Characterization of Kif2a in Xenopus embryos identifies new roles for the Kif2a microtubule depolymerase in coordinating cytokinesis and centrosome coalescence. In addition, defects in mitosis can inhibit large-scale developmental movements in vertebrate tissues.
Abstract
Kif2a is a member of the kinesin-13 microtubule depolymerases, which tightly regulate microtubule dynamics for many cellular processes. We characterized Kif2a depletion in Xenopus animal caps and embryos. Kif2a depletion generates defects in blastopore closure. These defects are rescued by removing the animal cap, suggesting that Kif2a-depleted animal caps are not compliant enough to allow gastrulation movements. Gastrulation defects are not rescued by a Kif2a mutated in an Aurora kinase phosphorylation site, suggesting that the phenotypes are caused by problems in mitosis. During animal cap mitoses, Kif2a localizes to the spindle poles and centromeres. Depletion of Kif2a generated multipolar spindles in stage 12 embryos. Kif2a-depleted animal caps have anaphase lagging chromosomes in stage 9 and 10 embryos and subsequent cytokinesis failure. Later divisions have greater than two centrosomes, generating extra spindle poles. Kif2a-depleted embryos are also defective at coalescing extra spindle poles into a bipolar spindle. The gastrulation and mitotic phenotypes can be rescued by either human Kif2a or Kif2b, which suggests that the two homologues redundantly regulate mitosis in mammals. These studies demonstrate that defects in mitosis can inhibit large-scale developmental movements in vertebrate tissues.
INTRODUCTION
Microtubule dynamics must be tightly regulated to allow chromosome segregation (Ems-McClung and Walczak, 2010). The kinesin-13 family of microtubule depolymerases comprises major regulators of catastrophe events at both plus and minus microtubule ends (Sanhaji et al., 2011). Most vertebrates have two kinesin-13s, known as Kif2a and mitotic centromere-associated kinesin/Kif2c (MCAK). Mammals have a third, known as Kif2b (Manning et al., 2007). These proteins localize to distinct structures of the spindle, controlling microtubule dynamics in time and space (Manning et al., 2007). Depletion of Kif2a generates profound mitotic defects in human cancer cells, yet kif2a−/− mice are born, albeit with brain abnormalities, and a human retardation syndrome maps to the Kif2a locus (Homma et al., 2003; Jaillard et al., 2011). There is growing evidence that kinesin-13s play important roles in both transformation and resistance to spindle poison chemotherapeutics, especially the derivatives of paclitaxel (Sanhaji et al., 2011).
The kinesin-13 microtubule depolymerase, Kif2a, is a critical regulator of the mitotic spindle in tissue culture cells, where the depletion of Kif2a produces monopolar spindles (Ganem and Compton, 2004). Kif2a is required for microtubule flux in both human cells and Xenopus extracts, where it is proposed to depolymerize the minus ends of microtubules at the pole, maintaining spindle length (Gaetz and Kapoor, 2004; Ganem et al., 2005). Kif2a is localized to spindle poles in HeLa cells but to both poles and centromeres/kinetochores of Xenopus cells and extracts (Ganem and Compton, 2004; Knowlton et al., 2009). However, the critical role of Kif2a in spindle regulation seen in tissue culture has been challenged by studies in mice. Kif2a−/− mice live to birth but have multiple brain abnormalities, caused in part because of suppression of collateral branch extension of neurons (Homma et al., 2003). It is unclear whether the lack of mitotic defects is because of redundant activities of the related Kif2b in mammals or tissue-specific roles of Kif2a for mitosis in brain tissues or whether Kif2a activity is essential for mitosis of cells grown in culture but not required in a tissue context.
Aurora B kinase is present at the inner centromere of each prometaphase/metaphase chromosome, whereas Aurora A kinase is localized to the spindle poles. These kinases are key regulators of microtubule dynamics in the spindle (Sampath et al., 2004; Carmena et al., 2009). The functions of kinesin-13 are regulated through phosphorylation by Aurora kinase and Plk1 (Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004; Rosasco-Nitcher et al., 2008; Knowlton et al., 2009; Ems-McClung and Walczak, 2010). Phosphorylation of a conserved serine in the neck of the kinesin domain by Aurora kinases inhibits depolymerase activity of both Kif2a and MCAK in vitro (Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004). Kif2a inhibited by Aurora B phosphorylation can be reactivated either by dephosphorylation or interaction with the protein ICIS (Knowlton et al., 2009). Aurora phosphorylation of the N-terminal centromere-targeting domain of MCAK regulates localization of Kif2a to spindles in a complex manner in Xenopus extracts (Zhang et al., 2007). The importance of the related site on Kif2a at T-70 has not been determined.
