A combination of quantitative live imaging of fluorescently tagged actin, N-cadherin, and myosin in primary neurons and computer modeling of actin dynamics shows that a clutch-like mechanism connecting N-cadherin–based transsynaptic adhesions and the actin/myosin network drives the stabilization of dendritic filopodia into spines.
Abstract
The morphology of neuronal dendritic spines is a critical indicator of synaptic function. It is regulated by several factors, including the intracellular actin/myosin cytoskeleton and transcellular N-cadherin adhesions. To examine the mechanical relationship between these molecular components, we performed quantitative live-imaging experiments in primary hippocampal neurons. We found that actin turnover and structural motility were lower in dendritic spines than in immature filopodia and increased upon expression of a nonadhesive N-cadherin mutant, resulting in an inverse relationship between spine motility and actin enrichment. Furthermore, the pharmacological stimulation of myosin II induced the rearward motion of actin structures in spines, showing that myosin II exerts tension on the actin network. Strikingly, the formation of stable, spine-like structures enriched in actin was induced at contacts between dendritic filopodia and N-cadherin–coated beads or micropatterns. Finally, computer simulations of actin dynamics mimicked various experimental conditions, pointing to the actin flow rate as an important parameter controlling actin enrichment in dendritic spines. Together these data demonstrate that a clutch-like mechanism between N-cadherin adhesions and the actin flow underlies the stabilization of dendritic filopodia into mature spines, a mechanism that may have important implications in synapse initiation, maturation, and plasticity in the developing brain.
INTRODUCTION
In neurons, dendritic spines are micrometer-scale structures protruding from the neurite shaft and forming the postsynaptic components of excitatory synapses. They are believed to originate from motile dendritic filopodia, which enlarge after contacting axons (Ziv and Smith, 1996; Fischer et al., 1998; Korobova and Svitkina, 2010). Spine morphology is a critical indicator of synapse development and function in the brain (Yuste and Bonhoeffer, 2004). For example, spines grow upon synaptic potentiation, shrink upon synaptic depression, and may exhibit abnormal shapes in disease-related conditions (Comery et al., 1997; Colicos et al., 2001; Matsuzaki et al., 2004; Zhou et al., 2004; Cingolani and Goda, 2008). In basal conditions, spines exhibit continuous motility or morphing, characterized by phases of membrane protrusions and retractions (Fischer et al., 1998).
All of these morphological states rely on a dynamic network of actin filaments, whose turnover is controlled by several actin-binding proteins, including capping, nucleating, elongating, and severing proteins, and small Rho/Rac GTPases (Star et al., 2002; Ackermann and Matus, 2003; Okamoto et al., 2004; Tashiro and Yuste, 2004; Schubert et al., 2006; Korobova and Svitkina, 2010; Chazeau et al., 2014). As a result of those various protein regulations, F-actin flows rearward from the tip to the base of the spine head, with a lower velocity than in immature filopodia (Honkura et al., 2008; Tatavarty et al., 2009, 2012; Frost et al., 2010). Spine shape is also influenced by myosins, since, for example, knockdown or pharmacological inhibition of myosin II induces filopodium-like morphologies (Zhang et al., 2005; Ryu et al., 2006; Hodges et al., 2011; Rubio et al., 2011). This raises the interesting possibility that myosin II could contribute to the actin retrograde flow in dendritic spines by pulling on and/or severing F-actin (Tatavarty et al., 2012; Koskinen et al., 2014), as demonstrated in other neuronal motile structures, such as growth cones (Medeiros et al., 2006; Wilson et al., 2010).
Besides intracellular components, dendritic spine shape is also regulated by transsynaptic structures, such as the cadherin/catenin complex (Takeichi and Abe, 2005; Arikkath and Reichardt, 2008). Perturbing N-cadherin adhesions in hippocampal neurons enhances spine motility and reduces spine length (Mysore et al., 2007), whereas down-regulating N-cadherin or β-catenin levels induces thin, filopodium-like spines (Togashi et al., 2002; Okuda et al., 2007; Mendez et al., 2010). Similarly, knockdown of p120-catenin reduces spine and synapse densities (Elia et al., 2006). Furthermore, in the absence of αN-catenin, neurons have abnormally motile spines and actively protruding filopodia, whereas neurons overexpressing αN-catenin show reduced spine turnover and higher spine density (Abe et al., 2004). The cadherin/catenin complex is also strongly affected by synaptic activation, which inhibits N-cadherin endocytosis, induces the dimerization of synaptic N-cadherin, resulting in protease resistance, and triggers a redistribution of β-catenin from neurite shaft to spine head (Bozdagi et al., 2000; Tanaka et al., 2000; Murase et al., 2002; Schuman and Murase, 2003; Mysore et al., 2007). Finally, spine head expansion induced by synaptic activation is blocked by expression of nonadhesive cadherin mutants (Okamura et al., 2004). Taken together, these studies point to a role for the connection between N-cadherin and the cytoskeleton in morphological spine plasticity.
Despite these advances, the question of whether and how the actin/myosin cytoskeleton and cadherin adhesions mechanically interact to produce spine morphing remains unclear. We previously demonstrated that a molecular clutch between N-cadherin adhesions and the actin flow drives growth cone locomotion (Bard et al., 2008) and suggested that a similar mechanism may control dendritic spine morphology (Giannone et al., 2009). To evaluate this model, we examined actin dynamics in dendritic spines and filopodia of primary hippocampal neurons expressing actin-GFP. We triggered N-cadherin adhesions with N-cadherin–coated beads or micropatterns, challenged the connection between N-cadherin and actin by expressing a dominant-negative N-cadherin mutant, and pharmacologically stimulated myosin II contractility. We further interpreted our experiments with quantitative computer simulations of actin dynamics. Overall we show that engagement of a mechanical connection between transsynaptic N-cadherin adhesions and the actin/myosin network promotes the transition of dendritic filopodia into spines by stabilizing the actin cytoskeleton.
RESULTS
Codistribution of N-cadherin, actin, and myosin in dendritic spines
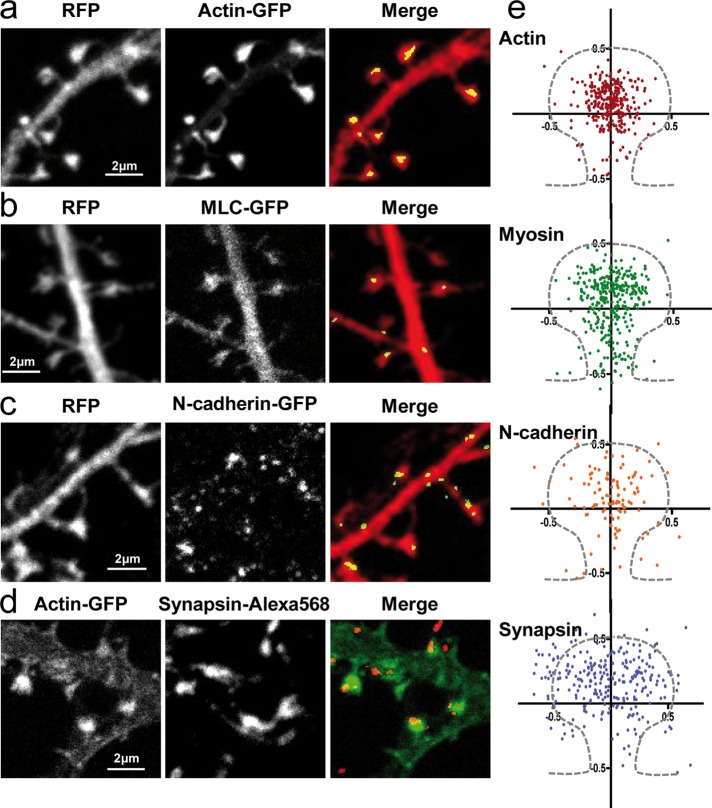
To first determine whether the spatial distribution of N-cadherin, actin, and myosin within dendritic spines reflected potential interactions between them, we examined the relative localization of these proteins in primary hippocampal neurons by confocal microscopy. Neurons were cotransfected with red fluorescent protein (RFP) and actin–green fluorescent protein (GFP), myosin light chain (MLC)–GFP, or N-cadherin–GFP constructs and processed at days in vitro (DIV) 18–21 for dual immunofluorescence labeling of GFP and RFP. The volume marker RFP provided the outline of the spine, whereas the GFP signals indicated subspine localizations of the three proteins. Actin most often distributed as a single large mass at the spine head (Figure 1, a and e), in agreement with high-resolution electron microscopy (EM), photoactivated localization microscopy (PALM), and stimulated emission depletion images (Frost et al., 2010; Korobova and Svitkina, 2010; Izeddin et al., 2011; Urban et al., 2011). Of importance, staining with anti-actin antibodies or phalloidin revealed that actin-GFP represents 15% of the total actin pool in transfected neurons and is able to assemble into native filaments (Supplemental Figure S1), thus serving as a suitable tool for further investigation. Besides actin, MLC was also detectable in spines, typically forming clusters distributed throughout the spine head, similarly to actin, but also present in the spine neck (Figure 1, b and e). N-cadherin was also detected as discrete puncta often localized at the edge of dendritic spines (Figure 1, c and e), reflecting its function in transsynaptic adhesion and consistent with immunogold EM data (Uchida et al., 1996; Korobova and Svitkina, 2010). As a control for these quantifications, we examined the presynaptic marker synapsin, which distributed mostly at the periphery of spines, as expected (Figure 1, d and e). Overall, these data show that actin, myosin, and N-cadherin are codistributed in spines, supporting a potential interaction between transsynaptic N-cadherin adhesions and the actin/myosin network.
FIGURE 1:
Localization of N-cadherin, actin, and myosin expressed in dendritic spines. (a–c) Neurons cotransfected with RFP and actin-GFP, MLC-GFP, or N-cadherin-GFP were fixed at DIV 18–21, dual-immunolabeled for GFP and RFP, and examined by confocal microscopy. Primary anti-DsRed and Alexa 568–conjugated secondary antibodies were used to enhance the RFP signal, whereas primary anti-GFP and ATTO-647–conjugated secondary antibodies were used to enhance the GFP signal. (d) Neurons transfected with actin-GFP were labeled for the presynaptic marker synapsin. (e) As shown in the merge panels, GFP images were segmented to yield the coordinates of actin, MLC, N-cadherin, and synapsin puncta relative to the centroid of the spine obtained from the RFP image. Those data were normalized by the width and length of the spine, providing a plot of the relative distributions of each protein. An average spine profile is indicated by dashes. Some of the synapsin staining that seems to appear within the spine is likely due to the three-dimensional architecture not resolved in these two-dimensional images and the diffraction limits of conventional microscopy. Nonetheless, the distribution of points for synapsin was significantly different from that of the three other proteins (data analyzed by Mann–Whitney test, ***p < 0.005).
