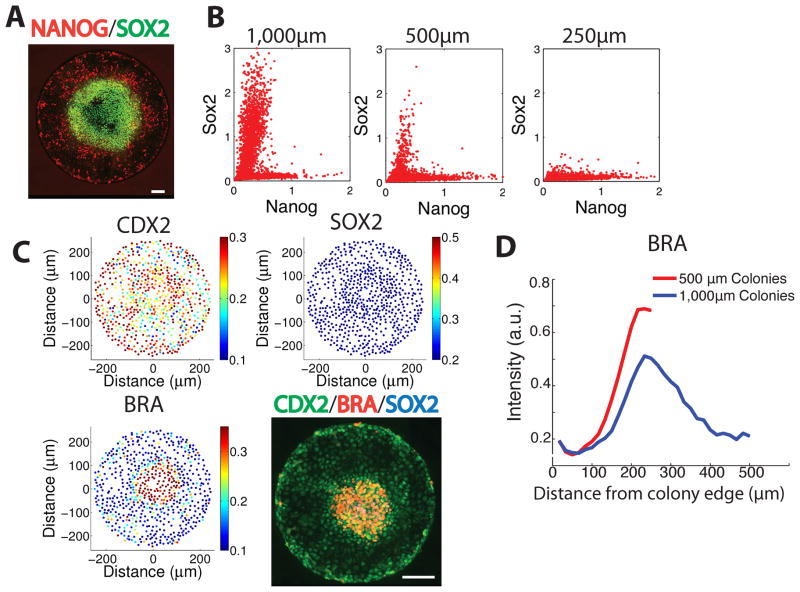
We next examined the differentiation patterns as a function of colony size. The edges of all colonies expressed the same markers at the same radial distance from the colony edge, irrespective of colony size. As the colony size decreased, the SOX2 expressing population on the interior of the colonies was lost (Fig. 4A–B) while the territory of mesodermal differentiation extended to the center of the colony (Fig. 4C–D), forming a disk rather than an annulus. Examined at the level of single cells, reductions in colony size caused a shift away from SOX2+ ectodermal cells and towards NANOG+ mesendodermal derivatives. In colonies 250μm and below, no SOX2 expression remained (Fig. 4B). Thus, our data show that cells confined to a disk-like geometry and presented with gastrulation-initiating signals form reproducible patterns under the control of a mechanism that senses the exterior edge of the colony.
Figure 4. Control of cell fate extends from the edge of the colony.
(A) Immunofluorescence for SOX2 and NANOG in a 1000μm colony following 42h BMP4 treatment. (B) Quantification of single cell expression of SOX2 and NANOG from immunofluorescence data showing a shift from the SOX2+ ectodermal population towards the NANOG+ mesendodermal population as the colony size is reduced. (C) Immunfluorescence in a 500 μm colony shows BRACHYURY expression at the center rather than in an annulus at fixed radius and the absence of staining for SOX2. Quantification of markers with single-cell resolution in a 500μm colony shows an absence of SOX2 and the expression of BRA in the colony center. (D) Comparison of BRACHYURY expression between 500μm and 1000μm colonies shows the spatial scale of expression is the same in the two sizes. Note the distance scale is inverted relative to previous panels to emphasize control from the boundary. All scale bars are 100μm.
We next examined the dynamics of how these patterns emerge in colonies of various sizes and found that the markers CDX2, BRA, SOX17, and EOMES all show initial expression at the colony border and then move inwards as a function of time to varying degrees (Supplementary Fig. 4). In contrast, SOX2 expression rises continually at the colony center while declining at the colony border. These data suggest that either the PS-like region moves through the colony in time, a process which may result from the spatial confinement, or that cells begin expressing differentiation markers before entering the PS-like region. Examining the dynamics of patterning in time for different colony sizes revealed identical dynamics as measured from the colony edge inwards for all markers examined in time – CDX2, BRA, EOMES, SOX17, SNAIL, and SOX2 (Supplementary Fig. 10). This demonstrates that patterning has a fixed length scale and proceeds identically in time, regardless of the size of the colony.