We investigated the role of Kif2a in spindle formation and chromosome segregation using embryonic animal caps and morpholino (MO)-mediated Kif2a depletions to explore a number of critical unanswered questions. Surprisingly, we find that Kif2a is required for the epiboly (spreading) of animal cap, and failure of epiboly has the mechanical effect of retarding or blocking blastopore closure and endoderm internalization on the other (vegetal) side of the embryo. In animal caps, Kif2a localizes to the spindle poles (centrosomes) and centromeres during mitosis and is required for spindle integrity and proper chromosome segregation, as suggested by tissue culture systems. In addition, we demonstrate the critical role of Aurora kinase phosphorylation of T70 in localizing Kif2a to both spindle poles and centromeres. We show that Kif2a is required for chromosome segregation in Xenopus tissues and that Kif2a depletion can be rescued by either human Kif2a or Kif2b. We also identify new requirements for Kif2a in both cytokinesis and pole coalescence.
RESULTS
Aurora phosphorylation of Kif2a on T70 in cells and frog egg extracts
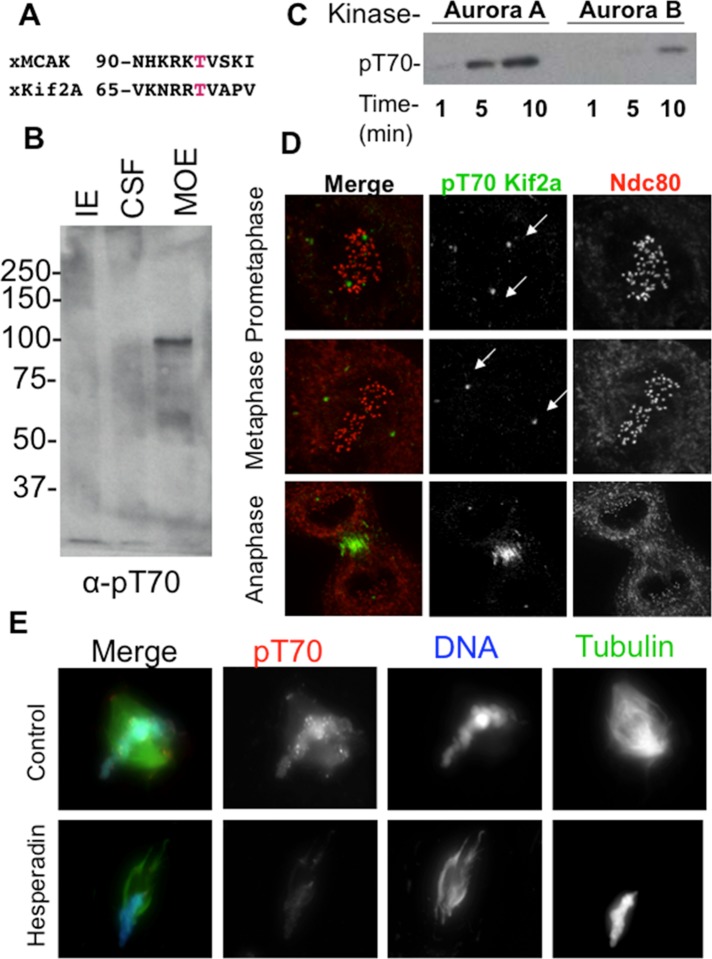
The localization of MCAK, a kinesin-13 protein related to Kif2a, is regulated by Aurora phosphorylation on the N-terminus (Zhang et al., 2007). We noted that a similar Aurora B consensus site could be seen on the N-terminus of Kif2a (Figure 1A). To characterize this phosphorylation event, we generated a phosphospecific antibody to the 15 amino acids around T70 of Kif2a, and the antibody specifically recognized a single band that corresponds to the molecular weight of Kif2a in Xenopus extracts. This band was found only in extracts treated to activate Aurora kinases (mature ooctye extract [MOE] in Figure 1B; Lan et al., 2004). The antibody also recognized recombinant Kif2a after phosphorylation with either Aurora A or Aurora B kinase (Figure 1C). The antibody recognized two foci in the correct location for centrosomes/spindle poles in Xenopus S3 tissue culture cells and poles and centromeres in Xenopus egg extract (Figure 1D). Note that we previously showed that Kif2a is localized to centromeres in Xenopus S3 cells (Knowlton et al., 2009), suggesting that centromere phosphorylation of Kif2a by Aurora B can be regulated in a cell-type manner. In extracts the signals were lost after treatment with the Aurora kinase inhibitor Hesperadin, demonstrating that the signal is dependent on Aurora kinases (Figure 1E).