A molecular linkage between N-cadherin and actin restrains dendritic spine motility
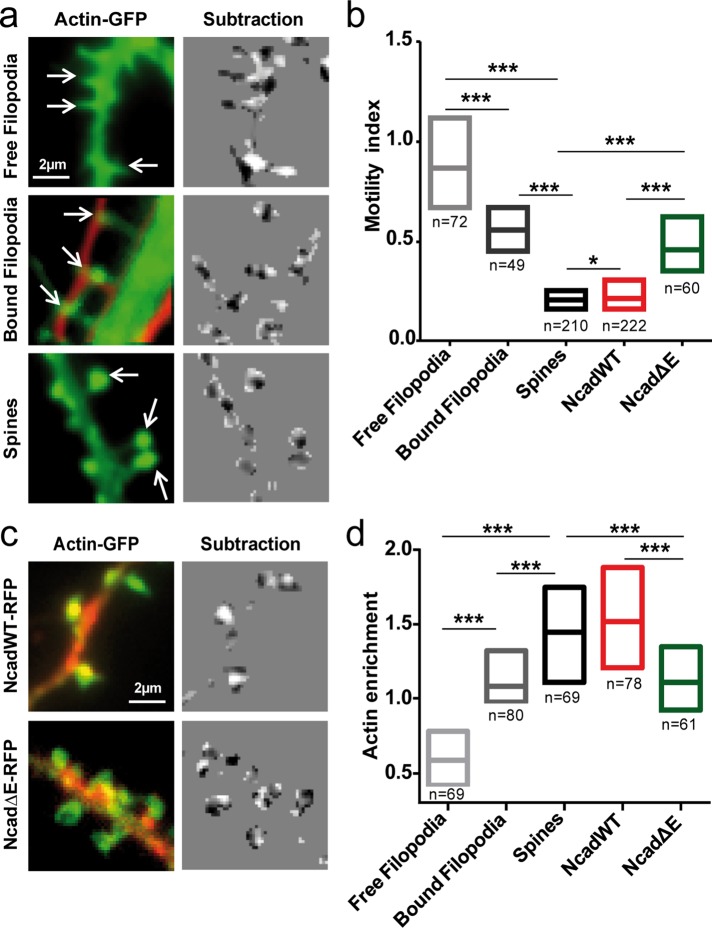
To determine whether dendritic spine dynamics relied on the coupling between F-actin and N-cadherin adhesions, we imaged dendritic filopodia versus mature spines in neurons expressing actin-GFP by time-lapse epifluorescence microscopy (Supplemental Movie 1). A motility index characterizing actin-dependent spine morphing was quantified as the normalized variance of the intensity distribution obtained from two-by-two image subtraction (Ibarretxe et al., 2007; Supplemental Figure S2a). The median motility index of dendritic spines in DIV 21 neurons was 0.21 (Figure 2, a and b). Addition of the actin-capping drug cytochalasin D significantly decreased the motility index to a steady-state value of 0.17 (Supplemental Figure S2, b and c), demonstrating that the observed changes in spine shape were driven by actin dynamics. Fixation of cells with paraformaldehyde (PFA) resulted in a significantly lower motility index of 0.15, a residual value due to image noise at such small length scales. In DIV 8–10 neurons, freely moving dendritic filopodia exhibited a fourfold higher motility index (0.91) than spines (Figure 2, a and b). Furthermore, dendritic filopodia that were contacting axons from cocultured RFP-expressing cells were significantly less motile than free filopodia (motility index, 0.56), showing that transsynaptic axon/dendrite interactions reduce actin-dependent motility. By measuring the alignment of presynaptic synapsin staining and postsynaptic spines, we showed that in young neurons, only 55% of dendritic filopodia were in contact with a presynaptic element, whereas this ratio increased to 85% for dendritic spines of older neurons (Supplemental Figure S3). These results further indicate that the formation of transsynaptic contacts induces filopodial stabilization. Of importance, dendritic filopodia in contact with axons often showed local actin accumulation at their tips, similar to what was observed in mature spines, but not in free filopodia (Figure 2a), suggesting that actin recruitment at contact sites is linked to spine stability. Indeed, quantification of actin levels at the tips of filopodia and spines normalized to shaft levels revealed that actin enrichment was inversely correlated with the motility index (Figure 2d).
FIGURE 2:
Actin-driven motility of dendritic filopodia and spines. (a) Two neuronal populations were electroporated with actin-GFP or RFP and imaged at DIV 8–10 for observation of free dendritic filopodia (top) and bound dendritic filopodia and axons (middle). Independently, neurons were transfected with actin-GFP and imaged at DIV 17–21 for observation of dendritic spines (bottom). Images of actin-GFP (left) were taken every 5 s for 5 min. Consecutive images were segmented and subtracted (right), making protrusions appear white and retractions appear dark. The normalized intensity variance of these images was calculated for each spine or filopodium and defined as the instantaneous motility index. (b) The individual motility index integrated over time is presented as interquartile distributions for filopodia and spines in all conditions. (c) Neurons were cotransfected with actin-GFP and either NcadWT-RFP or NcadΔE-RFP and observed at DIV 17-21. Left, the actin-GFP images (green) were merged to the RFP images (red) to show the distribution of the N-cadherin constructs. Right, corresponding subtracted images of actin-GFP. (d) Interquartile distributions of the motility index for the Ncad constructs. The number of filopodia or spines examined is given under each bar. Data were analyzed by one-way analysis of variance (ANOVA) and compared two by two using Dunnett's posttest. *0.01 < p < 0.05; ***p < 0.001.
To investigate this relationship further, we perturbed the molecular linkage between N-cadherin adhesions and F-actin by cotransfecting mature neurons with an RFP-tagged N-cadherin construct lacking the full extracellular domain (NcadΔE-RFP; Riehl et al., 1996). This deleted cadherin is unable to form homophilic adhesions and thereby acts as a competitor for the binding between endogenous N-cadherin and F-actin by sequestering catenin partners (Togashi et al., 2002; Okamura et al., 2004; Bard et al., 2008). As a positive control, we coexpressed actin-GFP with wild-type N-cadherin (NcadWT-RFP). Dendritic spines from cells expressing NcadΔE displayed a significantly higher motility index (0.46) than cells expressing NcadWT-RFP (0.22) or cells expressing endogenous N-cadherin (0.21) (Figure 2, b and c), indicating that an intact connection between N-cadherin and actin is required to stabilize dendritic spines. Furthermore, neurons expressing NcadΔE showed lower actin enrichment at dendritic spines than neurons expressing endogenous N-cadherin or recombinant NcadWT (Figure 2, c and d), confirming the positive relationship between actin accumulation and spine stability.
Actin turnover is lower in dendritic spines than in filopodia and is dependent on the transsynaptic binding of N-cadherin
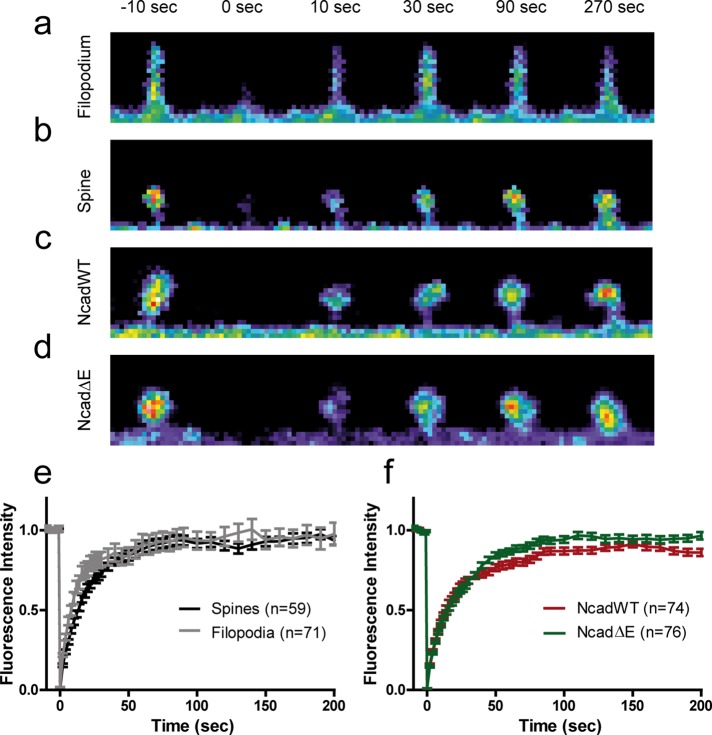
To examine whether the intrinsic actin turnover was different in dendritic filopodia and spines, we performed fluorescence recovery after photobleaching (FRAP) experiments in neurons expressing actin-GFP at DIV 10 and 18 (Figure 3 and Supplemental Movie 2). Actin-GFP fluorescence recovered to 92 and 90% in filopodia and spines, respectively, within ∼100 s (Figure 3, a and b), indicating a small (<10%) stable actin pool, in agreement with previous reports (Star et al., 2002; Honkura et al., 2008; Koskinen et al., 2014). However, the recovery occurred significantly faster in dendritic filopodia than in spines (Figure 3e), consistent with a more dynamic actin network, faster retrograde flow rate (Tatavarty et al., 2012), and higher overall motility (Figure 2, a and b). To assess whether disrupting the link between endogenous N-cadherin and actin affected actin turnover in spines, we compared actin-GFP recovery in neurons coexpressing either NcadWT or NcadΔE. Actin-GFP recovered to significantly higher levels in spines expressing NcadΔE than in those expressing NcadWT (94 vs. 83%, respectively; Figure 3, c, d, and f), suggesting that NcadΔE expression diminishes the stable actin pool. In line with the effects of NcadΔE on spine motility, this result further indicates that the connection of N-cadherin to the actin cytoskeleton stabilizes the actin network in spines.
FIGURE 3:
FRAP experiments on actin-GFP in dendritic filopodia and spines. (a, b) Images of a dendritic filopodium and spine, respectively, in which actin-GFP was photobleached at time 0 and allowed to recover for 300 s. (c, d) Images of dendritic spines coexpressing NcadWT-RFP or NcadΔE-RFP, respectively, in which actin-GFP was photobleached at time 0 and allowed to recover for 200 s. (e) Recovery of actin-GFP fluorescence in dendritic filopodia and spines. The average curves were fitted by a single-exponential function, m[1 − exp(−κt)], where m is the mobile fraction and κ a characteristic rate constant, yielding the values m = 0.92 ± 0.01 and 0.90 ± 0.01 (not significantly different) and κ = 5.5 ± 0.4 and 3.2 ± 0.2 min−1 (p < 0.001, by extra sum of squares F test) for filopodia and spines, respectively. (f) Recovery of actin-GFP fluorescence in dendritic spines from neurons coexpressing NcadWT-RFP or NcadΔE-RFP. Parameters from exponential fits were m = 0.83 ± 0.01 and 0.94 ± 0.01 (p < 0.001), and κ = 3.5 ± 0.1 and 2.5 ± 0.1 min−1 (p < 0.001 by extra sum of squares F test) for NcadWT and NcadΔE, respectively. The number of filopodia/spines examined in each condition is given in the keys.
Myosin II–driven F-actin retraction is altered by N-cadherin mutants
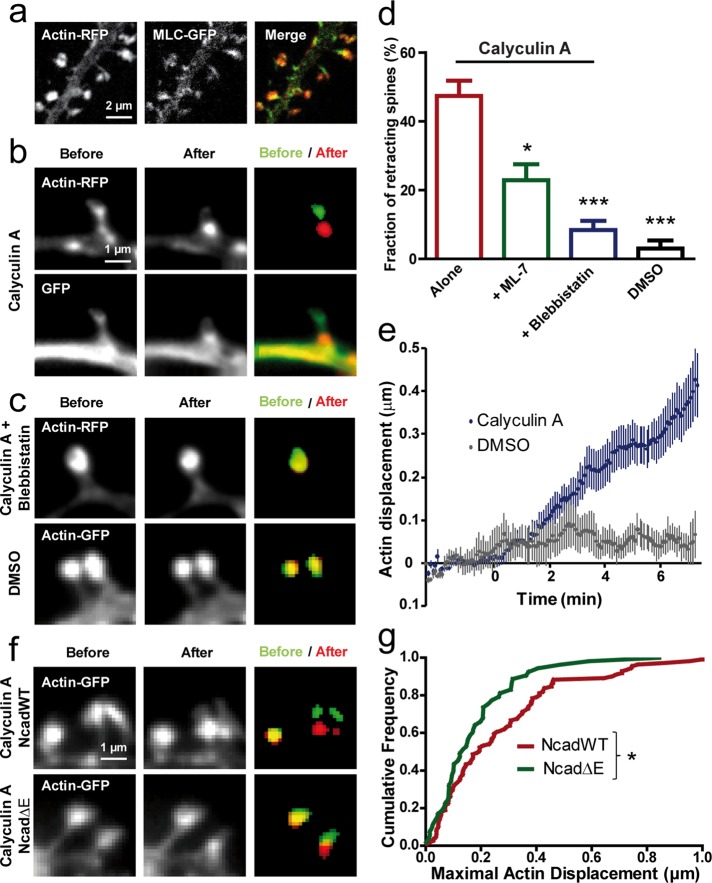
To understand further the molecular mechanism underlying spine morphing, we addressed the potential role of myosin II in regulating actin dynamics in relation to N-cadherin adhesion. Of interest, MLC-GFP clusters were often detected downstream of actin-RFP distribution at the base of the spine head (Figure 4a), in agreement with previous reports (Korobova and Svitkina, 2010; Tatavarty et al., 2012) and raising the possibility that myosin exerts tension on the actin network. To examine this hypothesis, we monitored actin-GFP redistribution in spines upon treating neurons with calyculin A, an inhibitor of PP1 and PP2A phosphatases that targets MLC phosphatase, thus enhancing myosin II contraction. Calyculin A is known to increase F-actin flow in keratocytes (Vallotton et al., 2004) and induce actin network contraction in Aplysia growth cones (Zhang et al., 2003) and neurite retraction in hippocampal neurons (Inutsuka et al., 2009). Calyculin A induced a major retraction of the actin-GFP network from the tip to the base in ∼50% of spines (Figure 4, b and d, and Supplemental Movie 3), indicating that myosin II was pulling rearward on actin filaments. As a control, addition of the vehicle (dimethyl sulfoxide [DMSO]) did not produce any significant actin retraction (Figure 4, c and d). Furthermore, preincubation of neurons with blebbistatin, a selective inhibitor of myosin II ATPase (Kovács et al., 2004), or ML-7, an inhibitor of MLC kinase activity (Zhang et al., 2003), significantly blocked calyculin A–induced actin retraction (Figure 4, c and d; 8 and 23% of spines retracting, respectively), demonstrating that the effect of calyculin A was mainly due to enhancement of myosin II activity in dendritic spines.
