Abstract
During vertebrate embryogenesis retinoic acid (RA) synthesis must be spatiotemporally regulated in order to appropriately stimulate various retinoid signaling pathways. Various forms of mammalian aldehyde dehydrogenase (ALDH) have been shown to oxidize the vitamin A precursor retinal to RA in vitro. Here we show that injection of Xenopus embryos with mRNAs for either mouse Aldh1 or mouse Raldh2 stimulates RA synthesis at low and high levels, respectively, while injection of human ALDH3 mRNA is unable to stimulate any detectable level of RA synthesis. This provides evidence that some members of the ALDH gene family can indeed perform RA synthesis in vivo. Whole-mount immunohistochemical analyses of mouse embryos indicate that ALDH1 and RALDH2 proteins are localized in distinct tissues. RALDH2 is detected at E7.5–E10.5 primarily in trunk tissue (paraxial mesoderm, somites, pericardium, midgut, mesonephros) plus transiently from E8.5–E9.5 in the ventral optic vesicle and surrounding frontonasal region. ALDH1 is first detected at E9.0–E10.5 primarily in cranial tissues (ventral mesencephalon, dorsal retina, thymic primordia, otic vesicles) and in the mesonephros. As previous findings indicate that embryonic RA is more abundant in trunk rather than cranial tissues, our findings suggest that Raldh2 and Aldh1 control distinct retinoid signaling pathways by stimulating high and low RA biosynthetic activities, respectively, in various trunk and cranial tissues.
Keywords: retinoic acid, aldehyde dehydrogenase, Aldh1, Raldh2, retinoid signaling
INTRODUCTION
Vitamin A (retinol) plays a role in the regulation of vertebrate embryonic development through its active metabolite retinoic acid (RA) [reviewed by Hofmann and Eichele, 1994; Gudas et al., 1994; Maden, 1994]. RA functions as a ligand for several nuclear retinoic acid receptors (RARs) which function as ligand-dependent transcription factors in conjunction with retinoid-X receptors (RXRs), with which they form heterodimers [reviewed by Kastner et al., 1994; Mangelsdorf et al., 1994]. Essential roles for these receptors in vitamin A function have been confirmed in mice carrying targeted mutations of RARs and RXRs [Lohnes et al., 1994; Mendelsohn et al., 1994; Luo et al., 1996; Mascrez et al., 1998]. A major challenge remaining in the study of retinoid signaling is the elucidation of how production of the ligand is spatiotemporally regulated in various tissues during development to provide the correct amount of RA for each discrete receptor-mediated signaling event.
The major biosynthetic pathway of RA from retinol involves two sequential steps catalyzed by several candidate enzymes [reviewed by Duester, 1996; Napoli, 1996]. First, reversible dehydrogenation of retinol into retinal is catalyzed by either cytosolic retinol dehydrogenases, which are members of the alcohol dehydrogenase (ADH) family, or by microsomal retinol dehydrogenases, which are members of the short-chain dehydrogenase/reductase family (SDR). Mice carrying a targeted Adh4 null mutation have defects in retinol utilization during vitamin A deficiency [Deltour et al., 1996b] and Adh1−/− mice display significant reductions in the ability to metabolize a dose of retinol to RA [Deltour et al., 1999a]. Thus, at least two forms of ADH are involved in metabolizing retinol to retinal in vivo.
In the second step of the metabolic pathway, retinal is irreversibly oxidized to RA by cytosolic retinal dehydrogenases, which are members of the aldehyde dehydrogenase (ALDH) family. Examination of the second step of RA synthesis in vitro originally led to the identification of calf liver ALDH as an enzyme able to oxidize retinal to RA [Futterman, 1962]. Subsequent studies revealed an extensive family of mammalian ALDHs, with the original form active for RA synthesis corresponding to class I ALDH (ALDH1) in the human [Dockham et al., 1992; Yoshida et al., 1992], as well as its mouse ALDH1 homolog previously known as AHD2 [Lee et al., 1991; McCaffery et al., 1992], and its rat ALDH1 homolog also known as RALDH or RalDH-I [Labrecque et al., 1995; Penzes et al., 1997; Kathmann and Lipsky, 1997]. Two additional classes of ALDH, i.e., mitochondrial ALDH2 and cytosolic ALDH3, have been shown to have no RA synthetic activity in vitro [Yoshida et al., 1992]. The discovery that mouse ALDH1 is localized during embryogenesis in the dorsal retina (a retinoid-target tissue) provided impetus to examine this enzyme as a potential embryonic RA synthetic enzyme [McCaffery et al., 1991]. Further studies in the mouse culminated in the discovery of an additional RA synthetic enzyme identified as a distinct class of ALDH sharing approximately 70% amino acid sequence identity with ALDH1; this form has been named RALDH2 in mouse [Zhao et al., 1996], human [Ono et al., 1998], and chick [Sockanathan and Jessell, 1998], and RalDH-II in the rat [Wang et al., 1996].
During early mouse embryogenesis, there exists a posterior preference for initial RA synthesis [Hogan et al., 1992]. This may help establish the anteroposterior axis during early development due to regulation of members of the Hox gene family, which are RA-inducible and differentially expressed along the anteroposterior axis with expression only in posterior tissues (trunk and posterior hindbrain) [Shimeld, 1996]. In addition to anteroposterior patterning, RA does play a role in cranial development, as demonstrated by embryonic vitamin A-deficiency studies [Maden et al., 1996; Dickman et al., 1997] as well as gene knockouts of RARs [Lohnes et al., 1994] and Raldh2 [Niederreither et al., 1999]. Thus, RA must exist in cranial locations as well as trunk locations. Localization of RA in mouse embryos has been determined using either a transgenic RA reporter mouse line [Rossant et al., 1991] or a whole-embryo explant RA bioassay [Ang et al., 1996a] which employs an RA reporter cell line that was originally used to identify RA in the neural tube [Wagner et al., 1992]. In both assays, RA is initially detected at E7.5, limited to posterior tissues, and then from E8.5–E10.5 RA is still abundant in the trunk, but it is now also detected in the optic vesicles and surrounding frontonasal region. Raldh2 mRNA transcripts are initially expressed in the posterior mesoderm of mouse embryos at E7.5, with additional expression in the heart at E8.25, transient expression in the optic vesicles at E8.5, and a continued high level of expression in the trunk from stages E8.5–E10.5 and in the spinal cord by E12.5 [Zhao et al., 1996; Niederreither et al., 1997; Moss et al., 1998]. Raldh2 −/− null mutant mice die at midgestation, displaying no detectable RA in the trunk and frontonasal region at late E8.5, but with some RA still detectable in the optic vesicles [Niederreither et al., 1999]. Thus, Raldh2 appears to be responsible for essentially all of the RA synthesis occurring in the trunk and frontonasal regions by late E8.5, but not for all RA synthesis occurring in the optic vesicles. A role for mouse Aldh1 in optic vesicle RA synthesis is suggested based on its expression in the dorsal retina at E9.5 [McCaffery et al., 1991]; however, a more complete analysis of Aldh1 is needed to determine the full extent of its role in RA synthesis.
A direct demonstration of the abilities of Aldh1 and Raldh2 to catalyze embryonic RA synthesis by overexpression in an in vivo setting has not been previously reported. Also, the localization of both ALDH1 and RALDH2 proteins during embryogenesis has not been adequately addressed in order to identify tissues where the enzymes actually exist and RA synthesis can thus be expected to occur. Here, we examined mouse Aldh1 and Raldh2 for their ability to function in RA synthesis in vivo by expression in Xenopus embryos. Our results provide firm evidence that both genes stimulate RA synthesis when expressed in Xenopus, and these findings provide insight as to their relative abilities. In addition, we used specific antibodies directed against ALDH1 and RALDH2 to demonstrate distinct patterns of protein localization for these two enzymes in mouse cranial and trunk embryonic tissues, thus defining distinct domains where Aldh1 and Raldh2 may influence retinoid signaling during development.
MATERIALS AND METHODS
ALDH cDNAs
A full-length cDNA for mouse Aldh1 (originally known as Ahd2) has been previously described [Hsu et al., 1999]. A full-length cDNA for human ALDH3 has also been described [Hsu et al., 1992].
A full-length cDNA for mouse Raldh2 was obtained as follows. Total RNA was isolated from adult mouse testis by the acid guanidinium thiocyanate-phenolchloroform method [Chomczynski and Sacchi, 1987]. cDNA cloning was performed by reverse transcription coupled by polymerase chain reaction (PCR). First strand cDNA synthesis was performed on 5 µg of total RNA using oligo-dT as a primer and AMV reverse transcriptase, as described in the cDNA synthesis kit supplied by Amersham Life Science Inc. (Arlington Hts., IL). The single-stranded cDNA product was subjected to PCR using standard methodology [Ausubel et al., 1989] with oligonucleotide primers corresponding to the 5′ and 3′ regions of the mouse Raldh2 sequence encompassing the start and stop codons, respectively [Zhao et al., 1996]. PCR products of approximately 1,600 bp were cloned into Bluescript IIKS (Stratagene Cloning Systems, La Jolla, CA) and a clone containing the complete open reading frame of mouse Raldh2 was verified by DNA sequence analysis.