FIGURE 1:
Aurora kinases phosphorylate Kif2a on T70. (A) Alignment of putative Aurora consensus sites on Kif2a with their homologous MCAK phosphorylation sites. The putative phosphorylated residue is shown in red. (B) Immunoblot of anti-pT70 Kif2a antibody against three Xenopus extracts. CSF, cytostatic factor–arrested egg extract; IE, interphase extract; MOE, mature ooctye extract. Note that that the antibody weakly recognizes a protein at the expected molecular weight of Kif2a at 85 kDa in CSF, but this is much stronger in the MOE, where phosphatases are inhibited by the addition of microcystin. Our interpretation is that only a small portion of Kif2a is phosphorylated in CSF, which lack centromeres. (C) Kinase assay using phosphoantibodies against pT70 shows that Aurora A in this case can phosphorylate Kif2a more efficiently than Aurora B/INCENPin box. This assay also shows that when these two kinases are mixed, the substrate is phosphorylated less than with Aurora A alone. (D) Immunofluorescence in Xenopus S3 cells shows localization of pT70 Kif2a to spindle poles during prometaphase and metaphase and to the spindle midzone during anaphase. Green, pT70; red, Ndc80 (kinetochores). Note that Kif2a localizes to centromeres in these cells (Supplemental Figure 8 in Knowlton et al., 2009), and therefore the absence of staining is not due to absence of Kif2a at these sites. (E) Sperm were added to Xenopus CSFs, and the resulting spindles were spun onto coverslips and stained for pT70. In extracts, pT70 Kif2a is localized to spindle poles and chromatin and enriched at centromeres. pT70 is dependent on Aurora activity, as this staining pattern is lost in the presence of Hesperadin. Red, pT70; blue, DNA; green, microtubules.
Depletion of Kif2a inhibits the completion of gastrulation in Xenopus laevis embryos
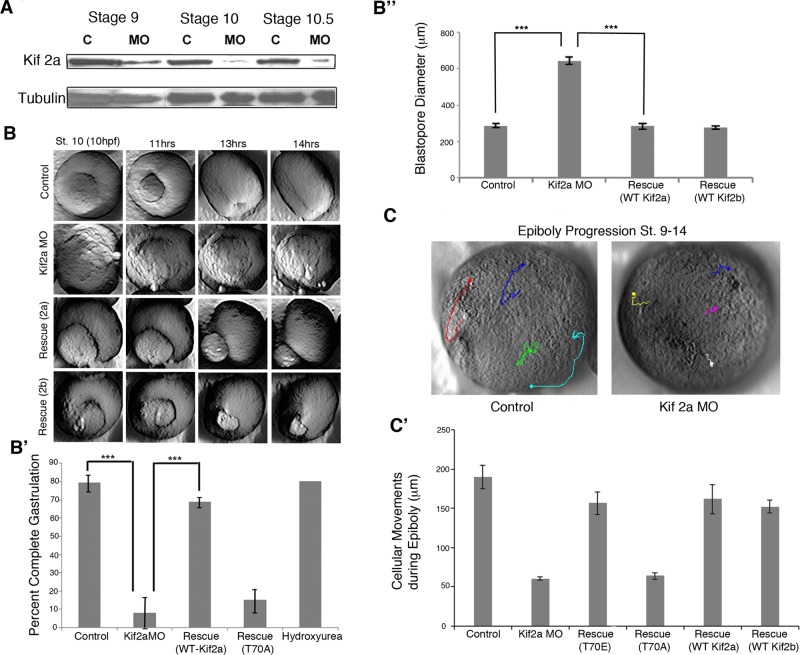
We characterized the phenotype of Kif2a knockdown as well as the role of pT70 phosphorylation in Xenopus embryos. We depleted Kif2a by injecting specific MOs in to one-cell Xenopus laevis embryos. Maternally loaded Kif2a was reduced at late blastula stages (stage 9) in MO-injected embryos, and nearly complete depletion of maternal Kif2a protein occurred by the time embryos undergo gastrulation at stage 10 (Figure 2A). The initial stages of gastrulation appeared normal in Kif2a-depleted embryos, as dorsal blastopore lips surrounded yolk plugs to generate stage 10.5 embryos with a normal external appearance. However, blastopore closure is significantly defective in Kif2a-depleted embryos, and large, round yolk plugs remain apparent at control stage 12 (Figure 2, B and B', Supplemental Figure S1A, and Supplemental Movies S1, A and B). We imaged gastrulation events of Kif2a-depleted embryos by time-lapse imaging. The movies show circumblastoporal constriction in the morphants, but it often results in exogastrulation—the expansion and extrusion of the yolk plug rather than its internalization—which suggests lack of space for the endoderm to move into the blastocoel rather than failure of convergence (and extension) of the marginal zone. Time-lapse movies of the animal cap confirmed that loss of Kif2a interferes with epiboly (Figure 2C and Supplemental Movie S2, A and B). By tracking and measuring the movement of nuclei of cells in the animal pole during gastrulation stages, we find that control embryos exhibit normal epiboly, moving vegetally from stage 9 to stage 14. Kif morphant embryos display delayed epiboly and less vegetal movement. This phenotype is recapitulated in embryos expressing the T70A mutant but rescued by injecting RNA encoding the full-length Kif2a or Kif2b or the T70E mutant (Figure 2C').