Patterns originate in self-organized signaling responses
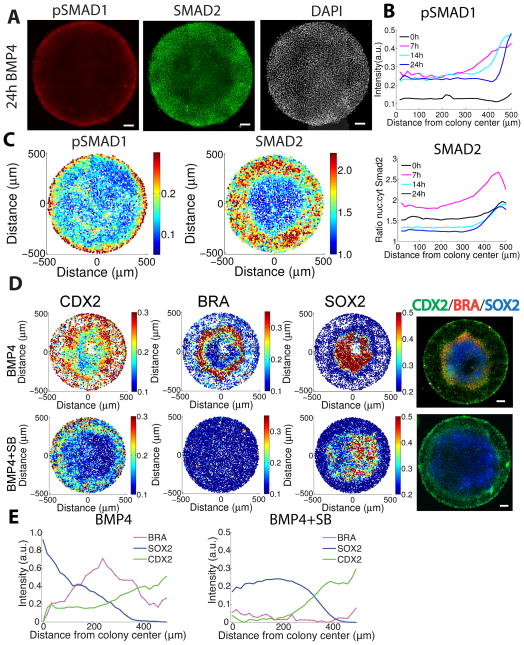
The ability of BMP4 ligand to generate these patterns is surprising since it is presented homogenously at high doses to all cells. To examine the origins of the self-organized differentiation, we determined SMAD1/5/8 status (a proximal readout of BMP signaling) as a function of time (Fig. 5A–C). The initial response was highest at the colony edge with some patches of response in the center. Signaling became increasingly restricted to the colony edge as a function of time. At 24 hours after the application of the BMP4 ligand, elevated signaling was largely confined to a narrow ring at the colony edge (Fig. 5A–C). Thus, prolonged BMP4 signaling at the colony border likely specifies the extraembryonic fates found there (consistent with the expression of BMP4 in extraembryonic tissue in the mouse embryo1).
Figure 5. Self-organized signaling responses in micropatterned colonies.
(A) Immunofluorescence for pSMAD1 and SMAD2 after 24h of BMP4 treatment showing the BMP4 response is sustained only at the colony border while Activin/Nodal signaling forms a broader gradient. (B) Quantification of average pSMAD1 intensity and SMAD2 nuclear:cytoplasmic ratio as a function of time after treatment with 50 ng/ml BMP4 in 1000um diameter colonies. (C) Quantification of pSMAD1 and SMAD2 responses in the colony shown in (A). Each dot represents a single cell. (D) Immunofluorescence of RUES2 colonies treated with 50 ng/ml BMP4 with or without 10μM SB431542 showing that Activin/Nodal signaling is required for mesendodermal differentiation. All scale bars are 100μm. (E) Profiles showing that the inhibition of Activin/Nodal signaling by SB eliminates BRA expression and also the portion of CDX2 that overlaps BRA.
As Activin/Nodal signaling functions downstream of BMP4 in the mouse embryo and is crucial for specifying mesendodermal derivatives, we reasoned that these signals may pattern the mesendoderm in our micropatterned colonies as well. Indeed, a gradient of nuclear SMAD2 formed across the colony and by 7 hours it spanned the range of the future mesendoderm, peaking in the endoderm (Fig. 5A–C). To determine the relevance of this gradient for patterning, we inhibited Activin/Nodal signaling with the small molecule SB43154229 during BMP4-mediated differentiation. SB431542 completely inhibited mesendodermal differentiation as reflected by an absence of BRA and SOX17 positive cells. Under these conditions, the entire colony could be divided into two territories: CDX2+ extraembryonic cells and SOX2 positive ectodermal cells (Fig. 5D). These results indicate that in the absence of Activin/Nodal signaling, cells make a binary fate choice likely based on the level of response to the BMP4 signal. BMP signaling thus specifies a range of fates by inducing CDX2 positive cells at the colony border while also inducing an Activin/Nodal signaling gradient that induces mesendodermal fates. Cells at the center of the colony that do not receive either of these signals adopt an ectodermal fate.