RNA Transcription and Translation
Full-length cDNAs for mouse Aldh1, mouse Raldh2, and human ALDH3 were subcloned into plasmid pSP65 (Promega, Madison, WI) in the sense direction for in vitro transcription from the SP6 promoter. Plasmids were linearized and subjected to in vitro transcription with SP6 RNA polymerase, 5′-capping with 7-methylguanosine, followed by 3′-polyadenylation with Escherichia coli poly(A) polymerase to produce full-length mRNAs [Vize et al., 1991]. The mRNAs generated in this fashion were brought to a concentration of 0.2 µg/µl in water, and 0.4 µg was used for in vitro translation in a rabbit reticulocyte lysate (Stratagene, CA) to monitor the ability of the mRNAs to produce full-length proteins.
Frog Embryo Microinjection and RA Detection
Xenopus laevis embryos were produced by artificial fertilization and staged as described previously by Nieuwkoop and Faber [1994]. Embryos were placed in 1× MMR, 3% Ficoll-400 at room temperature for microinjection of mRNA as described previously [Kay, 1991]. Various amounts of mRNA (4.6–23.0 nl at a concentration of either 0.2 ng/ml in water) was injected into the vegetal pole of embryos at the 2–4 cell stages using a Nanoject microinjection apparatus (Drummond Scientific, Broomall, PA) attached to the micropipet prefilled with light mineral oil. Four hours after injection, embryos were transferred to 0.1× MMR and incubated to stage 8 (blastula).
RA was detected using a bioassay that employs the RA reporter cell line F9-RARE-lacZ [Wagner et al., 1992]. This cell line was derived from stable transfection of mouse F9 cells with a transgene containing an RA response element driving expression of lacZ. F9-RARE-lacZ detects the presence of RA in cultured embryo explants by detecting diffusion of this lipid-soluble molecule from the embryo to the surrounding reporter cells. Bioassay of Xenopus RA was performed as previously described for mouse embryos [Ang et al., 1996a]. Briefly, F9-RARE-lacZ cells were grown until 80–90% confluent, at which time stage 8 Xenopus embryos were placed on top of the reporter cell monolayer, incubated for 18 h, then fixed in 1% glutaraldehyde and assayed for β-galactosidase activity produced by lacZ expression. Xenopus embryos placed upon the reporter cells and incubated under standard mammalian cell culture conditions did not develop further, but remained intact for the duration of the assay.
Production of Antibodies and Western Blot Analysis
Mouse ALDH1 and RALDH2 proteins were expressed in the E. coli strain BL-21 as N-terminal fusions to Schistosoma glutathione-S-transferase (GST) using pGEX expression vectors (Pharmacia Biotech, Uppsala, Sweden). A 1.6 kb full-length cDNA for mouse Aldh1 [Hsu et al., 1999] was cloned downstream of GST in the EcoRI site of pGEX-4T-2. Likewise, a 1.6 kb full length cDNA for mouse Raldh2 (described above) was cloned into the EcoRI site of pGEX-4T-3. Both constructions resulted in fusion proteins with approximate molecular weights of 84,000 (GST ~29 kDa plus ALDH ~55 kDa) after induction of transformed BL-21 cultures with isopropyl-β-D-thiogalactopyranoside (IPTG) at 0.1%. Inclusion body preparations of ALDH1-GST and RALDH2-GST fusion proteins prepared as described [Domingo et al., 1992] were solubilized in Laemmli sample buffer and resolved on preparative SDS-PAGE gels using standard techniques [Sambrook et al., 1989]. After staining the gel with 0.05% coomassie brilliant blue in water for 1 h, the 84 kDa fusion proteins were excised and divided into 200 µg aliquots. Each of these were mascerated in Freund’s adjuvant and used to perform four monthly injections into rabbits using standard techniques [Harlow and Lane, 1988]. Antibodies were affinity-purified from antisera using 100 µg aliquots of the specific ALDH-GST fusion protein electroblotted to PVDF membrane, and were quantitated as previously described [Haselbeck and Duester, 1997].
Western blot analysis was performed on homogenates of adult mouse tissues (FVB/N) as previously described [Haselbeck and Duester, 1997].Western blots were subjected to immunochemical detection using the ECL chemiluminescence detection kit (Amersham). ALDH1 and RALDH2 primary antibodies were used at a concentration of 0.02 µg/ml. Electroblotted membranes were stripped and reanalyzed using each affinity-purified antibody.
Immunohistochemical Analysis of Mouse Embryos
Mouse embryos (FVB/N) were staged by vaginal plug appearance with noon on the day of plug detection being considered embryonic day 0.5 (E0.5). Further staging of mouse embryos was according to Kaufman [1992]. For all immunohistochemical analyses, ALDH1 and RALDH2 primary antibodies were used at a concentration of 1.0 µg/ml.
Whole-mount immunohistochemistry was performed on mouse embryos from stages E7.5–E10.5 using the above affinity-purified ALDH antibodies as described previously [Hogan et al., 1994]. Color detection was performed by incubation with diaminobenzidine/nickel chloride for 30 min at room temperature followed by addition of hydrogen peroxide and further incubation for up to 60 min. Prior to photography, embryos were cleared in benzyl alcohol:benzyl benzoate (1:2) for 10 min.
Additional immunohistochemical studies were performed on mouse embryos embedded in paraffin and sectioned at 6 µm using standard methodology [Kaufman, 1992]. ALDH localization was performed by incubation for 30 min with purified antibody serially diluted in normal blocking solution, followed by detection using the Vectastain Elite avidin-biotin-horseradish peroxidase ABC kit (Vector Laboratories, Burlingame, CA). Incubation with diaminobenzidine for color detection was performed for 3 min essentially as recommended by the Vectastain kit, followed by dilution in tap water to stop the reaction. Slides were lightly counterstained with hematoxylin prior to mounting and observation by bright-field microscopy.