FIGURE 2:
Kif2a is required to complete gastrulation. (A) MO (Kif2a)-injected and MilliQ H2O–injected (control) embryos were collected at stages 9, 10, and 10.5, and lysates were examined by immunoblot analysis using Kif2a and tubulin antibodies. Nearly complete depletion of maternal Kif2a occurred by stage 10, and partial depletion was observed at stage 9. (B) Two cell embryos were injected with water (control), Kif2a morpholino (MO), the MO plus RNA encoding human Kif2a (Rescue 2a), or the MO plus RNA encoding human Kif2b (Rescue 2b), and progression of blastopore closure was monitored by time lapse, starting around stage 10 to stage 15. Still frames of the movie are shown at the indicated times postfertilization. (B′) Quantification of completed gastrulation in embryos injected with water (control), Kif2a morpholino (MO), the MO plus RNA encoding human Kif2a (Rescue WT Kif2a), or the MO plus RNA encoding Kif2a phospho-null mutant (T70A) or treated with hydroxyurea and aphidicolin at stage 9 (hydroxyurea). ***p < 0.001 (B′′) Blastopore diameter was measured at control stage 12 of embryos injected with Kif2a morpholino, morpholino with human Kif2a RNA (Rescue WT Kif2a), or morpholino with human Kif2b RNA (Rescue WT Kifb). ***p < 0.001. (C) Time-lapse movies of epiboly were made from stages 9–14. Representative images; the dot indicates the ending point of the track. (C′) Animal cap cells were tracked using the Manual Tracking plug-in of ImageJ (National Institutes of Health, Bethesda, MD). Quantification of the distances spread is shown for control embryos, Kif morphant embryos, and Kif MO rescues (T70A and T70E mutants and human Kif2a and 2b). n = 4.
To learn how the animal cap and vegetal endodermal defects are related, we performed targeted injections of the Kif2a MO at the 32-cell stage of development. We injected the Kif2a MO into the A-tier blastomeres to specifically target the animal cells (Animal Cap Targeted MO, Supplemental Figure S1B, Supplemental Movie S5C) or into the B1/2 and C1/2 blastomeres to target the dorsal marginal zone cells (Dorsal MZ Targeted MO, Supplemental Figure S1B, Supplemental Movie S5D). Targeting the animal cells recapitulated the whole embryo phenotype, whereas embryos gastrulated normally if the MO was targeted to the dorsal marginal zone (Supplemental Figure S1B, Supplemental Movie S5). This suggests that lack of Kif2a function in the animal cap prevents gastrulation movements on the other side of the embryo by a mechanism unique to the animal cap.
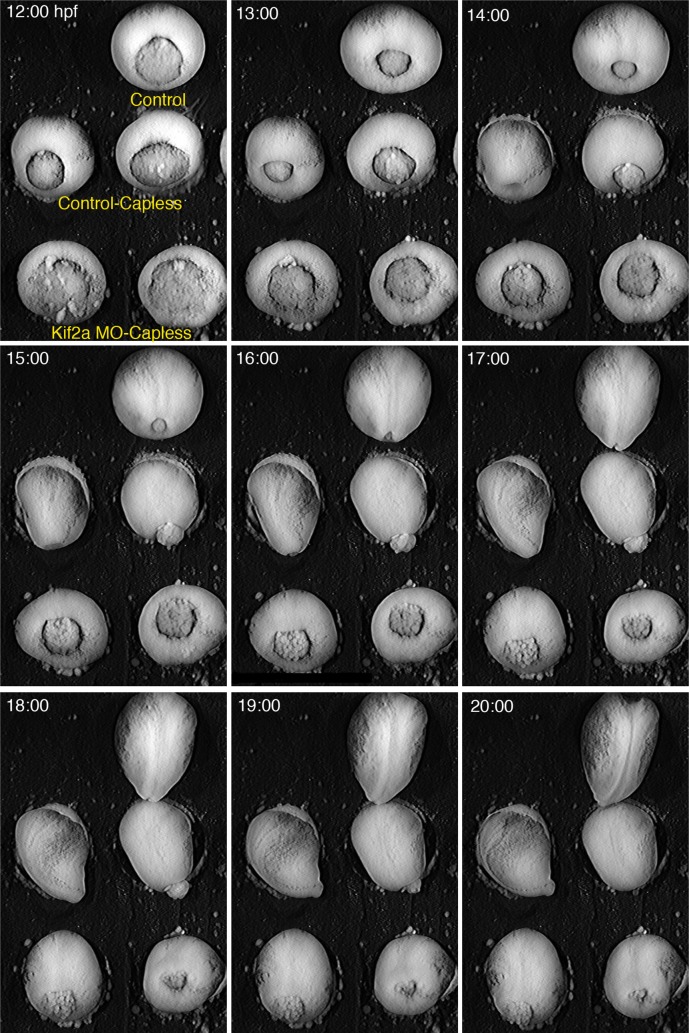
To test this hypothesis, we injected Kif2a MO into embryos at two-cell stage and then removed the animal caps and imaged the gastrulation movements. The animal cap in the Xenopus blastula and gastrula is an epithelial tissue above the blastocoel that can be easily isolated by dissection. The removal of the animal cap from Kif2a morphant embryos largely rescues blastopore closure (Figure 3 and Supplemental Movie S3). This suggests that the blastopore closure defects are due to failure of the spreading movements of epiboly of the animal cap epithelium, probably by interfering with the radial intercalation that drives it (Keller, 1980), and this, in turn, can mechanically interfere with the vegetal internalization movements during gastrulation (Keller and Jansa, 1992, Petridou et al., 2012; also see Discussion).