These results also shed light on discrepancies in previous data regarding the outcome of BMP-mediated differentiation16,20. Note that the range of CDX2 expression extends from the colony border and encompasses BRA+ cells as well as the trophoblast-like cells at the border (Supplementary Fig. 11). Thus, there are at least two populations of CDX2+ cells at spatially distinct locations: a population that overlaps with BRA expression and represents embryonic mesoderm or primitive streak and a BRA negative population at the edge of the colonies that is suggestive of its extraembryonic expression1. Notably, treatment with SB431542 both abolishes BRA staining and diminishes the range of CDX2 expression (Fig. 5D–E), further confirming that the CDX2+ population that overlaps with BRA expression is mesodermal while that at the colony edge is not. The ability to segregate and manipulate these two populations highlights the power of the micropatterning approach for understanding differentiation by providing spatial context.
TGF-β and BMP inhibitors are required for pattern formation
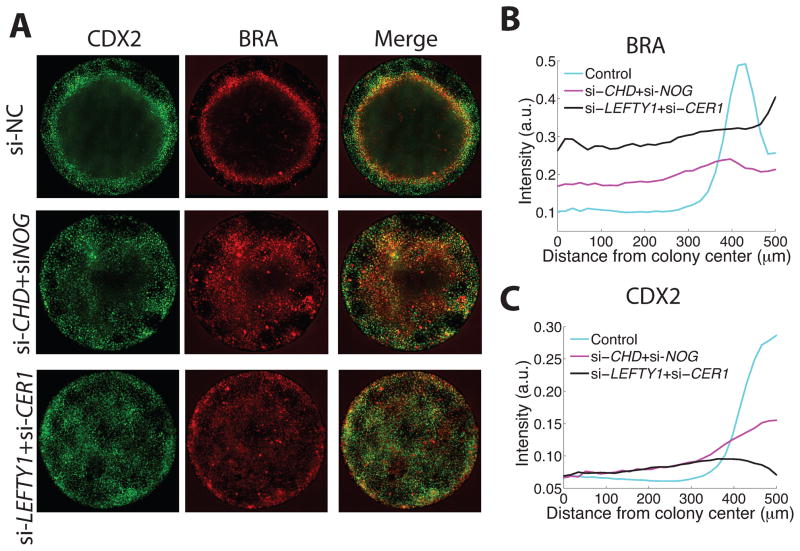
How is the differential response to the exongenously supplied BMP established within the colony? The fact that signaling is highest at the colony edge suggested that a diffusible inhibitor that is lost from the colony edge and thus assumes its highest level in the center, might be responsible for forming these patterns. In the mouse embryo the double knockout of BMP inhibitors Chordin and Noggin shows a loss of the forebrain as a result of expanded BMP signaling30. To test whether these genes are involved in the differential response to BMP, we used siRNA to reduce their expression. Consistent with the knockout phenotype, knockdown caused expansion of mesodermal markers into the ectodermal territory in the center (Fig. 6A–C). We used a similar strategy to determine whether Activin/Nodal inhibitors are necessary to restrict activity to the primitive-streak like region. In the mouse embryo, the combined activities of Lefty1 and Cer1 are required to restrict the primitive streak to one side of the embryo31. In micropatterned culture, knockdown of LEFTY1 and CER1 with siRNA also caused an expansion of the BRA expressing cells into the ectodermal territory (Figure 6A–B). In contrast to the knockdown of the BMP inhibitors, the region of BRA expression also expanded into the region of CDX2+BRA- cells that typically forms at the colony border (Fig. 6A–C). These results demonstrate that inhibitors to the BMP pathway are necessary to preserve the ectodermal character of the colony center while those of the Activin/Nodal branch are necessary to restrict the PS-like region from both sides. These experiments also provide proof of principle that micropatterned culture can be used to study the effects of genetic perturbations on spatial patterning in the human system. Consistent with a role for diffusible inhibitors in establishing patterns, blocking diffusion out of the colony by growing cells at the bottom of a PDMS microwell, prevented the formation of boundary fates. Inside the microwells, we observed an absence of staining for trophoectodermal or mesendodermal markers CDX2, BRA, EOMES, and SOX17 and instead nearly all cells stained positively for SOX2 (Supplementary Fig. 12). Control cells in the same culture dish but outside of the microwells established patterns containing all three germ layers. These results suggest that geometric control of paracrine signaling could be used to enhance the purity of stem cell derivatives.