RESULTS
Aldh1 and Raldh2 mRNAs Stimulate RA Synthesis in Xenopus Embryos
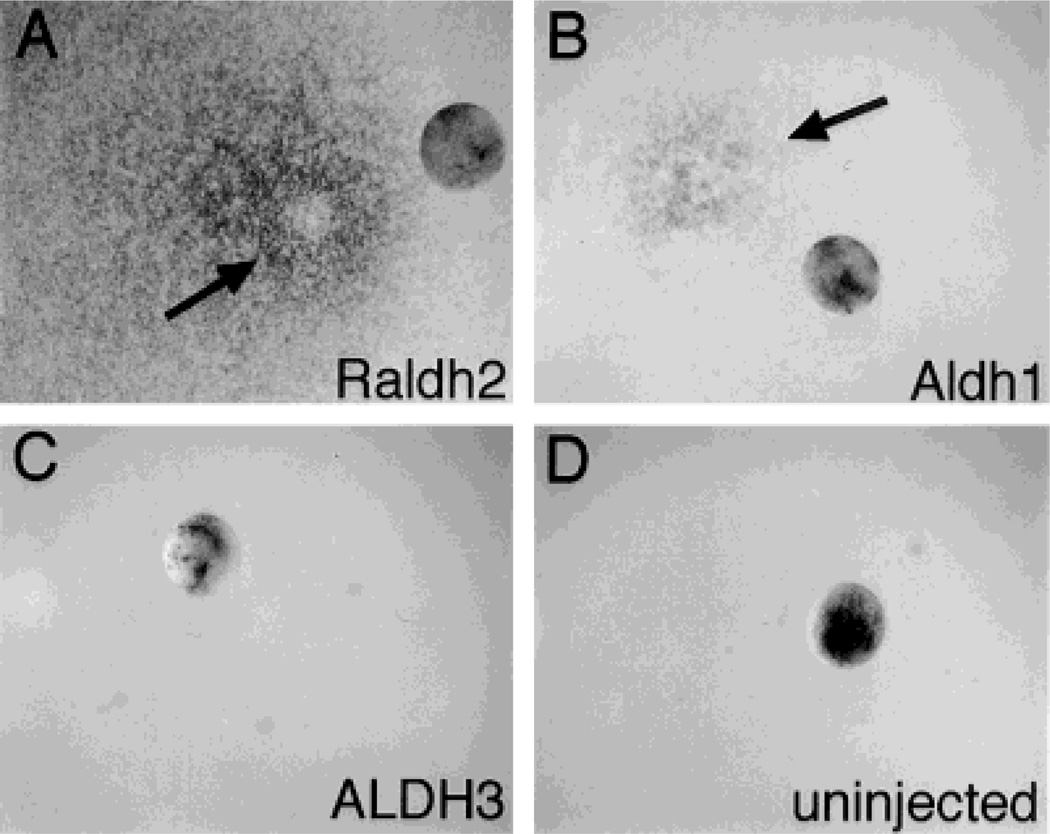
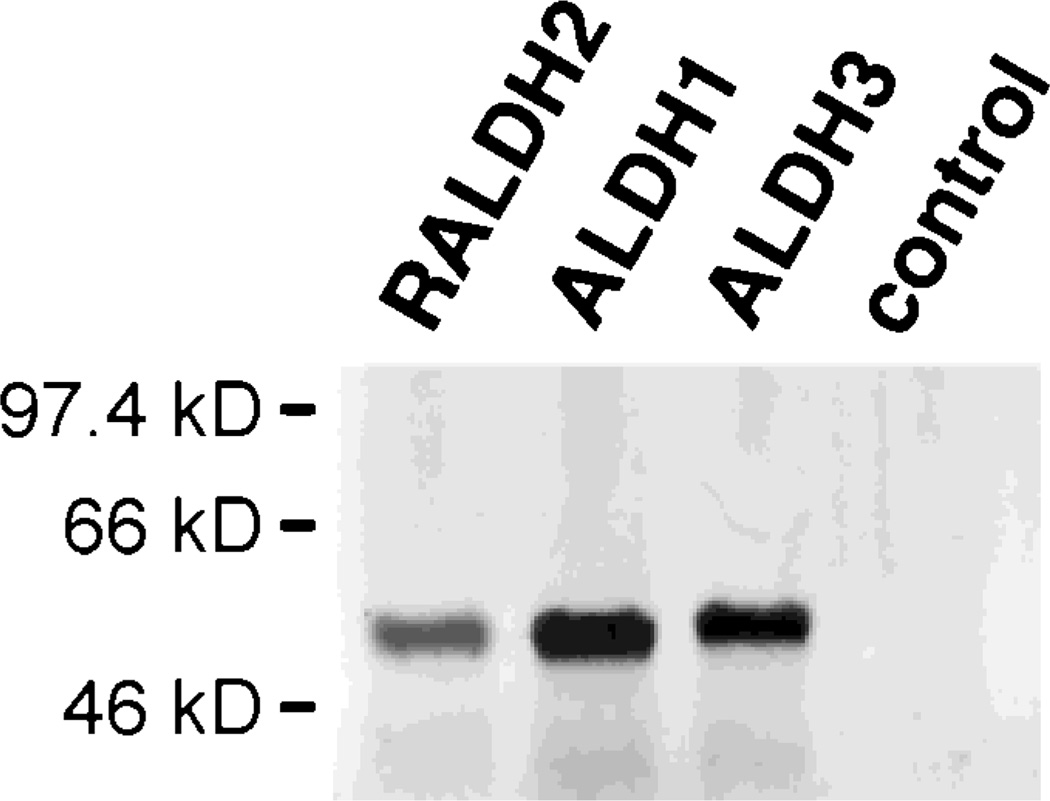
In order to explore the ability of certain ALDH genes to regulate RA synthesis in vivo, we injected mouse Aldh1, mouse Raldh2, or human ALDH3 mRNAs into Xenopus embryos at the 2–4 cell stage, incubated the embryos to the blastula stage, then monitored RA levels in individual embryos using a lacZ-linked bioassay. Whereas RA was not detectable in uninjected embryos (0 out of 45 examined) nor in embryos injected with ALDH3 mRNA (0 out of 47 embryos injected), a significant amount of RA was detectable in embryos injected with either Raldh2 mRNA (36 out of 36 embryos injected) or Aldh1 mRNA (47 out of 61 embryos injected) (Fig. 1). Raldh2 mRNA was more efficient in this regard than Aldh1 mRNA, as evidenced by a much larger area of lacZ detection around the embryo, but both of these mRNAs were clearly distinguishable from ALDH3 mRNA, which completely lacked the ability to stimulate detectable RA synthesis. The mRNAs prepared from Raldh2, Aldh1, and ALDH3 were each able to be translated into a polypeptide of approximately 55 kDa, corresponding to the expected size of ALDH subunits (Fig. 2).
Fig. 1.
Aldh1 and Raldh2 mRNA transcripts stimulate RA synthesis in Xenopus embryos. Xenopus embryos at the 2–4 cell stages were injected with 23 nl of mRNA(0.2 µg/µl) transcribed from either mouse Raldh2 (A), mouse Aldh1 (B), or human ALDH3 (C), or were uninjected as a control (D). Following growth to blastula stage 8, embryos were subjected to an RA bioassay which involves incubating the embryo on a lawn of RA-reporter cells followed by assay for lacZ expression. In each photograph the embryo has been moved aside from its site of incubation, and arrows point to sites of lacZ expression.
Fig. 2.
Relative efficiency of protein production from ALDH mRNAs. Mouse Raldh2, mouse Aldh1, and human ALDH3 mRNA samples were examined for the ability to produce protein by translation of 0.4 µg of each sample in the presence of [35S]methionine followed by SDS-polyacrylamide gel electrophoresis. Each mRNA produced a protein of approximately 55 kDa, the expected size of an ALDH subunit, and a control sample containing no mRNA produced no protein.
Distribution of ALDH1 and RALDH2 Proteins in Adult Mouse Tissues
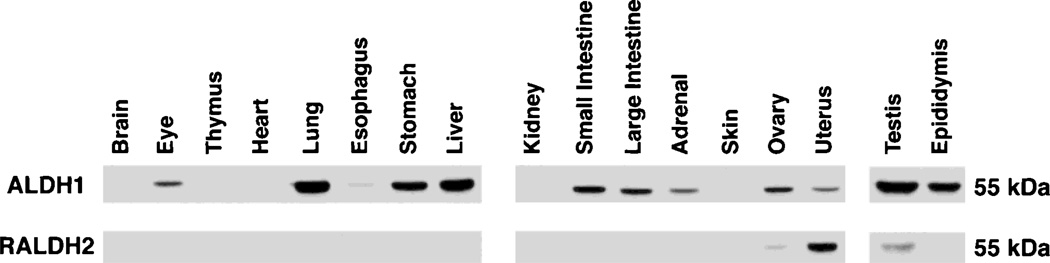
Since forced expression of mouse Aldh1 and Raldh2 genes in Xenopus embryos resulted in RA synthesis, we generated antibodies against both of these enzymes to examine their distribution patterns in the mouse. Polyclonal antibodies raised against full-length proteins for mouse ALDH1 and RALDH2 were used to probe adult mouse tissue extracts by Western blot analysis. Both antibodies detected proteins of 55 kDa, the expected size of an ALDH subunit, but the distribution of each protein was quite distinct. ALDH1 immunodetection was observed in extracts of the whole eye, lung, esophagus (weak), stomach, liver, small intestine, large intestine, adrenal, ovary, uterus, testis, and epididymis, with no detection observed in extracts of the whole brain, thymus, heart, kidney, or skin (Fig. 3). On the other hand, after examination of all the above tissues with RALDH2 antibodies, immunodetection was observed in only a very limited set of tissues, including the ovary (weak), uterus, and testis (Fig. 3). Based on our observed widespread appearance of ALDH1 in numerous adult organs containing retinoid-dependent epithelia, but a much more restricted localization of RALDH2 to a few reproductive organs,ALDH1 is likely to play a much wider role than RALDH2 in RA synthesis for adult tissues.
Fig. 3.
Western blot of adult mouse tissues showing specificity of antibodies for ALDH1 and RALDH2 proteins. Tissue extracts were loaded at 20 µg of total protein per lane. Blots were sequentially probed with each antibody, and proteins detected were 55 kDa in each case.
Our distribution of ALDH1 protein agrees with the expression pattern of Aldh1 mRNA, which has previously been observed in mouse liver, intestine, lung, and testis, but not in kidney, brain or heart [López-Fernández and Del Mazo, 1997; Hsu et al., 1999]. The specificity of the ALDH1 antibody is also shown by the presence of a strong signal in the lung, liver, and testis, but a complete absence of a signal in the kidney (Fig. 3), results which match previous findings on the distribution of mouse ALDH1 enzyme activity in adult organs [Rout and Holmes, 1985]. The specificity of the RALDH2 antibody is shown by our detection of a protein signal in the testis but not liver (Fig. 3), thus matching previous studies on Raldh2 mRNA distribution in these adult tissues [Zhao et al., 1996]. Since the adult mouse liver contains abundant ALDH1 enzyme activity [Rout and Holmes, 1985] and Aldh1 mRNA [López-Fernández and Del Mazo, 1997], the absence of a signal in the liver indicates that the RALDH2 antibody does not detect ALDH1. Also, the ALDH1 and RALDH2 antibodies do not recognize another related cytosolic enzyme, ALDH-PB, as evidenced by the lack of a signal in the adult kidney (Fig. 3), which has previously been shown to contain abundant ALDH-PB [Hsu et al., 1999]. As these results demonstrate that the ALDH1 and RALDH2 antibodies have a high degree of specificity and do not display significant cross-reaction with each other or with ALDH-PB, they are thus suitable for studies on embryonic protein localization.
Distinct Cranial and Trunk Localization of ALDH1 and RALDH2 Proteins During Early Mouse Embryogenesis
The overall results of our whole-mount immunohistochemical analyses of ALDH1 and RALDH2 during stages E7.5–E10.5 of mouse embryogenesis are summarized in Table 1. Embryos older than E10.5 were not examined in detail, but selected organs were examined.
TABLE 1.