FIGURE 4:
Stimulating myosin contractility induces actin retraction in spines. (a) Neurons at DIV 17–21 coexpressing actin-RFP and MLC-GFP were dual-immunolabeled for GFP and RFP and examined by confocal microscopy. The merge image reveals the downstream localization of MLC-GFP compared with actin-RFP in spine heads. (b) Neurons coexpressing GFP and actin-RFP were imaged live in two colors and treated with calyculin A (1 μM). Note the retraction of spines in both channels. Segmented actin-GFP signal before (green) and at the end of the treatment (red) is represented on the merged image. (c) Neurons expressing actin-RFP were treated with calyculin A in the continuous presence of blebbistatin (50 μM), which blocked spine retraction. No retraction was detected either in spines expressing actin-GFP when the vehicle DMSO was added. (d) Fraction of retracting spines when calyculin A was added alone (178 spines, 7 neurons) or in the presence of ML-7 (279 spines, 6 neurons) or blebbistatin (357 spines, 9 neurons). As a control, DMSO was added alone (183 spines, 3 neurons; p < 0.0001 by one-way ANOVA). *0.01 < p < 0.05; ***p < 0.001. (e) Relative displacement of segmented actin-GFP spots over time. Neurons expressing actin-GFP were imaged every 5 s for 10 min, and calyculin A or the vehicle DMSO were added 2.5 min after the start of the recording (average ± SEM for 8 spines). (f) Representative actin-GFP images for cells coexpressing NcadWT or Ncad∆E, with similar calyculin A treatment as shown in b. (g) Cumulative distribution of the maximal displacement of actin-GFP spots within 10 min after drug addition for NcadWT or Ncad∆E conditions. Data were analyzed by nonparametric Mann–Whitney test. *0.01 < p < 0.05.
Dual-color experiments in neurons coexpressing GFP and actin-RFP revealed a concomitant retraction of the GFP signal (Figure 4b), indicating partial collapse of the spine potentially associated with a rip-off of axon–dendrite contacts and consistent with a strong mechanical connection between the actin/myosin network and transsynaptic membrane adhesions. The displacement of segmented actin spots with calyculin A was roughly linear over time, allowing calculation of a retraction velocity of the order of 0.001 μm/s (Figure 4e), on the lower end of values reported for the actin retrograde flow in spines (Honkura et al., 2008; Tatavarty et al., 2009; Frost et al., 2010; Chazeau et al., 2014). The fact that the actin retraction induced by calyculin A was not faster than the basal actin flow rate suggests that transsynaptic adhesions are resisting the rearward motion of the actin network triggered by the enhancement of myosin II activity.
To study further whether the link between N-cadherin and actin potentially resisted such myosin-driven traction, we treated neurons cotransfected with actin-GFP and NcadWT or the NcadΔE mutant with calyculin A (Figure 4, f and g). The induced displacement of F-actin spots was significantly lower in cells cotransfected with NcadΔE than in cells transfected with NcadWT. One explanation for this finding is that upon overexpression of the catenin-sequestering NcadΔE mutant, the actin network becomes loosely connected to endogenous transsynaptic adhesions. Thus, in the presence of NcadΔE, the adhesive forces cannot counteract enough myosin II activity, and such disorganized actin network is susceptible to be dragged over shorter distances by myosin II. Taken together, these observations support a model in which, under normal conditions, the actin-network is bound to N-cadherin adhesions at the tip of spines and myosin II exerts tension on actin dynamics, thus contributing to the retrograde flow of actin.
N-cadherin–coated micropatterns promote the morphological maturation of dendritic filopodia
To determine whether exogenous N-cadherin adhesions could stabilize dendritic spines by locally organizing the actin network, we cultured neurons for 8 DIV on micropatterned substrates containing a hexagonal array of 2-μm dots coated with recombinant N-cadherin (Ncad-Fc) surrounded by a cytophobic background. These substrates stimulated the growth of neurites compared with control substrates coated with only the Fc fragment (Czöndör et al., 2013), reflecting specific recognition between immobilized Ncad-Fc and endogenous N-cadherin at the neuronal membrane. Neurons grown on these substrates often formed structures resembling dendritic spines, with a neck originating from dendritic shafts and a head making specific contacts with neighboring Ncad-Fc–coated dots, eventually allowing branching between dendrites (Figure 5a).
FIGURE 5:
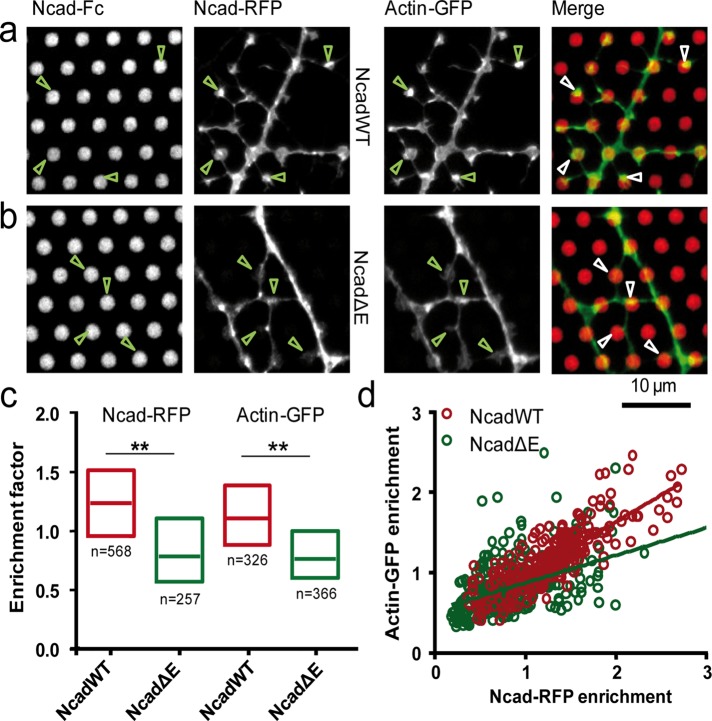
Formation of actin-rich dendritic filopodia on N-cadherin coated micropatterns. (a, b) Neurons coexpressing actin-GFP and either NcadWT-RFP or NcadΔE-RFP, respectively, were cultured on micropatterned substrates coated with Ncad-Fc and immobilized through Cy5-conjugated anti-Fc antibodies. Images show (from left to right) the Cy5, RFP, GFP, and merge between actin-GFP (green) and micropatterns (red). (c) Enrichment levels of Ncad-RFP and actin-GFP at Ncad-Fc micropatterns in NcadWT-RFP and NcadΔE-RFP conditions. The two conditions were compared by a Mann–Whitney test separately for each staining (actin-GFP or Ncad-RFP). **0.001 < p < 0.01. The number of micropatterns analyzed was 257 for NcadWT-RFP and 291 for NcadΔE-RFP (four cells in each condition). (d) Plot of actin-GFP vs. Ncad-RFP signals for the NcadWT and NcadΔE conditions, respectively. The correlation coefficient was r = 0.83 for NcadWT (p < 0.0001, n = 257 dots) and 0.45 for NcadΔE (p < 0.0001, n = 291 dots).
To examine the role of N-cadherin/actin coupling in these processes, we cotransfected neurons with actin-GFP and NcadWT-RFP or NcadΔE-RFP and quantified the GFP and RFP signals at dendritic filopodia in contact with Ncad-Fc–coated dots. NcadWT-RFP displayed a significant accumulation at the micropatterns, reflecting specific adhesion to Ncad-Fc. In contrast, neurons transfected with NcadΔE-RFP showed little accumulation at the dots, demonstrating that the NcadΔE mutant was indeed defective in making homophilic interactions (Figure 5, b and c). Similarly, actin-GFP enrichment at Ncad-Fc micropatterns was high for NcadWT-RFP and lower for NcadΔE-RFP, and neurons expressing NcadΔE did not form spine-like structures at micropatterns as observed in the NcadWT condition (Figure 5, a and b). Moreover, the correlation between Ncad-RFP and actin-GFP enrichment measured at the same micropatterns was stronger than between NcadΔE-RFP and actin-GFP enrichments (Figure 5d). Overall these data suggest that transsynaptic interaction of N-cadherin is required for actin accumulation in dendritic spines and structural maturation of dendritic filopodia into dendritic spines.
N-cadherin–coated microspheres stabilize dendritic filopodia into spine-like structures
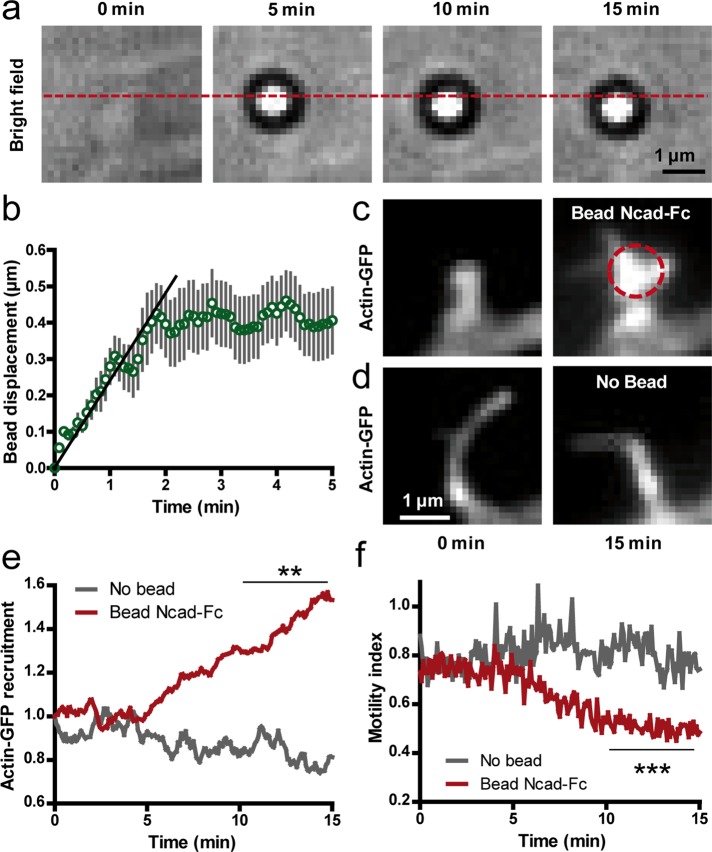
To study the structural changes triggered by N-cadherin adhesions in a dynamic manner, we placed Ncad-Fc–coated microspheres at the tip of dendritic filopodia of 10 DIV neurons using optical tweezers. We chose dendritic filopodia that were not engaged in adhesive interactions with axons and thus were accessible to external manipulation. We then monitored the bead motion in bright field and the distribution of actin-GFP in epifluorescence for 15 min while the trap was continuously turned on. In ∼50% of the cases, beads escaped the optical trap and moved rearward at a velocity of 0.07 ± 0.02 μm/s (n = 7 beads) before stopping at the base of the filopodium (Figure 6, a and b). This velocity was close to the speed of the rearward actin flow in dendritic filopodia and spines measured by local photoactivation or single-molecule tracking (Honkura et al., 2008; Tatavarty et al., 2009, 2012; Frost et al., 2010) and thus may represent transmembrane coupling between engaged N-cadherin receptors at the bead contact and the actin flow, as observed in growth cones (Suter and Forscher, 2000; Bard et al., 2008; Giannone et al., 2009). In parallel, actin-GFP progressively accumulated at bead contacts and dendritic filopodia often adopted mushroom shapes resembling spines (Figure 6, c and e, and Supplemental Movie 4). In addition, the motility index of these filopodia decreased over time (Figure 6f), reaching values characteristic of dendritic filopodia contacting axons (Figure 2, a and b). Control dendritic filopodia not in contact with beads kept constant actin levels and high motility (Figure 6, d and e). Taken together, these results indicate that N-cadherin adhesions physically couple to the actin network in dendritic filopodia and this connection promotes rapid actin recruitment and filopodium stabilization.