Figure 6. TGF-β inhibitors are required for pattern formation.
(A) Immunofluorescence showing the expansion of the mesodermal territory at the expense of the ectoderm when either BMP inhibitors or Activin/Nodal inhibitors are knocked-down using siRNA. (B) Quantification showing that loss of BMP inhibitors increases BRA expression at the colony center, while loss of Activin/Nodal inhibitors increases BRA expression on both sides of the PS-like region. (C) Quantification of CDX2 expression, showing that loss of either BMP or Activin/Nodal inhibitors causes expansion of mesodermal CDX2 expression, and loss of Activin/Nodal inhibitors also causes a decrease in trophoblast-like CDX2 expression at the colony border.
Discussion
Our results show that simple confinement of hESCs to a disk shape region is sufficient to recapitulate much of germ layer patterning. The human epiblast is a disk-shaped epithelium at gastrulation and the cup-like mouse embryo is often approximated as a disk32,33. The number of cells in mammalian embryos at this stage is comparable to our larger disks. Thus cells grown on patterned substrates are a sensible approximation to the early gastrula and more appropriate than a solid embryoid body.
A number of groups have attempted to control spatial aspects of stem cell organization during differentiation and have shown that colony size can have an influence on the proportions of fates achieved10,11. However, these studies mostly focused on alternate differentiation protocols (e.g. withdrawal of pluripotency conditions) and did not find spatial ordering upon differentiation. Three dimensional culture systems have also been devised with no reports of spontaneous, reproducible spatial organization34.
Colony structure is shaped by inhibitors much as in the embryo. They plausibly leak from the colony boundary, as evidenced by the effects of confinement, and thus define a reference point from where the signaling pathways measure distance and define germ layer territories. The micropattern has a secondary consequence of increasing cell density and thus favoring cell communication over response to the external differentiation stimulus. In contrast to micropatterned culture, the SOX2 levels at the colony center never increase over their pluripotent values for BMP4 differentiation under standard culture conditions, a fact we attribute to lower levels of inhibitors produced or transmitted at lower cell densities. It will be interesting to probe the physical mechanisms of cell communication with a gentle flow over the colonies35.
Recently, self-organized organogenesis has received much attention with reports of the ability to generate optic cups36, brain-like organoids37, and mini-guts38–40 in vitro starting from embryonic stem cells. These studies employ primarily chemical cues rather than geometric confinement. Thus it will be interesting to see what effects spatial confinement has on later stages of differentiation. Quantitative models of signaling derived from disk geometery could potentially be used to engineer more reproducible organoids.