ALDH1 and RALDH2 Immunohistochemical Localization During Mouse Embryogenesis
| Stage/Tissue | ALDH1 | RALDH2 |
|---|---|---|
| E7.5–E8.0 | ||
| Anterior (headfolds) | − | − |
| Posterior (paraxial mesoderm) | − | + |
| E8.5 | ||
| Neural folds/tube | − | − |
| Optic vesicle | − | + |
| Otic vesicle | − | − |
| Somites | − | + |
| Paraxial mesoderm | − | + |
| Coelomic cavity adjacent to heart | − | + |
| Allantoic mesenchyme | − | + |
| E9.0–E10.5 | ||
| Cranial | ||
| Forebrain | − | − |
| Midbrain (mesencephalon) | + | − |
| Hindbrain | − | − |
| Optic vesicle (dorsal retina) | + | − |
| Optic vesicle (ventral/frontonasal) | − | + |
| Otic vesicle | + | − |
| Thymic primordia | + | − |
| Trunk | ||
| Neural tube | − | − |
| Somites | − | + |
| Limb buds | − | + |
| Coelomic cavity mesenchyme | − | + |
| Pericardium | − | + |
| Cloaca | − | + |
| Midgut | − | + |
| Mesonephros | + | + |
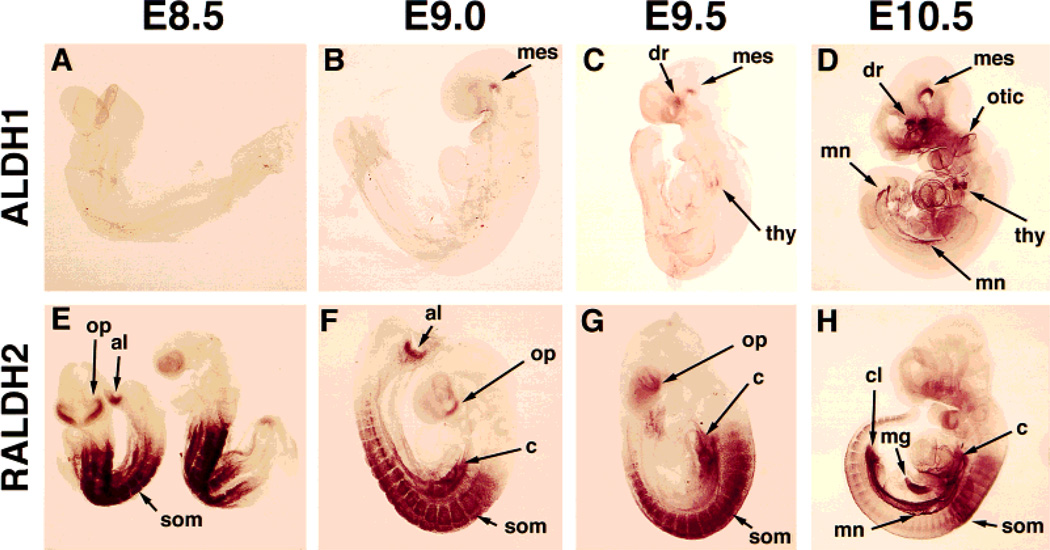
ALDH1 protein was not detected at E7.5 (data not shown) or E8.5 (Fig. 4A), but was first detected at E9.0 in a cranial location, particularly in the ventral flexure of the mesencephalon (Fig. 4B). By E9.5, ALDH1 protein was detected in several cranial derivatives, including the ventral mesencephalon, the dorsal retina, and the thymic primordia (Fig. 4C). At E10.5, ALDH1 protein was observed cranially in the ventral mesencephalon, dorsal retina, thymic primordia, and otic vesicles, and in the one trunk tissue, the mesonephric ducts (Fig. 4D). Interestingly, along the anteroposterior axis of the mesonephros, ALDH1 localization was much higher in those regions just posterior to the forelimb and hindlimb buds (Fig. 4D).
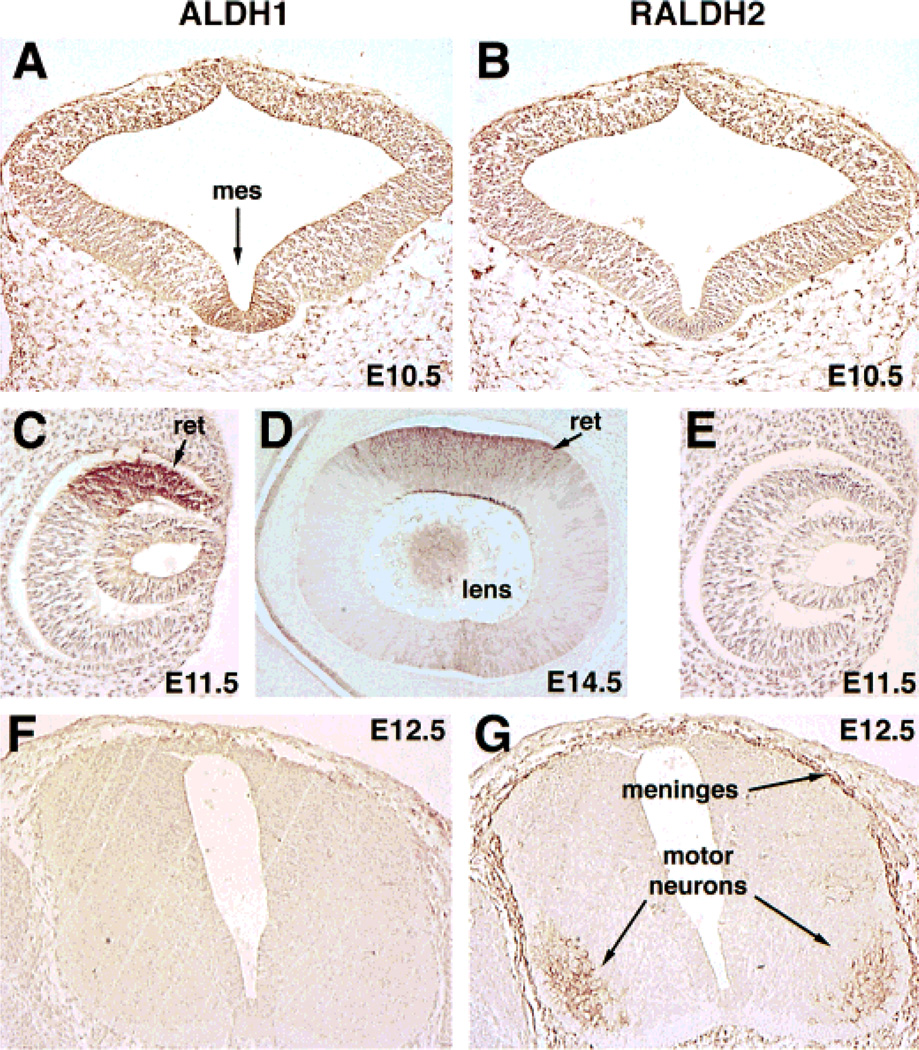
Fig. 4.
Localization of ALDH1 and RALDH2 proteins during stages E8.5–E10.5 of mouse embryogenesis. Embryos were subjected to whole-mount immunohistochemistry and prior to photography were cleared in benzyl alcohol:benzyl benzoate (1:2). Due to the transparent nature of the cleared embryos, both portions of bilateral structures are observed simultaneously. al, allantoic mesenchyme; c, coelomic cavity mesenchyme; cl, cloaca; dr, dorsal retina; mes, ventral flexure of mesencephalon; mg, midgut; mn, mesonephric ducts; op, optic vesicle/frontonasal mass; otic, otic vesicle; som, somite; thy, thymic primordia.
RALDH2 localization was for the most part nonoverlapping with that of ALDH1, occurring primarily in trunk tissues. RALDH2 protein was detected at E7.5–E8.0 in paraxial mesoderm up to the base of the headfolds (data not shown). At E8.5–E9.5, RALDH2 protein was detected in the developing somites, paraxial mesoderm, coelomic cavity mesenchyme adjacent to the heart, and allantoic mesenchyme, with transient detection in the ventral optic vesicle and surrounding frontonasal region (Fig. 4E–G). By E10.5, RALDH2 localization was observed in several trunk tissues, including somites, coelomic cavity mesenchyme, pericardium, cloaca, midgut, and mesonephric ducts (Fig. 4H). At E10.5 there was no longer an RALDH2 signal in the optic vesicles or frontonasal region (Fig. 4H).
Localization of ALDH1 and RALDH2 Proteins in Embryonic Neural Tissues
Immunohistochemical analyses of sections indicated that ALDH1 protein was clearly present in the midbrain localized to the ventral mesencephalon by E9.0 (data not shown) and continued to be localized there at E10.5, whereas RALDH2 protein was absent from this location (Fig. 5A,B). Neither ALDH1 nor RALDH2 were expressed in the forebrain, hindbrain, or dorsal midbrain through stage E10.5 (Fig. 4). Sections also demonstrated an abundance of ALDH1 protein in the dorsal retina at E11.5 (Fig. 5C) and in the lens plus the dorsal retina at E14.5 (Fig. 5D), but RALDH2 was not detected in either of these tissues (Fig. 5E, data not shown).ALDH1 protein was also localized in the medial aspect of the otic vesicles by E11.5, whereas RALDH2 was not detected in this tissue (Fig. 6A, B).
Fig. 5.
Immunohistochemical localization of ALDH1 and RALDH2 proteins in neural tissues. In the mesencephalic ventral flexure (mes) immunohistochemical analyses of E10.5 embryos sectioned coronally shows the presence of ALDH1 (A) but not RALDH2 (B). In coronal sections through the eye, ALDH1 immunodetection occurs in the dorsal retina (ret) at E11.5 (C) and in both dorsal retina and the lens at E14.5 (D), with no detection of RALDH2 (E). In transverse sections through the brachial spinal cord at E12.5, ALDH1 protein is absent (F), but RALDH2 protein is detected in the motor neurons and meninges (G).