FIGURE 6:
Stimulation of dendritic filopodia by N-cadherin–coated beads manipulated by optical tweezers. (a) Bright-field images of an Ncad-Fc–coated microsphere placed at the tip of a dendritic filopodium with optical tweezers. Note the rearward motion of the bead with respect to the position of the optical trap (dashed line). (b) Rearward displacement of Ncad-Fc–coated beads on filopodia over time (mean ± SEM of 14 beads). The initial slope (plain line) was defined as the rearward velocity of the bead. (c) Corresponding actin-GFP images at the start and end of the optical tweezers experiment. The dashed circle indicates the bead position. Note the shape change of the filopodium and accumulation of actin-GFP. (d) Control filopodia without beads did not show such a response. (e) Actin-GFP enrichment vs. time at filopodia in contact with Ncad-Fc–coated beads compared with control filopodia without beads (average ± SEM of 14 filopodia from 12 cells). (f) Corresponding measurements of motility index over time performed on the same filopodia. Data integrated over time between 10 and 15 min were compared by unpaired t tests (**p < 0.01; ***p < 0.002).
Computer simulations predict the relationship between actin turnover and enrichment at dendritic spines
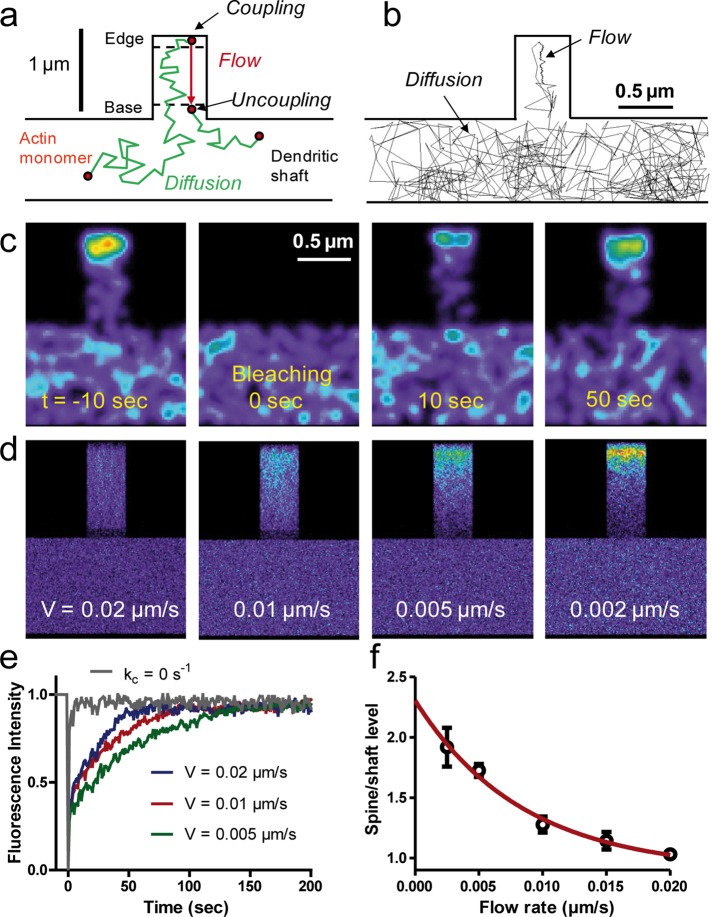
To integrate these various results into a global framework, we generated Monte Carlo simulations of actin dynamics in a model geometry representing a dendritic filopodium and a piece of dendritic shaft (Figure 7, a and b). Single actin monomers were assumed to diffuse freely in the dendritic shaft and in the filopodium, until they reached the filopodium tip, where they assembled into filaments through a polymerization reaction governed by a kinetic rate kc (where the subscript c signifies coupling). The filamentous pool was then set to slowly move rearward with constant velocity V. Actin monomers were allowed to dissociate from the F-actin network by severing or disassembling, either spontaneously with a reaction rate ku (the inverse of the actin filament lifetime; u signifies uncoupling) or when they reached the base of the filopodium, and then diffuse freely again. The actin flow rate V was varied between 0.002 and 0.02 μm/s, corresponding to values previously measured for spines and filopodia, respectively (Tatavarty et al., 2009, 2012; Chazeau et al., 2014). Reconstruction of image stacks based on the superimposition of 1000 individual trajectories was used to map the overall distribution of actin over time under resting conditions or changes in certain parameters, allowing us to predict various experiments.
FIGURE 7:
Computer simulations of actin dynamics in model dendritic structures. (a) Model description. Actin monomers transit between fast random diffusion in the shaft and filopodium and slow directional flow originating from the filopodium tip. (b) Example of a 450-s trajectory for a single actin monomer with time step 50 ms. Arrows indicate diffusion and trapping in the F-actin flow. (c) Simulated FRAP experiment for a flow velocity of 0.01 μm/s. Each actin molecule is represented by a Gaussian distribution of fluorescence coded in false colors. (d) Images of actin distribution obtained from integrating 1000 computer simulations of actin dynamics. Examples with different flow velocities. (e) FRAP curves for different flow velocities, each curve being an average of four independent sets of 1000 simulations. Note that increasing V accelerates recovery. The gray curve corresponds to actin monomer diffusion only (kc = 0, no trapping). (f) Relationship between actin enrichment and actin flow rate. Each point represents an average ± SD of eight spines from four independent sets of simulations. The red line is a fit with an exponentially decreasing function.
To find the relevant model parameters, we first mimicked FRAP experiments by setting the intensity of actin molecules present in the filopodium to zero after an initial equilibrium was reached, then building fluorescence-like images and monitoring the redistribution of unbleached molecules over time (Figure 7c and Supplemental Movie 5). We adjusted the kinetic rates of actin polymerization and depolymerization (parameters kc and ku) in the simulations to obtain the best agreement with experimental FRAP curves. To isolate the contribution of actin monomer diffusion with respect to monomer/polymer exchange on overall actin turnover in spines, we performed FRAP simulations using kc = 0, thus preventing actin monomers from entering the filamentous pool and resulting in a single population of freely diffusing actin monomers. These simulations yielded an extremely fast recovery (2–3 s; Figure 7e), indicating that the slower recovery of actin-GFP in filopodia/spines is mainly due to exchange between actin monomers and filaments and is not diffusion limited. This was compatible with FRAP experiments performed on spines expressing soluble enhanced GFP (Star et al., 2002), further validating the coefficients chosen in the model to characterize the diffusion of free actin monomers. When kc was increased to allow actin filament assembly, the predicted FRAP curves matched experiments performed on actin-GFP (Star et al., 2002; Koskinen et al., 2014). Of importance, increasing the actin flow rate V accelerated recovery after photobleaching (Figure 7e), in quantitative agreement with the faster turnover of actin-GFP in dendritic filopodia than in spines (Figure 3e). Using the same type of program, we generated simulations mimicking photoactivation of actin at the tip of spines, which was followed by retrograde motion of the illuminated spot and a gradual decrease of fluorescence due to actin depolymerization (Supplemental Figure S4 and Supplemental Movie 6), in excellent agreement with published observations (Honkura et al., 2008; Koskinen et al., 2014).
To check whether this set of parameters also quantitatively reproduced actin enrichment at spines, we then built heat maps integrating the number of actin molecules over time for different values of the parameters V, kc, and ku. These images first showed a longitudinal expansion of actin from the tip of the filopodium (Figure 7d), consistent with the theoretical filament length, equal to the ratio between the actin rearward velocity and the depolymerization rate (V/ku). For example, for a value ku = 0.02 s−1 and V between 0.002 (spines) and 0.02 μm/s (filopodia), the mean filament length would range between 0.1 and 1 μm, as observed in the simulated images (Supplemental Figure S5, a and b) and compatible with experimental findings (Figure 2a). The simulations also revealed a gradient of actin from tip to base, best seen at low velocities (Figure 7d) and high polymerization rate kc (Supplemental Figure S5a) and originating from the limited lifetime of actin filaments. Actin enrichment at spines can be seen as the ratio between total actin (monomer plus polymer) and free monomeric actin. As expected, actin enrichment was increased at high polymerization rates and decreased at high depolymerization rates (Supplemental Figure S5, c and d). Of importance, these simulations revealed that actin accumulation at the tips of filopodia/spines decreased exponentially with retrograde actin flow rate (Figure 7, d and f). Such behavior is due to the fact that the trapping of individual actin molecules in a slowly moving filamentous pool translates into actin accumulation at the ensemble level. Together with the cytochalasin D experiments demonstrating that spine motility is intrinsically linked to actin turnover (Supplemental Figure S2), these predictions agree with our live-cell imaging data showing a negative correlation between actin enrichment and spine motility (Figure 2, b and d).
Interpreting modulation of actin dynamics by changing model parameters
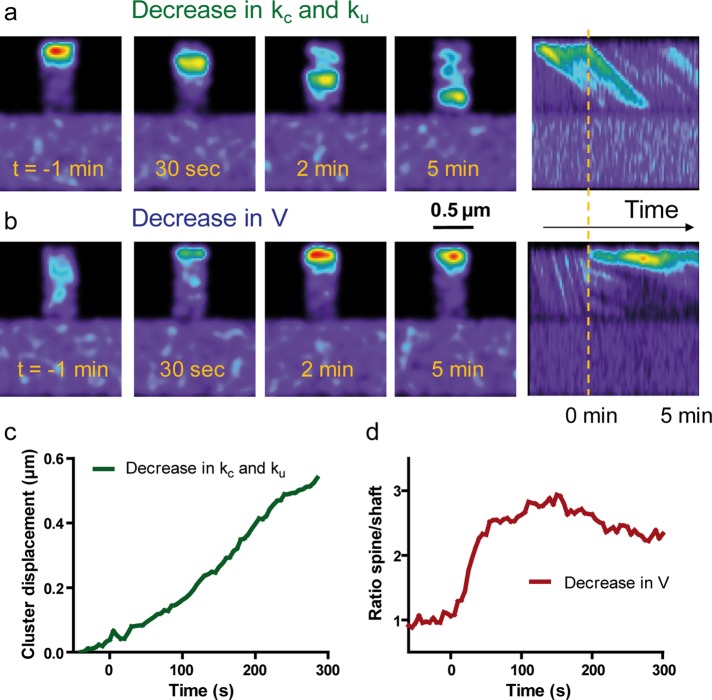
We then attempted to reproduce various experimental conditions by introducing changes in the model parameters. For example, we predict that a sudden decrease in the polymerization rate kc would rapidly remove actin filaments from spines (Supplemental Figure S6, a and c, and Supplemental Movie 7), as observed experimentally by sequestering actin monomers with latrunculin A (Star et al., 2002; Cingolani and Goda, 2008). In contrast, reducing kc and ku simultaneously to mimic the capping of actin filaments at barbed ends with cytochalasin D (Carlier et al., 1986) and the concomitant increase in local G-actin concentration would cause a mass of actin to move toward the base of the spines (Figure 8, a and c) and not a complete disappearance of actin from spines, compatible with experimental observations (Supplemental Figure S2). On the other hand, changing the depolymerization rate and/or flow velocity would directly alter filament length. For example, decreasing both ku and V to mimic inhibition of the actin-severing activity of myosin II and the reduction in actin flow caused by blebbistatin (Medeiros et al., 2006; Tatavarty et al., 2012) would cause actin filaments to grow longer (Supplemental Figure S6, b and c), thus inducing a more-filopodial phenotype (Ryu et al., 2006). The addition of blebbistatin induced in some spines the assembly of a very long actin network within 15 min, spanning from the spine tip to the dendritic shaft (Supplemental Figure S6d), suggesting that under physiological conditions, myosin II inhibits the penetration of the polymerizing actin network throughout the spine neck.