Developmental biology has made great strides by connecting genetic perturbations with defects in spatial patterning. In the future, micropatterned differentiation will allow the same manipulations to be performed for the early stages of hESC differentiation. We provide proof of principle for this approach by determining the spatial patterning phenotypes of knocking down gene products with siRNA. The advent of CRISPR technology will allow for the same assay to be performed with complete gene knockouts. The micropatterned colonies will facilitate time-lapse imaging of reporters for dynamic studies and provide an assay for mechanistic questions that are difficult to address in a mammalian embryo. When do the mesodermal and endodermal populations begin to diverge? What combination of geometric or genetic spatial symmetry breaking is needed to induce the anterior-posterior embryonic axis? Can extraembryonic tissue be replaced with signals applied directly on the epiblast? None of the current standards of ESC culture are capable of addressing these issues quantitatively. Other applications of micropatterned differentiation include interspecies comparisons under similar conditions (do mESCs and hESCs generate the same patterns?) and comparison between hiPSC and hESCs in a pattern-forming assay. As the patterns arise in a self-organized manner, micropatterned stem cell culture also provides a novel, controlled platform for studying how signaling generates developmental patterns. We thus propose that geometrically controlled cell culture should become standard practice for ES cell differentiation.
Online Methods
Cell culture
All experiments were performed with the RUES2 hESC line derived in our laboratory and described previously. For routine culture for maintenance, RUES2 cells were grown in HUESM medium that was conditioned by mouse embryonic fibroblasts (MEF-CM) and supplemented with 20ng/ml bFGF. Cells were tested for mycoplasma prior to beginning experiments and then again at two-month intervals. Cells were grown on tissue culture dishes coated with Matrigel (BD Biosciences 1:40 dilution). Dishes were coated in Matrigel overnight at 4 C and then incubated at 37 C for 1 hour immediately prior to seeding the cells on the surface.
For micropatterned cell culture, micropatterned glass coverslips (CYTOO) were first coated with 50 μg/ml Poly-D-Lysine in H2O (PDL; Millipore) for 2 hours. The PDL was then removed by serial dilutions without allowing the coverslip to dry (dilution 1:4 in H2O, six times), before performing two complete washes with H2O. Coverslips were then incubated with Matrigel (1:100 dilution in DMEM-F12) overnight at 4 C. Before cell seeding, the Matrigel was removed with serial dilutions in ice-cold PBS (dilution 1:4, six times) before 2 complete washes in ice cold PBS. Cells already resuspended in growth medium were seeding onto the coverslips immediately following the removal of the PBS. We found it was important to take care to keep the coverslips at 4 C at all times when in Matrigel solution and to ensure that the coverslips were not allowed to dry at any time after the application of the Matrigel. Both polymerization and drying of the Matrigel lead to inconsistent cell adhesion with cells more likely to detach from the surface during the experiment.
Cell seeding onto micropatterned coverslips was performed as follows: Cells growing in MEF-CM and FGF were pretreated with the Rock-inhibitor Y27632 (Rock-I; 10uM) for 1 hour, washed once with PBS, and dissociated with Trypsin. Cells were centrifuged and 5×105 cells were resuspended in 2.5 ml growth medium contain Rock-I and the entire solution placed over the coverslip in a 35mm tissue culture dish. After 2 hours, the medium was replaced with MEF-CM without Rock-I and cells were incubated overnight.
We also tested the effects of using the chemically defined mTeSR1 culture medium rather than MEF-CM and obtained similar patterns upon treatment with BMP4 to those presented here (Supplementary Fig. 9), however, we found that MEF-CM better promoted adhesion to the micropatterned surface and it was therefore used in all subsequent experiments. Finally we tested the effects of growing cells on recombinant Laminin521 (Biolamina) rather than Matrigel, and again obtained similar patterns (Supplementary Fig. 9). Laminin521 coating of coverslips was performed as follows – coverslips were coated with 5 ug/ml of Laminin521 diluted in PBS with calcium and magnesium for 2 hours at 37 C. Laminin was then removed with six serial dilutions in warm PBS (dilution 1:4) before two complete washes in PBS. Cells were then seeded as described above for the PDL-Matrigel coated coverslips.
siRNA Experiments
Cells were passaged as single cells in Rock-I into 35mm dishes at a density of 200000 cells/dish. The next day, cells were transfected with siRNA (Ambion Silencer select) using RNAiMax (Invitrogen). The final concentration of siRNA was 20nM and 2.5 μl of RNAiMax were used for each dish. The following day, cells were seeded onto micropatterned coverslips and differentiation experiments were performed as described above.