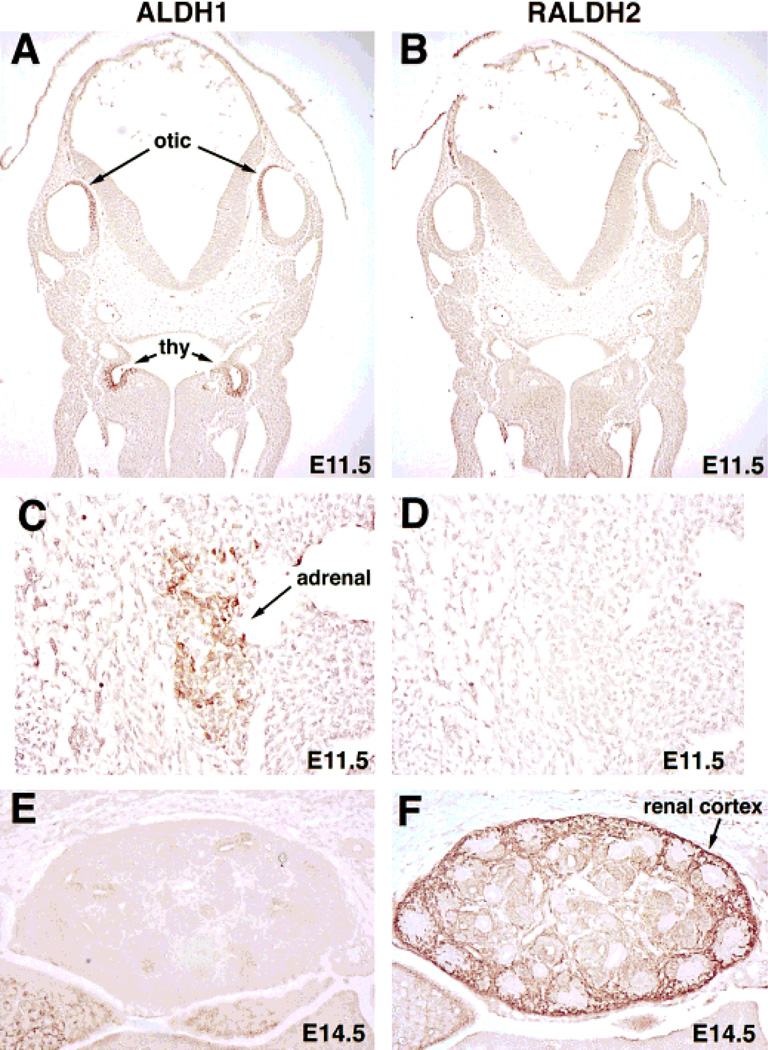
Fig. 6.
Organ-specific localization of ALDH1 and RALDH2 proteins revealed by immunohistochemistry. Analyses of E11.5 embryos sectioned coronally demonstrates ALDH1 protein in the medial portions of the otic vesicles and bilaterally in the thymic primordia (thy) (A), whereas RALDH2 is absent in these tissues (B). In sagittal sections of the E11.5 adrenal blastema, ALDH1 protein is detected (C), but not RALDH2 protein (D). At E14.5, sagittal sections through the developing kidney shows that ALDH1 protein is absent (E), but that RALDH2 protein is abundant and localized primarily in the renal cortex (F).
Although RALDH2 protein was not detected in cranial neural tissue following its disappearance from the optic vesicles at E9.5, nor in the neural tube up to E10.5, it was detected in trunk neural tissue later in development, particularly the spinal cord meninges and the motor neurons innervating the forelimbs at E12.5 (Fig. 5G). RALDH2 expression within the spinal cord motor neurons was limited to only the brachial and lumbar regions adjacent to the developing forelimbs and hindlimbs, respectively (data not shown). ALDH1 protein was not detected in the spinal cord (Fig. 5F).
ALDH1 and RALDH2 Protein Localization in Embryonic Thymus, Adrenal, and Metanephros
ALDH1, but not RALDH2, was detected in the thymic primordia at E9.5 (Fig. 4C) and was more abundant in this tissue at E10.5 (Fig. 4D). Serial sections provided evidence of ALDH1 protein localization bilaterally in the developing thymic primordia (third branchial pouches) at E11.5, with no detection of RALDH2 (Fig. 6A, B). Also, ALDH1 protein was localized to the adrenal blastemas at E11.5, the stage when this organ first develops, with RALDH2 once again being absent (Fig. 6C, D).
As described above, both ALDH1 and RALDH2 were found in the mesonephros by E10.5, suggesting a role in early genitourinary tract development. Examination of the metanephros (embryonic kidney) at E14.5 demonstrated RALDH2 protein in the developing renal cortex, with ALDH1 being nearly undetectable (Fig. 6E,F).
DISCUSSION
Regulated production of the ligand RA is undoubtedly a key event controlling the function of retinoid receptors during vertebrate development. Differences in the rate of RA synthesis from one embryonic tissue to another is likely to contribute to the establishment of RA gradients that affect the overall control of development by retinoids. Although it is clear that some embryonic tissues contain more RA than others, it is less clear how these differences are generated. Here we have explored the role of the aldehyde dehydrogenase gene family in embryonic RA synthesis. We found that mouse Aldh1 and Raldh2 both stimulate both stimulate RA synthesis when expressed in Xenopus embryos, with Raldh2 being more efficient than Aldh1. The distinct localization patterns of ALDH1 and RALDH2 proteins in mouse embryos and adult tissues suggest that these two RA synthetic enzymes initially establish differential cranial and trunk RA synthesis and function in distinct retinoid signaling events during development.
Aldh1 and Raldh2 Both Function in Embryonic RA Synthesis
We tested whether mouse Aldh1 and Raldh2, as well as human ALDH3, could stimulate RA synthesis in vivo by ectopic expression in Xenopus embryos. Using the RA bioassay described here, endogenous RA is undetectable in Xenopus embryos from fertilization through blastula stage 8, thus providing a large time window during which embryos can be injected with ALDH mRNAs and examined for effects on RA synthesis with essentially no background detection. This assay further relies on the fact that Xenopus eggs and embryos contain large amounts of the vitamin A substrate retinal, the immediate precursor of RA [Plack and Kon, 1961; Creech Kraft et al., 1994; Blumberg et al., 1996]. Based on our findings, we conclude that expression of either mouse Aldh1 or Raldh2 stimulates RA synthesis in this in vivo assay, but that expression of human ALDH3 does not lead to this result. These observations demonstrate that the enzymes encoded by Aldh1 and Raldh2 can catalyze RA synthesis in Xenopus embryos using endogenous concentrations of substrate, coenzyme, and any other factors which may effect enzyme activity.
Our assays suggest that RALDH2 is more active than ALDH1 as a RA synthetic enzyme in Xenopus embryos. In vitro enzyme assays indicate that rat RALDH2 has a catalytic efficiency approximately 15-fold higher than that of rat ALDH1 [Wang et al., 1996; Penzes et al., 1997]. Thus, both physiological and biochemical analyses of ALDH1 and RALDH2 provide evidence that these enzymes indeed function as RA synthetic enzymes, with RALDH2 having higher activity. The lack of RA synthetic activity by human ALDH3 matches previous findings indicating that this enzyme lacks in vitro activity for oxidation of retinal to RA [Yoshida et al., 1992]. This demonstrates that not all ALDHs are designed to function as RA synthetic enzymes.
Localization of ALDH1 and RALDH2 Proteins Relative to RA Distribution in Early Embryos
A posterior preference for initial RA synthesis during mouse embryogenesis has previously been reported [Hogan et al., 1992], and may function to differentially regulate Hox genes, which help establish the anteroposterior axis during early development [Lufkin, 1996; Shimeld, 1996]. Previous studies on mouse embryos have shown that RA is initially present only in the trunk at E7.5–E8.0; from E8.5–E10.5 there is still a high level of RA in the trunk, but RA is now detectable in the optic vesicles and surrounding frontonasal region [Rossant et al., 1991; Ang et al., 1996a]. Here we found that the more active RA synthetic enzyme RALDH2 is initially localized in the trunk at E7.5–E8.0 and continues to be highly localized in trunk tissues from E8.5–E10.5 except transiently in the ventral optic vesicle and surrounding frontonasal region from E8.5–E9.5. Previous studies have shown that Raldh2 mRNA is also localized primarily in trunk tissues from E7.5–E10.5, with transient expression in the optic vesicle region [Niederreither et al., 1997]. In contrast, the less active RA synthetic enzyme ALDH1 is initially localized in cranial tissue (ventral mesencephalon) at E9.0 and then in several cranial tissues from E9.5–E10.5, including ventral mesencephalon, dorsal retina, otic vesicles, and thymic primordia. We previously reported that mouse Aldh1 mRNA is detectable from E7.5 onwards [Ang and Duester, 1997], but since our present results show that ALDH1 protein is not detectable until E9.0, this enzyme is unlikely to contribute to RA synthesis until E9.0.
The expression patterns of the ALDH genes examined here are consistent with RALDH2 functioning to produce the RA seen in the trunk and frontonasal region, and with both RALDH2 and ALDH1 contributing to the RA observed in the optic vesicles. Indeed, Raldh2 −/− knockout mouse embryos at late E8.5 (equivalent to our E9.0) exhibit a total elimination of RA detection in the trunk and frontonasal region, but some RA is still detectable in the optic vesicles [Niederreither et al., 1999], perhaps produced by ALDH1. In addition, we suggest that ALDH1 may produce RA in several other cranial derivatives where this enzyme is detected, but where RA has not previously been detected, as discussed below.
Initial Localization of ALDH1 Is Primarily in Embryonic Cranial Derivatives
Previous studies indicated that embryonic dorsal retina (from E9.5 onwards) is a major site of protein accumulation for ALDH1 (formerly called AHD2) during mouse embryogenesis [McCaffery et al., 1991]. There is convincing evidence that ALDH1 contributes to RA synthesis during retina development and in the adult retina [McCaffery et al., 1991, 1993, 1999]. In those studies, the antibody used to localize ALDH1 was prepared against a phenobarbitol-inducible rat cytosolic ALDH [Lindahl and Evces, 1984] which is now known to be distinct from ALDH1, sharing 90% identity [Hsu et al., 1999]. Evidently there was some cross-reaction of this rat ALDH-PB antibody with mouse ALDH1 in the dorsal retina, which contains particularly high levels of this enzyme, but other sites of ALDH1 localization may have been missed with this antibody. The mouse ALDH1 antibody we have described here does indeed detect ALDH1 in the dorsal retina as well as the lens, but our studies indicate that ALDH1 protein is also localized in several other cranial structures outside the eye not previously reported. Also, the mouse ALDH1 antibody described here does not cross-react with mouse ALDH-PB as described above.