FIGURE 3:
Removal of animal cap tissue rescues Kif2a MO gastrulation phenotypes. Embryos were injected with water (Control) or Kif2a morpholino (Kif2aMO) at two-cell stage. At stage 8, animal caps of each treatment were removed microsurgically, and the blastopore closure was monitored through low-light time-lapse microscopy (Control-Capless, Kif2aMO-Capless). One embryo was left intact for staging purposes (Control). Failure of blastopore closure in the morphant phenotypes were rescued by removal of the cap, although closure was slightly delayed compared with the control.
Although we posit that loss of Kif2a prevents both animal cap cell division and epiboly, we do not think that the gastrulation phenotype seen after depletion of Kif2a is caused solely by a lack of cell proliferation. Blocking DNA replication by immersion of embryos into hydroxyurea and aphidicolin starting at stage 10 does not disrupt blastopore lip closure (Figure 2B and Supplemental Figure S1A; Cooke, 1973). We confirmed that proliferation was inhibited in drug-treated embryos, as visualized by larger cells (Supplemental Figure S2). These embryos gastrulated normally, indicating that a loss of cellular proliferation cannot fully account for the Kif2a-depletion phenotype; instead, disruption of radial cell polarity and intercalation is likely (see Discussion).
Xenopus Kif2a has 87% identical amino acids as human Kif2a and 53% identical amino acids as human Kif2b. Because there is no Kif2b homologue in nonmammalian vertebrates, we asked whether the phenotypes could be rescued by coinjecting RNA encoding human Kif2a or Kif2b with the MO. Both Kif2a- and Kif2b-rescued embryos continued development to tadpole stages (unpublished data). This argues strongly that Kif2a and Kif2b are redundant for their early embryonic roles. In contrast, injection of similar amounts of human Kif2A(T70A) RNA did not rescue blastopore lip closure or embryonic lethality (Figure 2B'). These data suggest that embryonic expression of Kif2a is required to complete gastrulation and that Aurora phosphorylation on T70 by Aurora B is critical for its function.
Kif2a is localized to mitotic centromeres and mitotic spindle poles in Xenopus animal cap epithelium
The fact that point mutants of Kif2a in a mitotic kinase site failed to rescue the gastrulation phenotypes argues strongly that the phenotypes are caused by defects in mitosis. We therefore examined whether there were defects in the animal cap mitoses that drove the lack of compliance of the tissue. Mitotic events can be imaged by dissecting animal cap tissues for fixation and immunofluorescence. All of the stages of mitosis are represented in a single field of view, and visualizing these spindles is comparable to tissue culture cells in fixed-cell imaging (Figure 4B and Supplemental Figure S3; Kieserman et al., 2008; Woolner et al., 2008).
FIGURE 4:
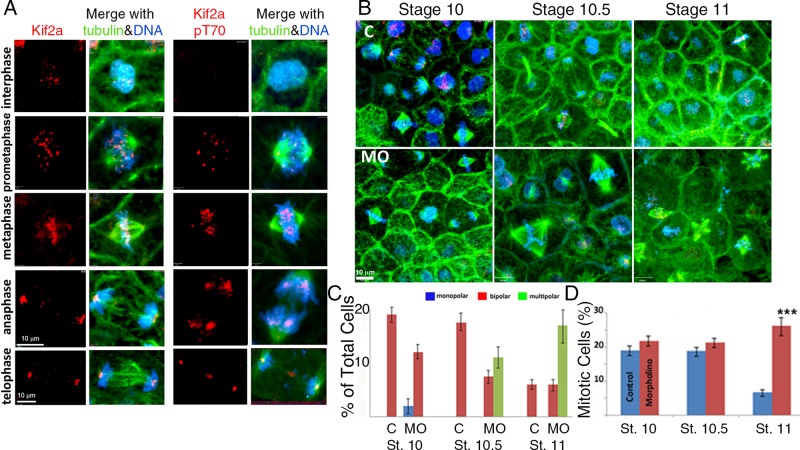
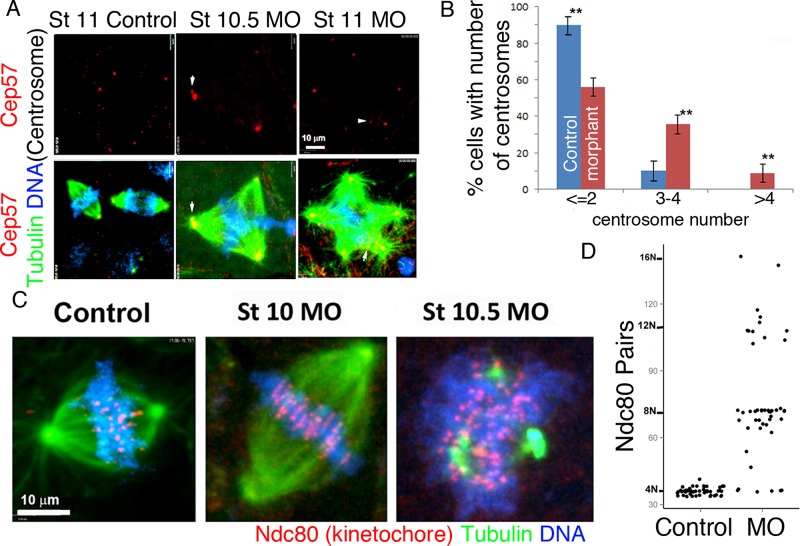
Kif2a localization and T70 phosphorylation during mitosis. Kif2a MO–treated caps at stages 10, 10.5, and 11 of development. (A) Confocal micrographs of interphase and mitotic cells in X. laevis animal caps double immunostained for Kif2a (red) and tubulin (green). DNA is stained blue with 4′,6-diamidino-2-phenylindole. For the phosphorylated form, the T70 protein is immunostained green and α-tubulin localized by a specific (DM1A) antibody red, and the DNA stains blue. During prophase and metaphase, Kif2a is localized to centromeres. During anaphase and telophase, Kif2a became localized to the spindle poles of the cell. The T70 phosphorylated form of Kif2a also localized to the centromeres during prophase and metaphase, and during anaphase, it is localized to the spindle. T-70 was also localized to the