PDMS microwells
Molds to create PDMS wells of controlled diameter and depth were designed using a 3D CAD software (Autodesk Inventor) and then 3D printed (3D Systems Projet 3510 HD Plus printer). The smallest wells this technique allowed us to make reliably were 250μm in diameter and 250μm in depth. We found that boiling the 3D printed parts in water containing 1% Triton X100 for four hours was necessary to allow the PDMS to cure on the 3D printed parts. Molds were filled with PDMS (10:1 base:reticulant ratio) and degased under vacuum. In order to create opened wells, the mold was placed between 2 glass slides on which pressure was applied, typically with a large paper clip. After PDMS curing at 80C for several hours, the PDMS wells were unmolded and boiled in distilled water to ensure sterility and that all PDMS was cured. PDMS wells were then washed with ethanol and dried in a cell culture cabinet to keep them sterile.
Wells were then stuck on the dry cell culture substrate (either glass coverslips, regular tissue culture dish, or optically clear plastic dishes (ibidi)). The wells were then coated with cell adhesion promoting proteins either in a one-step protocol (Laminin521) or a two-step protocol (PDL followed by Matrigel). To remove bubbles trapped in wells, the PDMS well filled with the coating solution were centrifuged (2000rpm, 2min). After the necessary incubation time the coating solution was aspirated and cells were seeded.
Immunofluorescence
Coverslips were rinsed once with PBS, fixed with 4% paraformaldehyde, rinsed twice with PBS, then blocked and permiabilized with 3% Donkey Serum, 0.1% Triton X-100 in PBS for 30 minutes. When performing immunofluorescence for pSmad1, cells were pretreated with 1% SDS in PBS for 30 min at 37 C before blocking. Coverslips were incubated with primary antibodies overnight at 4 C (for primary antibodies and dilutions see Supplementary Table 1), washed three times in PBS for 30 minutes each wash, incubated with secondary antibodies (Alexa488, Alexa555 or Alex647 conjugated (Molecular probes); dilution 1:500) and DAPI nuclear counterstain for 30 minutes, and then washed twice with PBS. Coverslips were mounted on slides using Fluoromount-G mounting medium (Southern Biotech).
Imaging
All widefield images were acquired on an Olympus IX71 inverted microscope with a 20X, 0.75 Na lens. We utilized tiled image acquisition to acquire images of the entire coverslip (approximately 2500 stage positions/coverslip) in four channels corresponding to DAPI and Alexa488, Alexa555, and Alexa647 conjugated antibodies. All confocal images were acquired on a Leica SP8 inverted confocal microscope with a 40X, 1.1 Na water immersion objective. Three-dimensional visualization and rendering was performed using Imaris software.
Image analysis
All image analysis was performed using custom software written in MATLAB. Since we imaged the entire coverslip, we utilized the fact that we also imaging the regions without cells to create background and normalization images as follows. We first took the pixel-by-pixel minimum and average across all images:
| (1) |
where IJ represents the jth image. We then used m as a background imaged and defined a normalization image according to:
| (2) |
where the max is across all pixels in the image. The mean and normalization images were then smoothed with a Gaussian filter several times the size of a cell diameter and each image was corrected according to:
| (3) |
Where the “./” represents pixel-wise division. In general, subtracting m removes the (typically spatially homogenous) camera noise, while dividing by n corrects for any inhomogeneities in the image due to the illumination or other factors.