We found localization of ALDH1 in the ventral aspect of the mesencephalic flexure from E9.0–E10.5. This finding suggests that the ventral mesencephalon may be a site of RA synthesis during brain development. However, analyses of transgenic RA reporter mouse embryos have not identified RA detection in the ventral mesencephalon [Rossant et al., 1991; Balkan et al., 1992]. If RA is indeed present in this tissue it may exist at a level too low to detect in these reporter mice or the reporter gene may be nonresponsive in the mesencephalon. Coinciding with the initial localization ofALDH1 at E9.0 in the ventral mesencephalon, this tissue initiates midbrain dopaminergic neuron development [Perrone-Capano and di Porzio, 1996], suggesting that ALDH1 may be functioning in this event. RA as well as retinoid receptors have been proposed to participate in dopamine signaling during late embryonic development and adulthood [Krezel et al., 1998; Valdenaire et al., 1998], and ALDH1 protein has been localized in axons and terminals of the mesostriatal and mesolimbic dopamine systems during late embryonic development and adulthood [McCaffery and Dräger, 1994a]. Interestingly, we previously reported that mouse Adh4 (encoding a retinol dehydrogenase able to convert retinol to retinal) is also expressed in the ventral aspect of the mesencephalic flexure from E9.0–E10.5 [Haselbeck and Duester, 1998b]. This suggests that ADH4 and ALDH1 may cooperate to locally convert retinol to RA in the ventral mesencephalon at the time when midbrain dopaminergic neurons first develop, but as of yet a direct demonstration of the presence of RA in this location is lacking.
The otic vesicles were found to contain ALDH1 protein at stages E10.5–E11.5. Expression was observed to be asymmetric, with immunodetection occurring only along the medial portion of the vesicle. Adh4 expression is also observed along the medial portions of the otic vesicles [Haselbeck and Duester, 1998b]. Since a role for RA in the development of the inner ear has been proposed [Corey and Breakefield, 1994], it is reasonable to propose that ADH4 and ALDH1 may function to locally convert retinol to RA for otic vesicle differentiation. However, transgenic RA reporter mouse embryos do not have RA detection in the otic vesicles [Rossant et al., 1991; Balkan et al., 1992], suggesting that the RA level may be low or that the reporter gene is nonresponsive in this tissue.
The thymic primordia develop as two symmetrical epithelial elongations in the ventral portion of the third brachial pouch, which then migrate caudally and fuse to form the thymus gland [Smith, 1965; Cordier and Haumont, 1980]. Our detection of ALDH1 protein, but not RALDH2 protein, in the thymic primordia from E9.5–E11.5 suggests that ALDH1 may catalyze local RA synthesis needed for thymus development. As the developing thymic primordia lie close to the boundary with trunk tissues that clearly contain RA, as observed in transgenic RA reporter mouse embryos, it is unclear if the thymic primordia themselves have detectable RA in such reporter embryos [Rossant et al., 1991; Balkan et al., 1992]. However, analyses of RAR null mutant mice have demonstrated the existence of persistent cervical thymus and other thymic abnormalities, indicating that retinoid signaling is required for proper development and caudal migration of the thymus [Mendelsohn et al., 1994].
Initial Localization of RALDH2 Is Primarily in Embryonic Trunk Tissues
Our immunohistochemical observations of RALDH2 protein localization in mouse embryos from E7.5–E10.5 correlate well for the most part with the results found previously for Raldh2 mRNA expression by in situ hybridization [Hiederreither et al., 1997]. From E7.5–E10.5, RALDH2 protein is quite widespread in many trunk tissues, such as paraxial mesoderm, somites, pericardium, coelomic cavity mesenchyme, and mesonephros and is absent from cranial tissues with the exception of transient expression in the ventral optic vesicles and frontonasal region. Examination of Raldh2 −/− mice has demonstrated that RALDH2 is in fact necessary for development of many trunk structures, including somites, heart, limb buds, and mesonephric ducts, but cranial defects are also seen, including truncation of the frontonasal region and reduction in size of the otic vesicles [Niederreither et al., 1999]. RALDH2 protein localization in the frontonasal region from E8.5–E9.5 could account for its involvement in RA production for frontonasal development. Raldh2 mRNA was reported in the lateral regions of the otic vesicles at E10.5 [Niederreither et al., 1997], which could account for local RA synthesis, but we observed no RALDH2 protein localization in the otic vesicles by this stage.
A further indication that RALDH2 functions to produce RA for trunk tissues comes from studies on the spinal cord where RALDH2, but not ALDH1, is localized. At E12.5 we found that RALDH2 protein localizes in the motor neurons which innervate the forelimbs and hindlimbs, but not in other regions of the spinal cord. Previous studies have shown that Raldh2 mRNA is highly localized by E12.5 of mouse development in a subpopulation of motor neurons innervating the limbs [Zhao et al., 1996], a pattern which is consistent with higher levels of RA in the brachial and lumbar regions of the embryonic spinal cord [McCaffery and Dràger, 1994b], and higher levels of retinoid-X receptor signaling in these regions [Solomin et al., 1998]. Furthermore, ectopic expression of Raldh2 in the chick spinal cord affects the specification of motor neuron subtype identity [Sockanathan and Jessell, 1998]. Together, these facts provide a compelling case for Raldh2 functioning to provide RA for a unique retinoid signaling event which specifies the identity of spinal cord motor neurons innervating the limbs.
Localization of ALDH1 and RALDH2 in Embryonic Mesonephros, Metanephros, and Adrenal
The high level of RALDH2 protein initially observed in the somites is much reduced by E10.5, but at this stage there is a high level of RALDH2 protein localized along the entire anteroposterior axis of the coelomic cavity and mesonephric ducts. Adh1, encoding a retinol dehydrogenase, is also expressed along the entire anteroposterior axis of the mesonephric ducts at both the mRNA level [Ang et al., 1996a] and the protein level (R.J. Haselbeck and G. Duester, unpublished data), thus potentially setting up a metabolic pathway to convert retinol to RA in the mesonephros. Interestingly, ALDH1 protein is also present in the mesonephric ducts at E10.5, but it is unevenly distributed along the anteroposterior axis such that localization is highest just posterior to the forelimb and hindlimb buds. This mesonephric pattern of ALDH1 localization could help boost the level of RA diffusing into the posterior regions of developing limb buds; however it is clearly not sufficient to support limb development, as Raldh2 knockout mice do not form limb buds [Niederreither et al., 1999].
It is likely that mesonephric localization of both ALDH1 and RALDH2 also provides the RA known to be needed for correct development of mesonephric derivatives of the genitourinary tract, which are sensitive to vitamin A deficiency or RAR null mutations [Wilson et al., 1953; Mendelsohn et al., 1994]. Raldh2 −/− mice lack mesonephric ducts, indicating that RA is needed for their initial development [Niederreither et al., 1999]. Examination of RAR null mutations have shown that the metanephros also does not develop properly when retinoid signaling is disrupted [Mendelsohn et al., 1994]. At E14.5, our studies indicate that RALDH2, but not ALDH1, is abundant in the metanephros. We have previously shown that Adh1 is expressed in the metanephros at E16.5 [Ang et al., 1996b], thus suggesting that ADH1 and RALDH2 may cooperate to convert retinol to RA in this organ.
Previous immunohistochemical studies have shown that ADH1 and ADH4 (both retinol dehydrogenases) localize to the adrenal blastema is it first develops [Haselbeck and Duester, 1998a], and that RA is detectable in isolated adrenal glands at stage E16.5 [Haselbeck et al., 1997]. Here we demonstrate that ALDH1 protein also localizes to the adrenal blastema, thus providing evidence that enzymes needed for both steps of the metabolic pathway converting retinol to RA exist in the adrenal gland.
Unique Functions for Aldh1 and Raldh2 in Retinoid Signaling
The bewildering array of retinoid signaling events which have been described over the years require intricately regulated pathways to supply RA at just the right time, place, and amount. The different efficiencies with which Aldh1 and Raldh2 are able to stimulate RA synthesis in Xenopus embryos, as well as their unique expression patterns in mouse embryos, suggests that these genes control distinct retinoid signaling pathways. Whereas Raldh2 undoubtedly functions as the predominant regulator of RA synthesis in midgestation mouse embryos, our findings suggest that Aldh1 is also likely to regulate embryonic RA synthesis in several discrete locations requiring low levels of RA. Also, the widespread occurrence of ALDH1, but not RALDH2, proteins in adult mouse organs argues that Aldh1 may function as the predominant regulator of RA synthesis in the adult organism.
ACKNOWLEDGMENTS
We thank L. Hsu and A. Yoshida for the human ALDH3 cDNA, M. Wagner for the F9-RARE-lacZ reporter cell line, and H.L. Ang, L. Deltour, and M. Foglio for critical discussions.
Contract grant sponsor: NIH; Contract grant number: AA07261.