poles during metaphase, with slight localization at the poles during anaphase. In addition, pT70 exhibits strong localization to the midbody during telophase. (B) Single-cell embryos were microinjected with MilliQ H2O or the antisense MO for Kif2a (40 nl at 1 mg/ml) and allowed to develop in 0.3× MMR. At stages 10, 10.5, and 11, both groups were dissected, fixed, and stained for Kif2a (red), α-tubulin (green), and DNA (blue). There was an accumulation of multipolar spindles, with development within the Kif2a morphant groups between stages 10 and 11. (C) Quantification of cellular mitotic spindle types (bipolar vs. multipolar) within different, staged animal caps demonstrated an increase or accumulation of multipolar spindles after stage 10 within the MO-injected embryos compared with controls. No multipolar spindles were observed within controls or stage 10 MO-injected groups. (D) The percentage of animal cap cells that are undergoing mitosis and the percentage of mitotic cells within each phase of mitosis are illustrated for each stage and experimental group. ***p < 0.001. Error bars indicate SEM.
We localized Kif2a from gastrula-stage embryos by performing immunofluorescence of animal caps, using a Kif2a antibody directed to the C-terminal region of human Kif2a (Figure 4A and Supplemental Figure S4; Knowlton et al., 2009). The caps were costained to detect microtubules and DNA. During prophase, there was punctate localization of Kif2a within the nucleus (Figure 4A). During prophase and metaphase, Kif2a localized to foci within chromosomes that are in the proper location for centromeres. Embryos stained with kinetochore marker have a similar staining patterns (see Figure 7C later in this article). There is also a faint localization within the spindle poles (Figure 4, A and B). During metaphase, there was Kif2a immunoreactivity on the whole spindle in addition to the strong centromere signal. During anaphase and telophase, Kif2a localized to the spindle poles (Figure 4, A and B). These staining patterns were all lost after morpholino depletion, demonstrating specificity of the antibodies (Figure 4B). The localization of Kif2a to centromeres appears cell type specific. Kif2a is found mostly at spindle poles in HeLa cells (Ganem and Compton, 2004; Knowlton et al., 2009). However Kif2a is also found at centromeres in Xenopus extracts, Xenopus S3 cells, and animal caps (Figures 1 and 4 and Supplemental Figure S4; Knowlton et al., 2009).
FIGURE 7:
Multipolar spindles have multiple centrosomes. Multipolarity within Kif 2a morphants results in an abnormal distribution of chromosomes to daughter cells. (A) Confocal images of animal caps stained with Cep57 (red), α-tubulin (green), and DNA (blue). Cep 57 is localized to the centrosome area within bipolar (control) and multipolar (MO) spindles. Note that the multipolar spindles generated by Kif2a depletion have additional centrosomes in each pole, and occasionally a pole has two centrosomes (white arrow tip). (B) The percentages of cells with two or fewer, three or four, or more than four centrosomes were quantified for both control and Kif2a morphants. A minimum number of 100 mitotic figures were counted from >10 different caps within each (control and MO-injected) group. **p < 0.01. (C) Confocal images of control, stage 10.5, and stage 11 morphant animal cap cells. Ndc80 immunostaining is red, α-tubulin staining is green, and DNA staining is blue. (D) Scatter plot of Ndc80 paired foci. Ndc80 paired foci increase in number in Kif2a multipolar morphants, indicating an increase in cellular chromosomes. Fifty percent of the Kif 2a morphant cells (n = 50) had Ndc80 foci number in excess of the average number of control cells (36 pairs) and exhibited a diploid (2N) increase in number.