FIGURE 8:
Predictions of experiments by changing parameters controlling actin dynamics. Different parameters in the simulations were changed after initial equilibrium is reached (150 s set as time 0), and then actin redistribution was monitored using the Gaussian representation. (a) The polymerization rate kc was decreased from 0.5 to 0.05 s−1, and the dissociation rate ku was decreased from 0.05 to 0.002 s−1. Note the gradual retraction of the actin cluster from the tip to the base of the filopodium, as observed in calyculin A experiments. (b) The actin flow rate was changed from V = 0.02 to 0.002 μm/s to mimic clutch engagement between N-cadherin adhesions and the actin/myosin network. Note a gradual accumulation of actin at the tip of the spines up to steady state. Right, kymographs represent actin intensity across the shaft and spine over time. Note changes in intensity and the diagonal slopes, reflecting actin flow rates. The dashed line indicates time 0. (c) Displacement of the actin cluster over time in response to a change in both kc and ku. (d) Actin enrichment vs. time in response to a change in V.
Paradoxically, the best way to model the effect of calyculin A on actin retraction (Figure 4) was to decrease kc to mimic a potential dissociation of the actin network from the plasma membrane where actin nucleators are located and simultaneously decreasing ku to prevent actin clusters from depolymerizing (Figure 8, a and c). However, those interpretations must be taken with caution, since F-actin turnover is regulated by many different proteins, and pharmacological agents may only target specific actin pools (Koskinen et al., 2014). Finally, to model the effect of the transsynaptic element, specifically the engagement of N-cadherin/actin connections induced by optical tweezers experiments, we performed simulations in which the actin flow rate was suddenly reduced from 0.02 to 0.002 μm/s (Figure 8, b and d, and Supplemental Movie 8). We then observed a progressive accumulation of actin at the tip of filopodia, quantitatively consistent with that observed at Ncad-Fc–coated beads (Figure 6, c and e). Thus these combined experimental and theoretical results indicate that actin trapping promoted by the transsynaptic engagement of N-cadherin adhesions may be explained by a reduction in the actin flow rate.
DISCUSSION
We demonstrated here that a mechanical connection between transsynaptic N-cadherin adhesions at axon/dendrite contacts and the postsynaptic actin/myosin network regulates dendritic spine morphology and stability. The codistribution of N-cadherin, F-actin, and myosin II in dendritic spines supported the possible interaction between these molecules, and the preferential localization of myosin at the base of spines further suggested that myosin might be pulling on the actin network. Indeed, stimulation of myosin II activity caused a retraction of the actin network from the tip to the base of the spine and an overall collapse of the spine, which reflects a strong connection between the contractile actin/myosin network and transsynaptic adhesions. The link between N-cadherin and the actin/myosin network was further supported by the expression of an N-cadherin construct unable to form transsynaptic binding, which drastically increased the motility of dendritic spines up to filopodia levels while simultaneously reducing actin enrichment at spines. Furthermore, expression of this mutant enhanced actin turnover as measured by FRAP and inhibited calyculin A–induced actin retraction. These results suggest that altering the physical connection between transsynaptic N-cadherin adhesions and actin filaments results in a highly dynamic and disorganized actin network associated with a filopodium-like phenotype (Togashi et al., 2002; Okamura et al., 2004; Mendez et al., 2010). In addition, the engagement of homophilic N-cadherin adhesions using purified N-cadherin immobilized on micropatterns or microspheres triggered the transition from actively moving dendritic filopodia into stable spines containing a dense actin network, further indicating an important role of N-cadherin adhesions in regulating actin-dependent spine initiation and maturation.
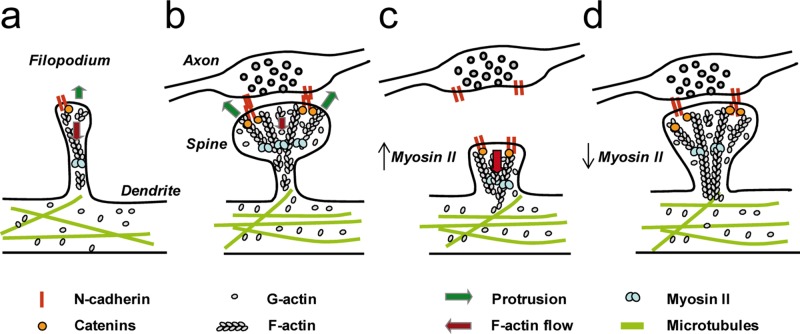
To interpret these data, we favor the molecular clutch model initially proposed to describe cell locomotion (Mitchison and Kirschner, 1988; Suter and Forscher, 2000) and previously demonstrated at the N-cadherin/actin interface in neuronal growth cones (Bard et al., 2008). According to this model, the clutch between N-cadherin adhesions and the actin flow in dendritic filopodia not contacting axons is in a slip mode, associated with fast rearward actin motion and high filopodium motility (Figure 9a). Actin-based protrusions and retractions of dendritic filopodia are likely to be dictated by a balance between the rate of actin cytoskeleton assembly at the tip and retrograde flow (Mallavarapu and Mitchison, 1999). On formation of axon/dendrite contacts, clutch engagement between N-cadherin adhesions and actin filaments leads to a reduction of the actin rearward flow and overall turnover, as observed by comparing dendritic spines and filopodia in PALM and FRAP experiments, respectively (Tatavarty et al., 2009, 2012; Chazeau et al., 2014; Koskinen et al., 2014). Such coupling causes actin polymerization forces to push against the plasma membrane, resulting in filopodia enlargement, actin accumulation, and structural stabilization (Figure 9b), as observed at optically restrained N-cadherin–coated microspheres and N-cadherin–coated micropatterns. The clutch concept applied here to describe spine initiation may further be extrapolated to the synaptic activity–dependent growth and stabilization of spines, which also rely on actin and N-cadherin adhesions (Matsuzaki et al., 2004; Okamoto et al., 2004; Okamura et al., 2004; Mendez et al., 2010). In addition, our results point to an important mechanical role for myosin II in spines. Indeed, increasing myosin II activity with calyculin A caused a retraction of the actin network from the tip to the base of the spine (Figure 9c), whereas decreasing myosin II contractility with blebbistatin led to the assembly of a longer actin network running to the spine neck through the myosin-rich region, suggesting that myosin II is severing and/or depolymerizing filaments at the base of spines (Figure 9d). These results are consistent with previous reports showing that blebbistatin decreases the treadmilling rate of the dynamic actin pool (Koskinen et al., 2014), resulting in a lower retrograde actin flow (Tatavarty et al., 2012), and promotes the formation of filopodium-like protrusions (Ryu et al., 2006). Overall our experiments suggest that at least two counteracting forces are exerted on the actin network in dendritic spines, one coming from transsynaptic N-cadherin adhesions at the membrane, and the other from myosin II activity at the base of spines. The balance between these antagonistic forces seems to be an important mechanism for regulating spine shape and stability (Supplemental Movie S9).
FIGURE 9:
Model of clutch engagement at dendritic filopodia and spines. (a) In motile filopodia, the actin flow is high, and protrusions are not stabilized. (b) On axon/dendrite encounter, the mechanical connection between homophilic N-cadherin adhesions and the actin network reduces the actin flow, stabilizes the structure, and allows spine enlargement by actin polymerization on the sides of the adhesive zone. (c) If myosin II activity is enhanced by calyculin A, adhesion forces cannot balance myosin tension exerted on actin filaments any longer, leading to an overall collapse of the spine. (d) In contrast, if myosin II activity is inhibited by blebbistatin, both the actin flow rate and the disassembly of actin filaments are decreased, promoting the elongation of an actin network into the spine neck through the myosin-rich region.
Computer simulations recapitulated the experimental data and provided a general framework to interpret actin dynamics in those motile structures. We suggest the important concept that reduction of actin flow rate is linked to accumulation of actin at spine tips, simply because actin molecules reside longer in a slowly moving filament network. Similarly, reducing the actin flow rate in growth cones by restraining N-cadherin–coated microspheres with a microneedle also induced punctual actin accumulation (Bard et al., 2008), suggesting that this represents a widespread biophysical mechanism. The fact that the stable actin pool identified in photoactivation and FRAP experiments increases during spine maturation and is correlated with spine volume (Honkura et al., 2008; Koskinen et al., 2014) might be related to such trapping of actin filaments. Overall our model, which considers a very polarized flow within a homogeneous actin population, provides a good quantitative interpretation of most of the experiments performed here, but the actual situation might be more complex (Pilo Boyl and Witke, 2014). Indeed, there is ongoing debate about the polarity of actin filaments within dendritic filopodia and spines. Experiments using photoactivation of actin-Dendra (Honkura et al., 2008) or tracking of single actin molecules (Frost et al., 2010) revealed a radial actin flow with decreasing velocity from the tip to the base of the spine head, suggesting the presence of actin filaments with barbed ends oriented toward the postsynaptic density and pointed ends toward the shaft. The myosin V–driven transport into the spine head of recycling endosomes moving on polarized actin filaments (Wang et al., 2008) agrees with this concept. On the other hand, electron micrographs revealed a dense network of small branched actin filaments of mixed polarities in spine heads, whereas in the spine neck, as in precursor dendritic filopodia, actin filaments were longer and longitudinally oriented but with few filament bundles (Korobova and Svitkina, 2010). This differential arrangement may explain the faster actin flow in dendritic filopodia compared with spines observed by single-molecule tracking (Tatavarty et al., 2009, 2012; Chazeau et al., 2014). Those studies further revealed both rearward and forward directional movements of individual actin molecules in spines, indicating that actin filaments exhibit a range of orientations and polarities. Our data showing a retraction of the actin network by calyculin A suggest that a fraction of actin filaments exist in an antiparallel configuration such that they can be contracted and/or severed by enhancement of myosin II activity, as shown in vitro (Reymann et al., 2012). In addition, superresolution microscopy showed that branched F-actin nucleation occurs at the vicinity of the postsynaptic density, whereas elongation occurs at the tip of finger-like protrusions, an organization that may not generate a completely fast and concerted actin rearward flow (Chazeau et al., 2014). Such additional factors regulating actin dynamics, together with variable filament orientations, might be incorporated into future versions of the model.
Although we provided mostly biophysical interpretations, there are several biochemical pathways that could regulate the association between N-cadherin adhesions and the actin/myosin network in spines. The strong phenotype of the NcadΔE mutant in inhibiting the connection to actin suggests that the intracellular tail of N-cadherin, which has several interacting partners, plays an important role. For example, N-cadherin may dynamically connect to actin via β-catenin and α-catenin (Maiden and Hardin, 2011), whose expression levels bidirectionally affect spine morphology (Abe et al., 2004; Takeichi and Abe, 2005; Okuda et al., 2007). N-cadherin adhesions may also recruit in a force-dependent manner the Arp2/3 complex (Verma et al., 2012), which controls actin branching in spines (Kim et al., 2013; Chazeau et al., 2014). The connection between N-cadherin and actin may also involve signaling molecules, including the small GTPase Rac-1, which controls the anchoring of N-cadherin to the actin flow in myogenic cells (Lambert et al., 2002), and RhoA, whose activity regulates N-cadherin function under the control of δ-catenin/p120 (Elia et al., 2006; Arikkath et al., 2009) and is linked to rapid spine enlargement (Murakoshi et al., 2011). Reverse signaling between actin and N-cadherin adhesion is also possible through afadin, a Rap-regulated actin-binding protein that promotes cadherin complex assembly and mediates spine growth (Xie et al., 2008; Beaudoin et al., 2012).
In conclusion, using a combination of quantitative live imaging and computer modeling, we showed that a mechanical connection between N-cadherin adhesions and the actin/myosin network stabilizes dendritic spines, a phenomenon that is likely to play an important role in regulating spine formation and morphology in the developing and mature nervous system.