REFERENCES
- Ang HL, Duester G. Initiation of retinoid signaling in primitive streak mouse embryos: spatiotemporal expression patterns of receptors and metabolic enzymes for ligand synthesis. Dev Dyn. 1997;208:536–543. doi: 10.1002/(SICI)1097-0177(199704)208:4<536::AID-AJA9>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Ang HL, Deltour L, Hayamizu TF, Zgombic-Knight M, Duester G. Retinoic acid synthesis in mouse embryos during gastrulation and craniofacial development linked to class IV alcohol dehydrogenase gene expression. J Biol Chem. 1996a;271:9526–9534. doi: 10.1074/jbc.271.16.9526. [DOI] [PubMed] [Google Scholar]
- Ang HL, Deltour L, Zgombic-Knight M, Wagner MA, Duester G. Expression patterns of class I and class IV alcohol dehydrogenase genes in developing epithelial suggest a role for alcohol dehydrogenase in local retinoic acid synthesis. Alcohol Clin Exp Res. 1996b;20:1050–1064. doi: 10.1111/j.1530-0277.1996.tb01946.x. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. Vol. 2. New York: John Wiley & Sons; 1989. [Google Scholar]
- Balkan W, Colbert M, Bock C, Linney E. Transgenic indicator mice for studying activated retinoic acid receptors during development. Proc Natl Acad Sci USA. 1992;89:3347–3351. doi: 10.1073/pnas.89.8.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg B, Bolado J, Jr, Derguini F, Craig AG, Moreno TA, Chakravarti D, Heyman RA, Buck J, Evans RM. Novel retinoic acid receptor ligands Xenopus embryos. Proc Natl Acad Sci USA. 1996;93:4873–4878. doi: 10.1073/pnas.93.10.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cordier AC, Haumont SM. Development of thymus, parathyroids, and ultimo-branchial bodies in NMRI and nude mice. Am J Anat. 1980;157:227–263. doi: 10.1002/aja.1001570303. [DOI] [PubMed] [Google Scholar]
- Corey DP, Breakefield XO. Transcription factors in inner ear development. Proc Natl Acad Sci USA. 1994;91:433–436. doi: 10.1073/pnas.91.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creech Kraft J, Schuh T, Juchau MR, Kimelman D. Temporal distribution, localization and metabolism of all-trans-retinol, didehydroretinol and all-trans-retinal during Xenopus development. Biochemistry J. 1994;301:111–119. [PMC free article] [PubMed] [Google Scholar]
- Deltour L, Foglio MH, Duester G. Metabolic deficiencies in alcohol dehydrogenase Adh1, Adh3, and Adh4 null mutant mice: overlapping roles of Adh1 and Adh4 in ethanol clearance and metabolism of retinol to retinoic acid. J Biol Chem. 1999a doi: 10.1074/jbc.274.24.16796. (in press). [DOI] [PubMed] [Google Scholar]
- Deltour L, Foglio MH, Duester G. Impaired retinol utilization in Adh4 alcohol dehydrogenase mutant mice. Dev Genet. 1999b;25:1–10. doi: 10.1002/(SICI)1520-6408(1999)25:1<1::AID-DVG1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Dickman ED, Thaller C, Smith SM. Temporally-regulated retinoic acid depletion produces specific neural crest, ocular and nervous system defects. Development. 1997;124:3111–3121. doi: 10.1242/dev.124.16.3111. [DOI] [PubMed] [Google Scholar]
- Dockham PA, Lee M-O, Sladek NE. Identification of human liver aldehyde dehydrogenases that catalyze the oxidation of aldophosphamide and retinaldehyde. Biochem Pharmacol. 1992;43:2453–2469. doi: 10.1016/0006-2952(92)90326-e. [DOI] [PubMed] [Google Scholar]
- Domingo A, Sarria AJ, Evans RM, Klymkowsky MW. Studying intermediate filaments. In: Carraway KL, Carraway CAC, editors. The cytoskeleton: a practical approach. Oxford: IRL Press; 1992. pp. 223–255. [Google Scholar]
- Duester G. Involvement of alcohol dehydrogenase, short-chain dehydrogenase/reductase, aldehyde dehydrogenase, and cytochrome P450 in the control of retinoid signaling by activation of retinoic acid synthesis. Biochemistry. 1996;35:12221–12227. doi: 10.1021/bi961176+. [DOI] [PubMed] [Google Scholar]
- Futterman S. Enzymatic oxidation of vitamin A aldehyde to vitamin A acid. J Biol Chem. 1962;237:677–680. [PubMed] [Google Scholar]
- Gudas LJ, Sporn MB, Roberts AB. Cellular biology and biochemistry of the retinoids. In: Sporn MB, Roberts AB, Goodman DS, editors. The retinoids: biology, chemistry, and medicine. 2nd ed. New York: Raven Press; 1994. pp. 443–520. [Google Scholar]
- Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Haselbeck RJ, Duester G. Regional restriction of alcohol/retinol dehydrogenases along the mouse gastrointestinal epithelium. Alcohol Clin Exp Res. 1997;21:1484–1490. [PubMed] [Google Scholar]
- Haselbeck RJ, Duester G. ADH1 and ADH4 alcohol/retinol dehydrogenases in the developing adrenal blastema provide evidence for embryonic retinoid endocrine function. Dev Dyn. 1998a;213:114–120. doi: 10.1002/(SICI)1097-0177(199809)213:1<114::AID-AJA11>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Haselbeck RJ, Duester G. ADH4-lacZ transgenic mouse reveals alcohol dehydrogenase localization in embryonic midbrain/hindbrain, otic vesicles, and mesencephalic, trigeminal, facial, and olfactory neural crest. Alcohol Clin Exp Res. 1998b;22:1607–1613. [PubMed] [Google Scholar]
- Haselbeck RJ, Ang HL, Deltour L, Duester G. Retinoic acid and alcohol/retinol dehydrogenase in the mouse adrenal gland: a potential endocrine source of retinoic acid during development. Endocrinology. 1997;138:3035–3041. doi: 10.1210/endo.138.7.5274. [DOI] [PubMed] [Google Scholar]
- Hofmann C, Eichele G. Retinoids in development. In: Sporn MB, Roberts AB, Goodman DS, editors. The retinoids: biology, chemistry, and medicine. 2nd Ed. New York: Raven Press; 1994. pp. 387–441. [Google Scholar]
- Hogan BLM, Thaller C, Eichele G. Evidence that Hensen’s node is a site of retinoic acid synthesis. Nature. 1992;359:237–241. doi: 10.1038/359237a0. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the mouse embryo. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Hsu LC, Chang W-C, Shibuya A, Yoshida A. Human stomach aldehyde dehydrogenase cDNA and genomic cloning, primary structure, and expression in Escherichia coli. J Biol Chem. 1992;267:3030–3037. [PubMed] [Google Scholar]
- Hsu LC, Chang WC, Hoffmann I, Duester G. Molecular analysis of two closely related mouse aldehyde dehydrogenase genes: identification of a role for Aldh1, but not Aldh-pb, in the biosynthesis of retinoic acid. Biochem J. 1999;339:387–395. [PMC free article] [PubMed] [Google Scholar]
- Kastner P, Chambon P, Leid M. Role of nuclear retinoic acid receptors in the regulation of gene expression. In: Blomhoff R, editor. Vitamin A in health and disease. New York: Marcel Dekker; 1994. pp. 189–238. [Google Scholar]
- Kathmann EC, Lipsky JJ. Cloning of a cDNA encoding a constitutively expressed rat liver cytosolic aldehyde dehydrogenase. Biochem Biophys Res Commun. 1997;236:527–531. doi: 10.1006/bbrc.1997.6998. [DOI] [PubMed] [Google Scholar]
- Kaufman MH. The atlas of mouse development. San Diego: Academic Press; 1992. [Google Scholar]
- Kay BK. Injection of oocytes and embryos. In: Kay BK, Peng HB, editors. Methods in cell biology. Vol. 36. San Diego: Academic Press; 1991. pp. 663–669. Xenopus laevis: practical uses in cell and molecular biology. [PubMed] [Google Scholar]
- Krezel W, Ghyselinck N, Samad TA, Dupé V, Kastner P, Borrelli E, Chambon P. Impaired locomotion and dopamine signaling in retinoid receptor mutant mice. Science. 1998;279:863–867. doi: 10.1126/science.279.5352.863. [DOI] [PubMed] [Google Scholar]
- Labrecque J, Dumas F, Lacroix A, Bhat PV. Anovel isoenzyme of aldehyde dehydrogenase specifically involved in the biosynthesis of 9-cis and all-trans retinoic acid. Biochem J. 1995;305:681–684. doi: 10.1042/bj3050681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M-O, Manthey CL, Sladek NE. Identification of mouse liver aldehyde dehydrogenases that catalyze the oxidation of retinaldehyde to retinoic acid. Biochem Pharmacol. 1991;42:1279–1285. doi: 10.1016/0006-2952(91)90266-8. [DOI] [PubMed] [Google Scholar]
- Lindahl R, Evces S. Rat liver aldehyde dehydrogenase. II. Isolation and characterization of four inducible isozymes. J Biol Chem. 1984;259:11991–11996. [PubMed] [Google Scholar]
- Lohnes D, Mark M, Mendelsohn C, Dollé P, Dierich A, Gorry P, Gansmuller A, Chambon P. Function of the retinoic acid receptors (RARs) during development. I. Craniofacial and skeletal abnormalities in RAR double mutants. Development. 1994;120:2723–2748. doi: 10.1242/dev.120.10.2723. [DOI] [PubMed] [Google Scholar]