We stained animal caps with antibodies specific to Aurora A and Aurora B to determine their localization (Supplemental Figure S4). Similar to other systems, Aurora A localized to spindle poles of metaphase spindles, and Aurora B localized to centromeres and sparsely to chromosome arms. These staining patterns were not changed by Kif2a depletion (Supplemental Figure S4). In animal caps, T70 is phosphorylated at centromeres and poles in prometaphase through anaphase (Figure 4A). During telophase, pT70 localization was restricted to the spindle poles and the midbody (Figure 4A). The tight colocalization of pT70 with Aurora kinases is consistent with the phosphorylation of pT70 by both kinases in animal cap mitoses.
Kif2a regulates mitotic spindle length, and depletion generates multipolar spindles in animal cap cells
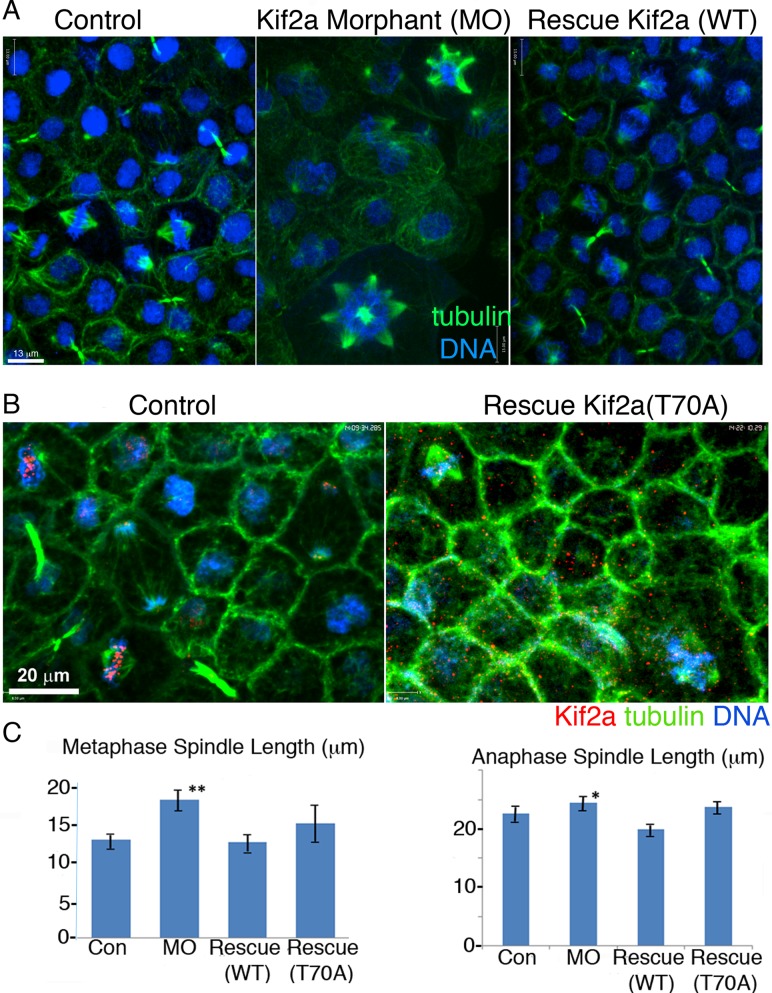
We injected MOs into single-celled embryos to determine the cellular phenotypes of Kif2a depletion. We processed animal caps from stage 9–11 embryos for immunofluorescence to visualize Kif2a, α-tubulin, and DNA (Figure 4, B–D). The Kif2a-depleted embryos exhibited longer metaphase and anaphase spindle lengths at each of the stages (Supplemental Figure S3B). The predominant phenotype seen in Kif2a depletion in cultured cells—monopolar spindles—was a minor phenotype in stage 10 animal caps. By stage 10.5, Kif2a-depleted embryos exhibited a multipolar cellular phenotype, which became even more prevalent at stage 11, when a large percentage of the mitotic figures were multipolar (Figure 4, B and C and Supplemental Figure S3, A and B). The number of mitotic cells was also largely increased in stage 11 embryos, suggesting that the multipolar spindles caused a mitotic arrest (Figure 4D). Coinjection of human Kif2a RNA with the Kif2a MO complemented bipolar spindles and the increased spindle lengths (Figure 5). We conclude that Kif2a is required to maintain proper spindle length and prevent multipolar spindles in Xenopus animal caps.