After each image was corrected in this way, we then identified and quantified the cells in each image using an algorithm which we described previously41 (see next section). Because larger colonies will span multiple images, we preformed image alignment and used the resulting overlap between images to put the coordinates of each cell in “coverslip coordinates” resulting in a list of the position of every cell on the coverslip. We then separated these cells into colonies by computing the alphavol of the points using the MATLAB function alphavol (http://www.mathworks.com/matlabcentral/fileexchange/28851-alpha-shapes) with a radius (rv) of 100 pixels. This function is similar in concept to a convex hull except that it will form separate boundaries for sets A and B if all points in set A are greater than rv away from all points in set B. Having identified colonies, we classified them based on their radius. For quantification of immunofluorescence, we either normalized the intensity to the DAPI intensity in the same cell, or, for proteins that translocate to the nucleus upon activation, we normalized the nuclear intensity to the cytoplasmic intensity in the same cell. Both of these normalizations serve to remove imaging artifacts.
Marker quantification
Individual cells were identified in images and quantified for markers as described previously41. All marker intensities were normalized to the DAPI intensity in the same cell with the exception of SMAD2 where we normalized the nuclear SMAD2 intensity to the cytoplasmic intensity in the same cell. We have found this nuclear to cytoplasmic ratio to be a sensitive metric for signaling activity41. Radial averages were performed over all colonies on the micropatterned coverslip (n=25 for 1000μm colonies, n=144 for 500μm colonies, and n=576 for 250μm colonies) and the error bars given shown are standard deviations between colonies. Each marker quantification was performed in at least two different independent experiments. For 500μm and 1000μm colonies, we manually excluded those colonies in which cell seeding was uneven (for example, large empty areas within the micropatterned patch).
Supplementary Material
Acknowledgments
The authors are grateful to S. Li and A. Yoney for technical assistance, C. Kirst for assistance with three-dimensional image segmentation, and members of the A.H.B. and E.D.S. laboratories, A.-K. Hadjantonakis and S. Nowotschin for helpful discussions. Funding supporting this work was provided by The Rockefeller University, NYSTEM, National Institutes of Health Grant R01 HD32105 (to A.H.B.) and R01 GM 101653 (to A.H.B. & E.D.S.), National Science Foundation Grant PHY-0954398 (to E.D.S.), and the Human Frontier Science Program LT000851/2011-L (to B.S.).
Footnotes
Author Contributions
A.W. designed and performed experiments, performed analysis, and wrote the paper. B.S. designed and performed experiments and contributed to writing the paper, F.E. performed experiments and contributed to writing the paper, E.D.S. designed experiments, performed analysis, and wrote the paper, A.H.B designed experiments and wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- 2.D’Amour KA, et al. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 3.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kattman SJ, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Ozair MZ, Noggle S, Warmflash A, Krzyspiak JE, Brivanlou AH. SMAD7 Directly Converts Human Embryonic Stem Cells to Telencephalic Fate by a Default Mechanism. Stem Cells. 2012;31:35–47. doi: 10.1002/stem.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Haim N, et al. The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev Cell. 2006;11:313–323. doi: 10.1016/j.devcel.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 7.ten Berge D, et al. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell. 2008;3:508–518. doi: 10.1016/j.stem.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang C, et al. isolation of primitive endoderm, mesoderm, vascular endothelial and trophoblast progenitors from human pluripotent stem cells. Nat Biotechnol. 2012;30:531–542. doi: 10.1038/nbt.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blauwkamp TA, Nigam S, Ardehali R, Weissman IL, Nusse R. Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nat Commun. 2012;3:1070. doi: 10.1038/ncomms2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peerani R, et al. Niche-mediated control of human embryonic stem cell self-renewal and differentiation. EMBO J. 2007;26:4744–4755. doi: 10.1038/sj.emboj.7601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauwens CL, et al. Control of human embryonic stem cell colony and aggregate size heterogeneity influences differentiation trajectories. Stem Cells. 2008;26:2300–2310. doi: 10.1634/stemcells.2008-0183. [DOI] [PubMed] [Google Scholar]