MATERIALS AND METHODS
Molecular constructs
Actin-GFP was a gift of A. Matus (Friedrich Miescher Institute, FMI, Basel, Switzerland; Fischer et al., 1998). MLC-GFP was a gift from Ann Bresnick (Albert Einstein College, New York, NY). The plasmid coding for C-terminal GFP-tagged chicken wild-type N-cadherin (NcadWT-GFP) was described earlier (Thoumine et al., 2006). To construct the plasmid expressing full-length chicken N-cadherin C-terminally fused to RFP (NcadWT-RFP), a 1400–base pair BamHI–HindIII DNA fragment encoding the monomeric DsRed sequence (Campbell et al., 2002) was subcloned into pCDNA 3.1 hygro (Invitrogen) to generate pCD mDsRed (Lambert et al., 2007). A XbaI–SacI 2500–base pair fragment encompassing the 5′ end of the chicken N-cadherin cDNA (Matsuzaki et al., 1990) was amplified by PCR using the oligonucleotides 5′GGAATTAAGAGAGCTCAAGGATC3′ and 5′CTAGTCTAGATCCATGTGCCGGATAGCGGG3′. A second PCR was performed to prepare a SacI–BamHI 100–base pair fragment corresponding to the 3′ end of the N-cadherin cDNA using the oligonucleotides 5′TCCTTGAGCTCTCTTAATTCC3′ and 5′CGCGGATCCCCGTCATCACCTCCACCGTAC3′. Each PCR fragment was digested to reveal their cohesive ends and cloned into the XbaI and BamHI sites of pCD mDsRed. N-terminal myc-tagged Xenopus laevis N-cadherin lacking the extracellular domain (Ncad∆E) was a kind gift of C. Holt (University of Cambridge, Cambridge, UK; Riehl et al., 1996). mRFP1 was amplified by PCR and inserted after the signal peptide in place of the myc tag using two BamHI sites. The fidelity of all constructs was verified by sequencing at the Genomic Transcriptomic Facility of Bordeaux (Bordeaux, France).
Substrate coating
The 22-mm square micropatterned glass coverslips were obtained from CYTOO (Grenoble, France) as a custom-designed “Ready to Coat” product (ref. 10-950-00), according to our specifications: hydrophilic dots of 1.5-μm diameter separated by 5 μm against a uniformly cytophobic environment. Substrates were then treated with 40 μg/ml polylysine (Sigma-Aldrich, St. Louis, MO), which adsorbed to the hydrophilic dots, dried, and stored at 4°C. Anti-human Fc antibodies (Jackson ImmunoResearch, West Grove, PA) were conjugated to Cy5 fluorophores using a kit from Amersham and recovered on a size exclusion column (Bio-Rad, Marnes-la-Coquette, France). One day before plating cells, a 9:1 ratio of unlabeled and Cy5-conjugated goat anti-human Fc antibodies (Jackson ImmunoResearch) was diluted at 0.023 mg/ml in borate buffer (boric acid 0.2 M, pH 8.5), and centrifuged for 10 min at 14,000 rpm and 4°C. Micropatterned substrates were flipped onto a 200-μl drop of the antibody mixture on Parafilm previously sterilized with ultraviolet and incubated at room temperature for 4–5 h. Substrates were then washed with borate buffer and incubated overnight at 4°C with recombinant rat N-cadherin fused to the constant fragment of human immunoglobulin G (Ncad-Fc; R&D Systems) diluted at 0.04 mg/ml in boric acid and previously centrifuged for 10 min at 14,000 rpm. For all other experiments, 18-mm glass coverslips were cleaned overnight with 6 M nitric acid, rinsed with deionized water, autoclaved, and marked with three paraffin dots. These coverslips were incubated for 2 h at 37°C with 1 mg/ml polylysine and then rinsed with water. Regular coverslips were placed in 60-mm dishes (four per dish) and micropatterned coverslips in six-well plates, washed with borate buffer, and left with MEM containing 10% horse serum for 1–2 h in the incubator (37°C, 5% CO2) before seeding the cells.
Cell culture and transfection
Dissociated hippocampal neurons from E18 rat embryos were plated on regular 18-mm coverslips at a density of 200,000 cells/dish or on micropatterned substrates (100,000 cells/coverslip). At 4–5 h after plating, coverslips were flipped onto dishes containing a feeder layer of astrocytes and cultured in Neurobasal medium (NB) supplemented with B27 (Kaech and Banker, 2006). Alternatively, micropatterned substrates were washed with NB supplemented with B27 (37°C) to remove unattached cells, and medium was replaced with conditioned medium previously incubated for 4 d in dishes containing a feeder glial cell layer. Neurons were cultured up to 3 wk at 37°C in a 5% CO2 atmosphere. Neurons were transfected at DIV 7 using lipofection, according to the manufacturer's protocol (Effectene kit; Qiagen, Courtaboeuf, France), using 1 μg of total DNA per micropatterned coverslip or 1 μg per regular dish (four coverslips). For cotransfections of actin-GFP and Ncad-RFP or RFP constructs, a 2:1 DNA ratio was used. Alternatively, 500,000 neurons taken right after dissection were electroporated with the Amaxa system (Lonza, Levallois-Perret, France) using 5 μg of DNA of RFP or actin-GFP. The two neuronal populations were then mixed and cultured on 18-mm coverslips for 8–10 DIV.
Immunocytochemistry
Neurons cotransfected with RFP plus N-cadherin-GFP, actin-GFP, or MLC-GFP at DIV 17–21 or just with actin-GFP were fixed for 10 min in warm 4% paraformaldehyde/4% sucrose in phosphate-buffered saline (PBS). Remaining active sites were saturated with 50 mM NH4Cl in PBS for 15 min, and then cultures were permeabilized with 0.1% Triton X-100 in PBS for 5 min, and nonspecific binding was blocked with PBS containing 1% bovine serum albumin (BSA) for 1 h at room temperature. Neurons were dual immunolabeled using mouse anti-GFP (1:200; Invitrogen, Life Technologies, St Aubin, France) plus rabbit anti-dsRed (1:200; Clontech, St-Quentin-en-Yvelines, France) for 1 h at room temperature, followed by Alexa 488–conjugated goat anti-mouse (2 mg/ml, 1:400; Invitrogen) and Atto647N-conjugated goat anti-rabbit (1:400; Sigma-Aldrich) secondary antibodies, respectively, for 30 min at room temperature. In some experiments, cells transfected with actin-GFP were immunolabeled with mouse anti-synapsin (Synaptic Systems, Goettingen, Germany) primary antibodies, followed by Alexa 568–conjugated goat anti-mouse (2 mg/ml, 1:400; Invitrogen). For quantification of actin-GFP overexpression, cultures were fixed for 5 min in −20°C methanol, blocked, and stained with mouse anti-actin (1:400; Millipore, Molshein, France), followed by Alexa 568–conjugated goat anti-mouse (2 mg/ml, 1:400; Invitrogen) Coverslips were mounted in Mowiol (Calbiochem, Îles-de-France, France).
Microscopy observation of fixed cells and subspine distribution of proteins
For observation of protein distribution within spines, cells were examined using a SP-5 Leica confocal microscope, using a 100×/1.40 numerical aperture (NA) objective and a 100-μm pinhole aperture. GFP images were acquired using the 488-nm argon laser line and emission band 500–550 nm, and RFP images were acquired using the 532-nm laser line and emission band 560–600 nm. GFP and RFP images were segmented using a custom macro written in MetaMorph software (Universal Imaging, Bedford Hills, NY), providing the coordinates of the GFP signals relative to the centroid of the spine obtained from the RFP signal. These values were normalized by the width and length of each spine, respectively. This semiautomatic image analysis performed for hundreds of dendritic spines provided a plot of the relative distributions of each protein within spines. To quantify the level of apposition between actin-GFP and synapsin puncta, we used a program written as a plug-in for ImageJ described previously (Mondin et al., 2011).
Live-cell imaging and motility index
Neurons were mounted in an open chamber containing 500 μl Tyrode solution (in mM: 120 NaCl, 5 KCl, 2 MgCl2, 2 CaCl2, 25 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, and 30 d-glucose, pH 7.4), and observed under an Olympus IX70 microscope thermostated at 37°C using a Plexiglas box filled with an air blower (World Precision Instruments, Sarasota, FL) and an objective heater (Bioptechs, Butler, PA). Fluorescence images were taken using a 100×/1.40 PlanApo oil objective and an electron-multiplying charge-coupled device (EMCCD) camera (QuantEM; Roper Scientific, Evry, France) upon illumination with a 150-W xenon lamp and filter cubes from Chroma (GFP: excitation HQ 480/30 nm, dichroic 86100bs, emission S528/38 nm; RFP: excitation HQ630/20 nm, dichroic 101848, emission S685/40 nm). Actin-GFP images were acquired every 5 s for 10 min using a shutter (Uniblitz, Rochester, NY) driven by MetaMorph, whereas axonal RFP and Ncad-RFP images were taken only once at the beginning of the acquisition. In some experiments, 1 μM cytochalasin D (Sigma-Aldrich), 1 μM calyculin A (Tocris), 50 μM blebbistatin (Tocris, Bristol, UK), 10 μM ML-7 (Millipore), or 1:2000 DMSO (Sigma-Aldrich) was added to the cells 2.5 min after beginning the recording. Stock solutions of cytochalasin D, calyculin A, and blebbistatin were 2 mM in DMSO. For blebbistatin treatment, neurons expressing actin-RFP were imaged, since blebbistatin is fluorescent and rendered ineffective when illuminated with blue light (Kolega, 2004).
Dendritic spine (or filopodium) motility was assessed by a motility index constructed as follows (Ibarretxe et al., 2007). An initial wavelet-based segmentation was performed on the images using a custom-made program provided by J. B. Sibarita (University Bordeaux Segalen, Bordeaux, France; Izeddin et al., 2012), giving very good delineation of the dendritic spine outline, and putting the background to zero. The area covered by the dendritic spine or filopodium during the recording period (up to 15 min) was determined by a threshold on the maximal image projection of the whole image stack over time. Pairs of consecutive frames were subtracted from each other with an added constant (1000) to avoid negative pixel values (Supplemental Figure S2a). In these images, pixels with newly appearing structures are white, and disappearing structures are black. Highly motile structures will thus appear as highly contrasted on this type of image. Such contrast is estimated by the SD of pixel values over the total area covered, divided by the average intensity over the total dendritic spine or filopodium area. This normalization corrects for different fluorescent protein expression levels and for photobleaching.
Actin and cadherin enrichments in filopodia and spines
For freely moving dendritic filopodia and filopodia in contact with RFP-expressing axons and spines, actin-GFP images were segmented with a custom program to isolate actin-rich regions in the spine head, and the ratio of fluorescence from this region to the fluorescence in neighboring shaft regions was computed. For the observation of filopodia in contact with Ncad-Fc micropatterns, neurons were visualized on a Leica DM R upright epifluorescence microscope (Leica Microsystems, Wetzlar, Germany) equipped with a 63×/1.32 NA objective and appropriate filter sets from Chroma (GFP: excitation S490/20 nm, dichroic 86100bs, emission S528/38 nm; tetramethylrhodamine isothiocyanate: excitation S555/28 nm, dichroic 101848, emission S617/73; Cy5: excitation S635/20, dichroic 101848, emission S685/40 nm). Images were acquired with a CCD camera (HQ CoolSnap; Roper Scientific), using MetaMorph software. The enrichment factors of actin-GFP and RFP at the N-cadherin–coated micropatterns were measured on three randomly chosen dendritic areas of each neuron and calculated using an automatic program written in MetaMorph (Mondin et al., 2011). Briefly, the outlines of the patterns from the Cy5 channel corresponding to the Cy5-conjugated anti-Fc antibody were transferred to the corresponding actin-GFP and RFP images, where the outline of the neurite was determined by a threshold function. The patterns that did not overlap with the threshold area were eliminated from further quantification. An enrichment index was calculated by dividing the GFP or RFP fluorescence intensity within each dot for the thresholded area by the corresponding average intensity measured from dendritic parts located outside patterns.
Microsphere preparation and optical tweezers experiments
A 5-μl amount of 1-μm sulfate latex microspheres (4.5 × 107 particles/μl; Polysciences, Eppelheim, Germany) was incubated overnight at 4°C with 10 μg of goat anti-Fc antibody (Jackson ImmunoResearch) in 50 μl of 0.2 M borate buffer, pH 8.5. Microspheres were rinsed in borate buffer containing 0.3% BSA, and 20 μl of the suspension was incubated at room temperature for 3 h with 3 μg of Ncad-Fc. Beads were rinsed again three times, resuspended in 100 μl, and kept on ice during the experiments. An Olympus IX70 inverted microscope was fed through its epifluorescence port by a fixed collinear Nd:YAG laser beam (1064 nm, 100 mW; Compass Coherent, Santa Clara, CA), filling the back aperture of a 100×/1.40 oil objective (Thoumine et al., 2006). Two shutters (Uniblitz) driven by MetaMorph and combined with appropriate filters and dichroics allowed dual bright-field and epifluorescence illumination of GFP. Images in each channel were taken every 5 s for 20 min with an EMCCD camera (QuantEM; Roper Scientific). A shutter controlled by a foot switch was used to activate the laser and capture microspheres in suspension. Beads were then moved using a motorized stage (MarzHauser, Wetzlar, Germany) and brought to the tip of motile dendritic filopodia. After an initial 5-min observation period to check for filopodial motility, the trap was maintained ON for 15 min. The bead position was monitored using the “track objects” function in MetaMorph, and the intensity of actin-GFP in the filopodium in contact with the bead relative to the baseline value was computed using macros developed in MetaMorph (Mondin et al., 2011).