- López-Fernández LA, Del Mazo J. The cytosolic aldehyde dehydrogenase gene (Aldh1) is developmentally expressed in Leydig cells. FEBS Lett. 1997;407:225–229. doi: 10.1016/s0014-5793(97)00352-9. [DOI] [PubMed] [Google Scholar]
- Lufkin T. Transcriptional control of Hoxgenes in the vertebrate nervous system. Curr Opin Genet Dev. 1996;6:575–580. doi: 10.1016/s0959-437x(96)80086-4. [DOI] [PubMed] [Google Scholar]
- Luo JM, Sucov HM, Bader JA, Evans RM, Giguère V. Compound mutants for retinoic acid receptors (RAR) β and RARα1 reveal developmental functions for multiple RARβ isoforms. Mech Dev. 1996;55:33–44. doi: 10.1016/0925-4773(95)00488-2. [DOI] [PubMed] [Google Scholar]
- Maden M. Role of retinoids in embryonic development. In: Blomhoff R, editor. Vitamin A in health and disease. New York: Marcel Dekker; 1994. pp. 289–322. [Google Scholar]
- Maden M, Gale E, Kostetskii I, Zile MH. Vitamin A-deficient quail embryos have half a hindbrain and other neural defects. Curr Biol. 1996;6:417–426. doi: 10.1016/s0960-9822(02)00509-2. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Umesono K, Evans RM. The retinoid receptors. In: Sporn MB, Roberts AB, Goodman DS, editors. The retinoids: biology, chemistry, and medicine. 2nd ed. New York: Raven Press; 1994. pp. 319–349. [Google Scholar]
- Mascrez B, Mark M, Dierich A, Ghyselinck NB, Kastner P, Chambon P. The RXRα ligand-dependent activation function 2 (AF-2) is important for mouse development. Development. 1998;125:4691–4707. doi: 10.1242/dev.125.23.4691. [DOI] [PubMed] [Google Scholar]
- McCaffery P, Dräger UC. High levels of a retinoic acid-generating dehydrogenase in the meso-telencephalic dopamine system. Proc Natl Acad Sci USA. 1994a;91:7772–7776. doi: 10.1073/pnas.91.16.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffery P, Dräger UC. Hot spots of retinoic acid synthesis in the developing spinal cord. Proc Natl Acad Sci USA. 1994b;91:7194–7197. doi: 10.1073/pnas.91.15.7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffery P, Tempst P, Lara G, Dräger UC. Aldehyde dehydrogenase is a positional marker in the retina. Development. 1991;112:693–702. doi: 10.1242/dev.112.3.693. [DOI] [PubMed] [Google Scholar]
- McCaffery P, Lee M-O, Wagner MA, Sladek NE, Dräger UC. Asymmetrical retinoic acid synthesis in the dorsoventral axis of the retina. Development. 1992;115:371–382. doi: 10.1242/dev.115.2.371. [DOI] [PubMed] [Google Scholar]
- McCaffery P, Posch KC, Napoli JL, Gudas L, Dräger UC. Changing patterns of the retinoic acid system in the developing retina. Dev Biol. 1993;158:390–399. doi: 10.1006/dbio.1993.1197. [DOI] [PubMed] [Google Scholar]
- McCaffery P, Wagner E, O’Neil J, Petkovich M, Dräger UC. Dorsal and ventral retinal territories defined by retinoic acid synthesis, break-down and nuclear receptor expression. Mech Dev. 1999;82:119–130. doi: 10.1016/s0925-4773(99)00022-2. [DOI] [PubMed] [Google Scholar]
- Mendelsohn C, Lohnes D, Dáecimo D, Lufkin T, LeMeur M, Chambon P, Mark M. Function of the retinoic acid receptors (RARs) during development. II. Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development. 1994;120:2749–2771. doi: 10.1242/dev.120.10.2749. [DOI] [PubMed] [Google Scholar]
- Moss JB, Xavier-Neto J, Shapiro MD, Nayeem SM, McCaffery P, Dräger UC, Rosenthal N. Dynamic patterns of retinoic acid synthesis and response in the developing mammalian heart. Dev Biol. 1998;199:55–71. doi: 10.1006/dbio.1998.8911. [DOI] [PubMed] [Google Scholar]
- Napoli JL. Retinoic acid biosynthesis and metabolism. FASEB J. 1996;10:993–1001. doi: 10.1096/fasebj.10.9.8801182. [DOI] [PubMed] [Google Scholar]
- Niederreither K, McCaffery P, Dräger UC, Chambon P, Dollé P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech Dev. 1997;62:67–78. doi: 10.1016/s0925-4773(96)00653-3. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Subbarayan V, Dollé P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) New York: Garland Publishing; 1994. [Google Scholar]
- Ono Y, Fukuhara N, Yoshie O. TAL1 and LIM-only proteins synergistically induce retinaldehyde dehydrogenase 2 expression in T-cell acute lymphoblastic leukemia by acting as cofactors for GATA3. Mol Cell Biol. 1998;18:6939–6950. doi: 10.1128/mcb.18.12.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Wang XS, Napoli JL. Enzymatic characteristics of retinal dehydrogenase type I expressed in Escherichia coli. Biochim Biophys Acta Protein Struct Mol Enzymol. 1997;1342:175–181. doi: 10.1016/s0167-4838(97)00102-7. [DOI] [PubMed] [Google Scholar]
- Perrone-Capano C, di Porzio U. Epigenetic factors and midbrain dopaminergic neurone development. BioEssays. 1996;18:817–824. doi: 10.1002/bies.950181008. [DOI] [PubMed] [Google Scholar]
- Plack PA, Kon SK. A comparative survey of the distribution of vitamin A aldehyde in eggs. Biochem J. 1961;81:561–570. doi: 10.1042/bj0810561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Zirngibl R, Cado D, Shago M, Giguère V. Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 1991;5:1333–1344. doi: 10.1101/gad.5.8.1333. [DOI] [PubMed] [Google Scholar]
- Rout UK, Holmes RS. Isoelectric focusing studies of aldehyde dehydrogenases from mouse tissues: variant phenotypes of liver, stomach and testis isozymes. Comp Biochem Physiol [B] 1985;81:647–651. doi: 10.1016/0305-0491(85)90380-3. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shimeld SM. Retinoic acid, HOX genes and the anterior-posterior axis in chordates. BioEssays. 1996;18:613–616. [Google Scholar]
- Smith C. Studies on the thymus of the mammal. XIV. Histology and histochemistry of embryonic and early postnatal thymuses of C57BL/6 and AKR strain mice. Am J Anat. 1965;116:611–630. doi: 10.1002/aja.1001160307. [DOI] [PubMed] [Google Scholar]
- Sockanathan S, Jessell TM. Motor neuron-derived retinoid signaling specifies the subtype identity of spinal motor neurons. Cell. 1998;94:503–514. doi: 10.1016/s0092-8674(00)81591-3. [DOI] [PubMed] [Google Scholar]
- Solomin L, Johansson CB, Zetterström RH, Bissonnette RP, Heyman RA, Olson L, Lendahl U, Frisén J, Perlmann T. Retinoid-X receptor signalling in the developing spinal cord. Nature. 1998;395:398–402. doi: 10.1038/26515. [DOI] [PubMed] [Google Scholar]
- Valdenaire O, Maus-Moatti M, Vincent JD, Mallet J, Vernier P. Retinoic acid regulates the developmental expression of dopamine D2 receptor in rat striatal primary cultures. J Neurochem. 1998;71:929–936. doi: 10.1046/j.1471-4159.1998.71030929.x. [DOI] [PubMed] [Google Scholar]
- Vize PD, Melton DA, Hemmati-Brivanlou A, Harland RM. Assay for gene function in developing Xenopus embryos. In: Kay BK, Peng HB, editors. Methods in cell biology. Vol. 36. San Diego: Academic Press; 1991. pp. 367–387. Xenopus laevis: practical uses in cell and molecular biology. [DOI] [PubMed] [Google Scholar]
- Wagner M, Han B, Jessell TM. Regional differences in retinoid release from embryonic neural tissue detected by an in vitro reporter assay. Development. 1992;116:55–66. doi: 10.1242/dev.116.1.55. [DOI] [PubMed] [Google Scholar]
- Wang XS, Penzes P, Napoli JL. Cloning of a cDNA encoding an aldehyde dehydrogenase and its expression in Escherichia coli. Recognition of retinal as substrate. J Biol Chem. 1996;271:16288–16293. doi: 10.1074/jbc.271.27.16288. [DOI] [PubMed] [Google Scholar]
- Wilson JG, Roth CB, Warkany J. An analysis of the syndrome of malformations induced by maternal vitamin A deficiency. Effects of restoration of vitamin A at various times during gestation. Am J Anat. 1953;92:189–217. doi: 10.1002/aja.1000920202. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Hsu LC, Dave V. Retinal oxidation activity and biological role of human cytosolic aldehyde dehydrogenase. Enzyme. 1992;46:239–244. doi: 10.1159/000468794. [DOI] [PubMed] [Google Scholar]
- Zhao D, McCaffery P, Ivins KJ, Neve RL, Hogan P, Chin WW, Dräger UC. Molecular identification of a major retinoic-acid-synthesizing enzyme, a retinaldehyde-specific dehydrogenase. Eur J Biochem. 1996;240:15–22. doi: 10.1111/j.1432-1033.1996.0015h.x. [DOI] [PubMed] [Google Scholar]