- 12.Sakai Y, Yoshiura Y, Nakazawa K. Embryoid body culture of mouse embryonic stem cells using microwell and micropatterned chips. J Biosci Bioeng. 2011;111:85–91. doi: 10.1016/j.jbiosc.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 13.James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 14.Xu RH, et al. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 16.Yu P, Pan G, Yu J, Thomson JA. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell. 2011;8:326–334. doi: 10.1016/j.stem.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amita M, et al. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci USA. 2013;110:E1212–21. doi: 10.1073/pnas.1303094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sudheer S, Bhushan R, Fauler B, Lehrach H, Adjaye J. FGF inhibition directs BMP4-mediated differentiation of human embryonic stem cells to syncytiotrophoblast. Stem Cells and Development. 2012;21:2987–3000. doi: 10.1089/scd.2012.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, et al. BMP4-directed trophoblast differentiation of human embryonic stem cells is mediated through a ΔNp63+ cytotrophoblast stem cell state. Development. 2013;140:3965–3976. doi: 10.1242/dev.092155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernardo AS, et al. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 2011;9:144–155. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loh KM, Lim B. A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell. 2011;8:363–369. doi: 10.1016/j.stem.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Thomson M, et al. Pluripotency Factors in Embryonic Stem Cells Regulate Differentiation into Germ Layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teo AKK, et al. Pluripotency factors regulate definitive endoderm specification through EOMESodermin. Genes Dev. 2011;25:238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Oron E, Nelson B, Razis S, Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 25.Li L, et al. Location of transient ectodermal progenitor potential in mouse development. Development. 2013;140:4533–4543. doi: 10.1242/dev.092866. [DOI] [PubMed] [Google Scholar]
- 26.Chawengsaksophak K, de Graaff W, Rossant J, Deschamps J, Beck F. CDX2 is essential for axial elongation in mouse development. Proc Natl Acad Sci USA. 2004;101:7641–7645. doi: 10.1073/pnas.0401654101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams M, Burdsal C, Periasamy A, Lewandoski M, Sutherland A. Mouse primitive streak forms in situ by initiation of epithelial to mesenchymal transition without migration of a cell population. Dev Dyn. 2012;241:270–283. doi: 10.1002/dvdy.23711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng VY, Ang SN, Chan JX, Choo ABH. Characterization of epithelial cell adhesion molecule as a surface marker on undifferentiated human embryonic stem cells. Stem Cells. 2010;28:29–35. doi: 10.1002/stem.221. [DOI] [PubMed] [Google Scholar]
- 29.Inman GJ, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 30.Bachiller D, et al. The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature. 2000;403:658–661. doi: 10.1038/35001072. [DOI] [PubMed] [Google Scholar]
- 31.Perea-Gomez A, et al. Nodal antagonists in the anterior visceral endoderm prevent the formation of multiple primitive streaks. Dev Cell. 2002;3:745–756. doi: 10.1016/s1534-5807(02)00321-0. [DOI] [PubMed] [Google Scholar]
- 32.Behringer RR, Wakamiya M, Tsang TE, Tam PP. A flattened mouse embryo: leveling the playing field. Genesis. 2000;28:23–30. doi: 10.1002/1526-968x(200009)28:1<23::aid-gene30>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 33.Tam PPL, Gad JM. In: Gastrulation: From Cells to Embryo. Stern CD, editor. CSHL Press; 2004. [Google Scholar]
- 34.Hwang YS, et al. Microwell-mediated control of embryoid body size regulates embryonic stem cell fate via differential expression of WNT5a and WNT11. Proc Natl Acad Sci USA. 2009;106:16978–16983. doi: 10.1073/pnas.0905550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moledina F, et al. Predictive microfluidic control of regulatory ligand trajectories in individual pluripotent cells. Proc Natl Acad Sci USA. 2012;109:3264–3269. doi: 10.1073/pnas.1111478109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eiraku M, et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 37.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 39.Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 40.Spence JR, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warmflash A, et al. Dynamics of TGF-β signaling reveal adaptive and pulsatile behaviors reflected in the nuclear localization of transcription factor SMAD4. Proc Natl Acad Sci USA. 2012;109:E1947–56. doi: 10.1073/pnas.1207607109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.