FRAP experiments on actin-GFP
FRAP experiments were performed on 10 DIV or 18 DIV hippocampal neurons transfected with actin-GFP on a Nikon Eclipse TiE microscope equipped with a four-wavelength laser bench plugged to a multispot FRAP head (Roper Scientific). After acquisition of a 10-s baseline, the actin-GFP signal at dendritic filopodia or spines, respectively, was bleached using the 488-nm laser (50 mW, 20 ms). This resulted in a 70–80% loss in fluorescence at time zero. Fluorescence recovery was then recorded for 300 s using an Evolve camera (Roper Scientific) with 50-ms acquisition times separated by 1 s (for 30 s), 2 s (for 1 min), and then 10 s (for 3 min). The fluorescence signals on filopodia/spines were subtracted from background, divided by the signals before bleaching, and normalized to zero at the time of bleaching. No further correction was applied, since the fluorescence of nearby unbleached filopodia showed no significant decay during the acquisition period. In some experiments, neurons were cotransfected with actin-GFP and either NcadWT-RFP or Ncad-ΔE-RFP and observed at DIV 18.
Computer simulations of actin dynamics
Computer simulations describe the life cycle of single actin monomers in the dendritic shaft and filopodia. The computer program was written within commercial software (Mathematica 4.1; Wolfram Research, Oxfordshire, UK) and run on a personal computer. The program structure is based on a previously published framework describing diffusion/trapping of glutamate receptors at synapses (Czöndör et al., 2012). Based on experimental observations, the geometry was chosen as a rectangular portion of dendrite of 1 μm × 6 μm, comprising two thin perpendicular filopodia of 0.4 μm × 1 μm (Figure 7, a and b). An individual actin molecule was characterized by its two-dimensional coordinates x and y over time t. The total length of the trajectories (450 s) was based on the durations of FRAP and photoactivation of fluorescence (PAF) recordings, and the time step of the simulations was ∆t = 50 ms, corresponding to a typical camera acquisition rate. The initial molecule position was chosen randomly in the shaft. At each time step, the (x, y) coordinates of the molecule were incremented by the distances (Δx, Δy), which depended on whether the molecule was free to diffuse or bound to retrograde flow.
In the diffusive regime, x(t) and y(t) coordinates were incremented by (2D∆t)1/2x1 and (2D∆t)1/2y1, respectively, where x1 and y1 were random numbers generated from a normal distribution to account for the stochastic nature of diffusion. Single actin monomers were taken to diffuse freely in the dendritic shaft and filopodium with a high diffusion coefficient (D = 1 μm2/s), taken from measurements of GFP diffusion in spines (Star et al., 2002). Actin monomers were allowed to cross the shaft/filopodium interface with a probability value of 0.3, mimicking the existence of the spine neck, which acts as a diffusion barrier (Bloodgood and Sabatini, 2005). When actin monomers reached a 0.1-μm distance to the edge of the filopodium or spine, they were allowed to assemble into filaments, governed by a coupling rate kc (0.02–1 s−1), and set to slowly move rearward with constant velocity V (0.005–0.02 μm/s), as measured from photoactivation experiments (Honkura et al., 2008; Tatavarty et al., 2009, 2012; Frost et al., 2010). In that regime, the y-coordinate was then incremented by −V∆t and the x-coordinate by (2D′∆t)1/2x1, accounting for a short-range lateral motion (D′ = 0.01 μm2/s). Actin filaments were allowed to spontaneously depolymerize into monomers with an uncoupling rate ku (0.005–0.04 s−1). If filaments reached a 0.2-μm region from the base of the spine, they were set to disassemble and diffuse randomly again, in agreement with the relative absence of polymerized actin in the shaft, which contains mostly microtubules (Korobova and Svitkina, 2010).
One thousand trajectories were generated and superimposed. The output text file (x, y, t) was entered into a custom program run in MetaMorph, allowing the construction of stacks of images representing actin distribution over time (Czöndör et al., 2012). A zoom factor determined the pixel size of the reconstructed image (typically 30–100 nm). To measure the enrichment of actin at filopodia/spines, the whole image stack was summed into a single image, and the ratio of intensities in the filopodium/spine versus that in the dendritic shaft was computed. It took ∼100 s for the system to reach steady state, as characterized by a fairly constant number of actin molecules within spines. To model photobleaching, an intensity parameter was set to 0 for actin molecules localized in one of the two filopodia at a given time after steady state (150 s) and kept null thereafter. Otherwise, the intensity was set to a constant arbitrary gray level (100). The number of molecules in the bleached filopodium was then computed over time and normalized by the number of molecules in the unbleached neighboring filopodium. Four different FRAP simulations were run for each flow rate (0.0025, 0.005, 0.01, and 0.02 μm/s) and averaged. To model actin photoactivation simulations, the intensity parameter was first set to 0 for all molecules and then switched to 100 only for actin molecules localized in a 0.3 μm × 0.3 μm area at the tip of one filopodia (at time 150 s) and kept at the value of 100 thereafter. The parameters kc, ku, and V were varied to find the best agreement with experimental data.
Supplementary Material
Acknowledgments
We acknowledge A. Bresnick, C. Holt, A. Matus, R. M. Mège, and R. Tsien for the generous gifts of reagents. We thank Z. Karatas, C. Breillat, A. Frouin, D. Bouchet, and N. Retailleau for cell culture and P. Gonzales, R. Sterling, and A. Barbet for technical assistance. We thank C. Poujol, S. Marais, M. Mondin, and P. Legros at the Bordeaux Imaging Center for help in microscopy and image analysis, J. B. Sibarita for his image reconstruction program, and the Genomic Transcriptomic Facility of Bordeaux for DNA sequencing. The research leading to these results has received funding from the Centre National de la Recherche Scientifique, the Agence Nationale pour la Recherche (grants Neuroligation, Synapse-2Dt, and Preciput), the Conseil Régional Aquitaine, and the Fondation pour la Recherche Médicale.
Abbreviations used:
- Arp2/3
actin-related protein 2/3
- DIV
days in vitro
- DMSO
dimethyl sulfoxide
- Fc
immunoglobulin constant fragment
- FRAP
fluorescence recovery after photobleaching
- ML-7
myosin light chain kinase inhibitor
- MLC
myosin light chain
- Ncad
N-cadherin
- PALM
photoactivation localization microscopy
- Rac-1
Ras-related C3 botulinum toxin substrate 1
- RhoA
Ras homologous member A.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-06-1086) on January 7, 2015.
*These authors contributed equally to this work.
†Present address: Cell Biology, Faculty of Science, Utrecht University, 3584 CH Utrecht, Netherlands.
REFERENCES
- Abe K, Chisaka O, Van Roy F, Takeichi M. Stability of dendritic spines and synaptic contacts is controlled by alpha N-catenin. Nat Neurosci. 2004;7:357–363. doi: 10.1038/nn1212. [DOI] [PubMed] [Google Scholar]
- Ackermann M, Matus A. Activity-induced targeting of profilin and stabilization of dendritic spine morphology. Nat Neurosci. 2003;6:1194–1200. doi: 10.1038/nn1135. [DOI] [PubMed] [Google Scholar]
- Arikkath J, Peng I-F, Ng YG, Israely I, Liu X, Ullian EM, Reichardt LF. Delta-catenin regulates spine and synapse morphogenesis and function in hippocampal neurons during development. J Neurosci. 2009;29:5435–5442. doi: 10.1523/JNEUROSCI.0835-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikkath J, Reichardt LF. Cadherins and catenins at synapses: roles in synaptogenesis and synaptic plasticity. Trends Neurosci. 2008;31:487–494. doi: 10.1016/j.tins.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard L, Boscher C, Lambert M, Mège R-M, Choquet D, Thoumine O. A molecular clutch between the actin flow and N-cadherin adhesions drives growth cone migration. J Neurosci. 2008;28:5879–5890. doi: 10.1523/JNEUROSCI.5331-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin GMJ, Schofield CM, Nuwal T, Zang K, Ullian EM, Huang B, Reichardt LF. Afadin, a Ras/Rap effector that controls cadherin function, promotes spine and excitatory synapse density in the hippocampus. J Neurosci. 2012;32:99–110. doi: 10.1523/JNEUROSCI.4565-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodgood BL, Sabatini BL. Neuronal activity regulates diffusion across the neck of dendritic spines. Science. 2005;310:866–869. doi: 10.1126/science.1114816. [DOI] [PubMed] [Google Scholar]
- Bozdagi O, Shan W, Tanaka H, Benson DL, Huntley GW. Increasing numbers of synaptic puncta during late-phase LTP: N-cadherin is synthesized, recruited to synaptic sites, and required for potentiation. Neuron. 2000;28:245–259. doi: 10.1016/s0896-6273(00)00100-8. [DOI] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Criquet P, Pantaloni D, Korn ED. Interaction of cytochalasin D with actin filaments in the presence of ADP and ATP. J Biol Chem. 1986;261:2041–2050. [PubMed] [Google Scholar]
- Chazeau A, Mehidi A, Nair D, Gautier JJ, Leduc C, Chamma I, Kage F, Kechkar A, Thoumine O, Rottner K, et al. Nanoscale segregation of actin nucleation and elongation factors determines dendritic spine protrusion. EMBO J. 2014;33:2745–2764. doi: 10.15252/embj.201488837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani La, Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci. 2008;9:344–356. doi: 10.1038/nrn2373. [DOI] [PubMed] [Google Scholar]
- Colicos M, Collins B, Sailor M, Goda Y. Remodeling of synaptic actin induced by photoconductive stimulation. Cell. 2001;107:605–616. doi: 10.1016/s0092-8674(01)00579-7. [DOI] [PubMed] [Google Scholar]
- Comery TA, Harris JB, Willems PJ, Oostra BA, Irwin SA, Weiler IJ, Greenough WT. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc Natl Acad Sci USA. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czöndör K, Garcia M, Argento A, Constals A, Breillat C, Tessier B, Thoumine O. Micropatterned substrates coated with neuronal adhesion molecules for high-content study of synapse formation. Nat Commun. 2013;4:2252. doi: 10.1038/ncomms3252. [DOI] [PubMed] [Google Scholar]
- Czöndör K, Mondin M, Garcia M, Heine M, Frischknecht R, Choquet D, Sibarita J-B, Thoumine OR. Unified quantitative model of AMPA receptor trafficking at synapses. Proc Natl Acad Sci USA. 2012;109:3522–3527. doi: 10.1073/pnas.1109818109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia LP, Yamamoto M, Zang K, Reichardt LF. p120 catenin regulates dendritic spine and synapse development through Rho-family GTPases and cadherins. Neuron. 2006;51:43–56. doi: 10.1016/j.neuron.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Kaech S, Knutti D, Matus A. Rapid actin-based plasticity in dendritic spines. Neuron. 1998;20:847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- Frost NA, Shroff H, Kong H, Betzig E, Blanpied TA. Single-molecule discrimination of discrete perisynaptic and distributed sites of actin filament assembly within dendritic spines. Neuron. 2010;67:86–99. doi: 10.1016/j.neuron.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G, Mège R-M, Thoumine O. Multi-level molecular clutches in motile cell processes. Trends Cell Biol. 2009;19:475–486. doi: 10.1016/j.tcb.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Hodges JL, Newell-Litwa K, Asmussen H, Vicente-Manzanares M, Horwitz AR. Myosin IIb activity and phosphorylation status determines dendritic spine and post-synaptic density morphology. PLoS One. 2011;6:e24149. doi: 10.1371/journal.pone.0024149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkura N, Matsuzaki M, Noguchi J, Ellis-Davies GCR, Kasai H. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 2008;57:719–729. doi: 10.1016/j.neuron.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Ibarretxe G, Perrais D, Jaskolski F, Vimeney A, Mulle C. Fast regulation of axonal growth cone motility by electrical activity. J Neurosci. 2007;27:7684–7695. doi: 10.1523/JNEUROSCI.1070-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inutsuka A, Goda M, Fujiyoshi Y. Calyculin A-induced neurite retraction is critically dependent on actomyosin activation but not on polymerization state of microtubules. Biochem Biophys Res Commun. 2009;390:1160–1166. doi: 10.1016/j.bbrc.2009.10.108. [DOI] [PubMed] [Google Scholar]
- Izeddin I, Boulanger J, Racine V, Specht CG, Kechkar A, Nair D, Triller A, Choquet D, Dahan M, Sibarita JB. Wavelet analysis for single molecule localization microscopy. Opt Express. 2012;20:2081–2095. doi: 10.1364/OE.20.002081. [DOI] [PubMed] [Google Scholar]
- Izeddin I, Specht CG, Lelek M, Darzacq X, Triller A, Zimmer C, Dahan M. Super-resolution dynamic imaging of dendritic spines using a low-affinity photoconvertible actin probe. PLoS One. 2011;6:e15611. doi: 10.1371/journal.pone.0015611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Kim IH, Racz B, Wang H, Burianek L, Weinberg R, Yasuda R, Wetsel WC, Soderling SH. Disruption of Arp2/3 results in asymmetric structural plasticity of dendritic spines and progressive synaptic and behavioral abnormalities. J Neurosci. 2013;33:6081–6092. doi: 10.1523/JNEUROSCI.0035-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolega J. Phototoxicity and photoinactivation of blebbistatin in UV and visible light. Biochem Biophys Res Commun. 2004;320:1020–1025. doi: 10.1016/j.bbrc.2004.06.045. [DOI] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell. 2010;21:165–176. doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen M, Bertling E, Hotulainen R, Tanhuanpää K, Hotulainen P. Myosin IIb controls actin dynamics underlying the dendritic spine maturation. Mol Cell Neurosci. 2014;61:56–64. doi: 10.1016/j.mcn.2014.05.008. [DOI] [PubMed] [Google Scholar]
- Kovács M, Tóth J, Hetényi C, Málnási-Csizmadia A, Sellers JR. Mechanism of blebbistatin inhibition of myosin II. J Biol Chem. 2004;279:35557–35563. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- Lambert M, Choquet D, Mège RM. Dynamics of ligand-induced, Rac1-dependent anchoring of cadherins to the actin cytoskeleton. J Cell Biol. 2002;157:469–479. doi: 10.1083/jcb.200107104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert M, Thoumine O, Brevier J, Choquet D, Riveline D, Mège R-M. Nucleation and growth of cadherin adhesions. Exp Cell Res. 2007;313:4025–4040. doi: 10.1016/j.yexcr.2007.07.035. [DOI] [PubMed] [Google Scholar]
- Maiden SL, Hardin J. The secret life of α-catenin: moonlighting in morphogenesis. J Cell Biol. 2011;195:543–552. doi: 10.1083/jcb.201103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu A, Mitchison T. Regulated actin cytoskeleton assembly at filopodium tips controls their extension and retraction. J Cell Biol. 1999;146:1097–1106. doi: 10.1083/jcb.146.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Honkura N, Ellis-Davies GCR, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki F, Mege RM, Jaffe SH, Friedlander DR, Gallin WJ, Goldberg JI, Cunningham BA, Edelman GM. cDNAs of cell adhesion molecules of different specificity induce changes in cell shape and border formation in cultured S180 cells. J Cell Biol. 1990;110:1239–1252. doi: 10.1083/jcb.110.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros NA, Burnette DT, Forscher P. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat Cell Biol. 2006;8:215–226. doi: 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
- Mendez P, De Roo M, Poglia L, Klauser P, Muller D. N-cadherin mediates plasticity-induced long-term spine stabilization. J Cell Biol. 2010;189:589–600. doi: 10.1083/jcb.201003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Cytoskeletal dynamics and nerve growth. Neuron. 1988;1:761–772. doi: 10.1016/0896-6273(88)90124-9. [DOI] [PubMed] [Google Scholar]
- Mondin M, Labrousse V, Hosy E, Heine M, Tessier B, Levet F, Poujol C, Blanchet C, Choquet D, Thoumine O. Neurexin-neuroligin adhesions capture surface-diffusing AMPA receptors through PSD-95 scaffolds. J Neurosci. 2011;31:13500–13515. doi: 10.1523/JNEUROSCI.6439-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi H, Wang H, Yasuda R. Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature. 2011;472:100–104. doi: 10.1038/nature09823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase S, Mosser E, Schuman EM. Depolarization drives beta-Catenin into neuronal spines promoting changes in synaptic structure and function. Neuron. 2002;35:91–105. doi: 10.1016/s0896-6273(02)00764-x. [DOI] [PubMed] [Google Scholar]
- Mysore SP, Tai C-Y, Schuman EM. Effects of N-cadherin disruption on spine morphological dynamics. Front Cell Neurosci. 2007;1:1. doi: 10.3389/neuro.03.001.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K-I, Nagai T, Miyawaki A, Hayashi Y. Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat Neurosci. 2004;7:1104–1112. doi: 10.1038/nn1311. [DOI] [PubMed] [Google Scholar]
- Okamura K, Tanaka H, Yagita Y, Saeki Y, Taguchi A, Hiraoka Y, Zeng L-H, Colman DR, Miki N. Cadherin activity is required for activity-induced spine remodeling. J Cell Biol. 2004;167:961–972. doi: 10.1083/jcb.200406030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda T, Yu LMY, Cingolani La, Kemler R, Goda Y. beta-Catenin regulates excitatory postsynaptic strength at hippocampal synapses. Proc Natl Acad Sci USA. 2007;104:13479–13484. doi: 10.1073/pnas.0702334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilo Boyl P, Witke W. Small, smaller… dendritic spine. EMBO J. 2014;33:2737–2739. doi: 10.15252/embj.201490137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymann A-C, Boujemaa-Paterski R, Martiel J-L, Guérin C, Cao W, Chin HF, De La Cruz EM, Théry M, Blanchoin L. Actin network architecture can determine myosin motor activity. Science. 2012;336:1310–1314. doi: 10.1126/science.1221708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehl R, Johnson K, Bradley R, Grunwald GB, Cornel E, Lilienbaum A, Holt CE. Cadherin function is required for axon outgrowth in retinal ganglion cells in vivo. Neuron. 1996;17:837–848. doi: 10.1016/s0896-6273(00)80216-0. [DOI] [PubMed] [Google Scholar]
- Rubio MD, Johnson R, Miller Ca, Huganir RL, Rumbaugh G. Regulation of synapse structure and function by distinct myosin II motors. J Neurosci. 2011;31:1448–1460. doi: 10.1523/JNEUROSCI.3294-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J, Liu L, Wong TP, Wu DC, Burette A, Weinberg R, Wang YT, Sheng M. A critical role for myosin IIb in dendritic spine morphology and synaptic function. Neuron. 2006;49:175–182. doi: 10.1016/j.neuron.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Schubert V, Da Silva JS, Dotti CG. Localized recruitment and activation of RhoA underlies dendritic spine morphology in a glutamate receptor-dependent manner. J Cell Biol. 2006;172:453–467. doi: 10.1083/jcb.200506136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuman EM, Murase S. Cadherins and synaptic plasticity: activity-dependent cyclin-dependent kinase 5 regulation of synaptic beta-catenin-cadherin interactions. Philos Trans R Soc Lond B Biol Sci. 2003;358:749–756. doi: 10.1098/rstb.2002.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star EN, Kwiatkowski DJ, Murthy VN. Rapid turnover of actin in dendritic spines and its regulation by activity. Nat Neurosci. 2002;5:239–246. doi: 10.1038/nn811. [DOI] [PubMed] [Google Scholar]
- Suter DM, Forscher P. Substrate–cytoskeletal coupling as a mechanism for the regulation of growth cone motility and guidance. J Neurobiol. 2000;44:97–113. [PubMed] [Google Scholar]
- Takeichi M, Abe K. Synaptic contact dynamics controlled by cadherin and catenins. Trends Cell Biol. 2005;15:216–221. doi: 10.1016/j.tcb.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Shan W, Phillips GR, Arndt K, Bozdagi O, Shapiro L, Huntley GW, Benson DL, Colman DR. Molecular modification of N-cadherin in response to synaptic activity. Neuron. 2000;25:93–107. doi: 10.1016/s0896-6273(00)80874-0. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Yuste R. Regulation of dendritic spine motility and stability by Rac1 and Rho kinase: evidence for two forms of spine motility. Mol Cell Neurosci. 2004;26:429–440. doi: 10.1016/j.mcn.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Tatavarty V, Das S, Yu J. Polarization of actin cytoskeleton is reduced in dendritic protrusions during early spine development in hippocampal neuron. Mol Biol Cell. 2012;23:3167–3177. doi: 10.1091/mbc.E12-02-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatavarty V, Kim E-J, Rodionov V, Yu J. Investigating sub-spine actin dynamics in rat hippocampal neurons with super-resolution optical imaging. PLoS One. 2009;4:e7724. doi: 10.1371/journal.pone.0007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoumine O, Lambert M, Mège R-M, Choquet D. Regulation of N-cadherin dynamics at neuronal contacts by ligand binding and cytoskeletal coupling. Mol Biol Cell. 2006;17:862–875. doi: 10.1091/mbc.E05-04-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M. Cadherin regulates dendritic spine morphogenesis. Neuron. 2002;35:77–89. doi: 10.1016/s0896-6273(02)00748-1. [DOI] [PubMed] [Google Scholar]
- Uchida N, Honjo Y, Johnson KR, Wheelock MJ, Takeichi M. The catenin/cadherin adhesion system is localized in synaptic junctions bordering transmitter release zones. J Cell Biol. 1996;135:767–779. doi: 10.1083/jcb.135.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban NT, Willig KI, Hell SW, Nägerl UV. STED nanoscopy of actin dynamics in synapses deep inside living brain slices. Biophys J. 2011;101:1277–1284. doi: 10.1016/j.bpj.2011.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallotton P, Gupton SL, Waterman-Storer CM, Danuser G. Simultaneous mapping of filamentous actin flow and turnover in migrating cells by quantitative fluorescent speckle microscopy. Proc Natl Acad Sci USA. 2004;101:9660–9665. doi: 10.1073/pnas.0300552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Han SP, Michael M, Gomez Ga, Yang Z, Teasdale RD, Ratheesh A, Kovacs EM, Ali RG, Yap AS. A WAVE2-Arp2/3 actin nucleator apparatus supports junctional tension at the epithelial zonula adherens. Mol Biol Cell. 2012;23:4601–4610. doi: 10.1091/mbc.E12-08-0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Edwards JG, Riley N, Provance DW Jr, Karcher R, Li XD, Davison IG, Ikebe M, Mercer JA, Kauer JA, Ehlers MD. Myosin Vb mobilizes recycling endosomes and AMPA receptors for postsynaptic plasticity. Cell. 2008;135:535–548. doi: 10.1016/j.cell.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CA, Tsuchida MA, Allen GM, Barnhart EL, Applegate KT, Yam PT, Ji L, Keren K, Danuser G, Theriot JA. Myosin II contributes to cell-scale actin network treadmilling through network disassembly. Nature. 2010;465:373–377. doi: 10.1038/nature08994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Photowala H, Cahill ME, Srivastava DP, Woolfrey KM, Shum CY, Huganir RL, Penzes P. Coordination of synaptic adhesion with dendritic spine remodeling by AF-6 and kalirin-7. J Neurosci. 2008;28:6079–6091. doi: 10.1523/JNEUROSCI.1170-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T. Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat Rev Neurosci. 2004;5:24–34. doi: 10.1038/nrn1300. [DOI] [PubMed] [Google Scholar]
- Zhang X, Schaefer AW, Burnette DT, Schoonderwoert VT, Forscher P. Rho-dependent contractile responses in the neuronal growth cone are independent of classical peripheral retrograde actin flow. Neuron. 2003;40:931–944. doi: 10.1016/s0896-6273(03)00754-2. [DOI] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J Neurosci. 2005;25:3379–3388. doi: 10.1523/JNEUROSCI.3553-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Homma KJ, Poo M. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44:749–757. doi: 10.1016/j.neuron.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Ziv NE, Smith SJ. Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron. 1996;17:91–102. doi: 10.1016/s0896-6273(00)80283-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.