FIGURE 5:
Kif2a morphant cellular phenotypes are rescued by Kif2a but not Kif2a(T70A). (A) Confocal micrographs of control, Kif2a-MO, and WT rescue morphants (Rescue Kif2a WT) stained for α-tubulin (green) and DNA (blue). In the control and MO-rescue animal caps, spindles were bipolar, whereas within the Kif2a-MO group, multipolar spindles were abundant. (B) Confocal images of control and Kif2a morphants rescued with a phospho-null Kif RNA (Kif2a T70A) show that Kif2a is mislocalized in the absence of T70 phosphorylation. Animal caps were stained for α-tubulin (green), Kif2a (red), and DNA (blue). (C) Confocal measurements of spindle lengths were calibrated as a pole-to-pole length (note spindle lines). The stage 10.5 Kif2a morphant cells had significantly greater metaphase and anaphase spindle lengths compared with the control and rescue animal cap cells.
We rescued Kif2a depletion with mRNA expressing Kif2a(T70A) to determine whether Kif2a phosphorylation by Aurora kinases is required for proper spindle formation in animal caps. Neither increased spindle length nor multipolar spindles were rescued by RNA coding for Kif2a(T70A), whereas the wild-type protein fully rescued all phenotypes (Figure 5). We localized the Kif2a(T70A) protein after coinjection of RNA with the MO. Kif2a was no longer specifically found at centromeres or poles. We conclude that the Aurora phosphorylation of Kif2a is required to localize Kif2a to centromeres in Xenopus animal caps. Moreover, this phosphorylation is critical for Xenopus development and normal mitosis in Xenopus animal caps.
Kif2a morphant phenotypes are caused by lagging chromosomes and failure of cytokinesis
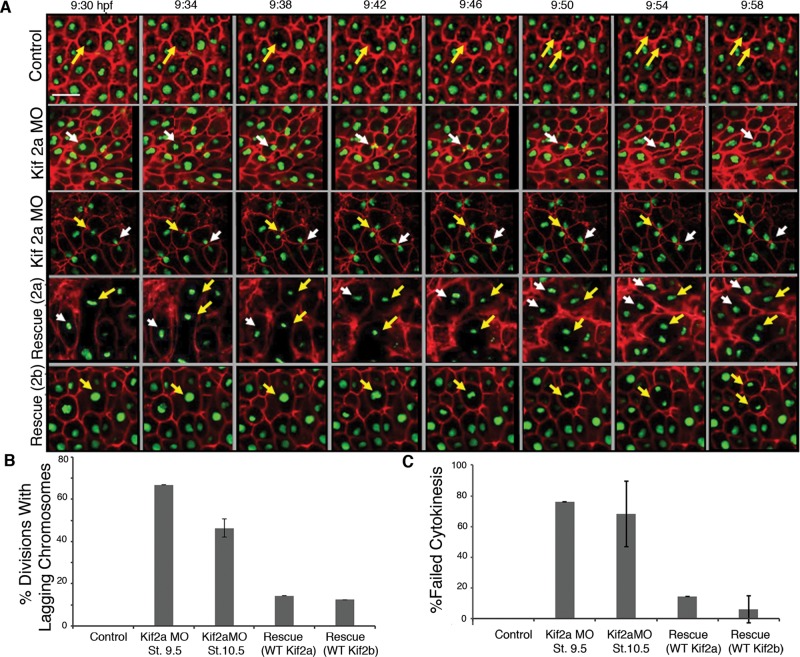
Multipolar spindles often occur because of cytokinesis failure, which generates both polyploidy and multiple centrosomes in the subsequent mitosis (Meraldi et al., 2002; Kwon et al., 2008). To determine whether Kif2a was required for cytokinesis, we injected RNA encoding green fluorescent protein (GFP)–histone H2b and red fluorescent protein (RFP)–GAP43, which acts as a plasma membrane marker, into Kif2a-depleted embryos, cut animal caps at stage 9, and imaged the layer of cells that make up the blastocoel roof by time-lapse imaging (Figure 6 and Supplemental Movie S4). In control cells, one sees faithful division of mitotic chromosomes, followed by cytokinesis between the segregated chromosomes. In >50% of the anaphase events of Kif2a-depleted embryos, we could identify chromosomes that failed to segregate and remained between the segregating masses (Figure 6, A and B). In all cases, there was a robust cytokinetic furrow that entrapped the lagging chromosomes in about half the divisions. In about half of these cells, the furrow eventually regressed, reforming a binucleate cell, and was scored as a failure in cytokinesis (Figure 6C). Cells with trapped chromosomes had a second outcome in which interphase nuclei reformed, although the cells gave the appearance that the chromatin between the cells was still attached, as they often had tear-shaped nuclei that pointed toward the sister nuclei, and these nuclei did not move to the center of the daughter cells but remained closely associated. These data demonstrate that Kif2a depletion generates anaphase-lagging chromosomes, which is likely the cause of failed cytokinesis. Moreover, note that these events happen at earlier stages than the generation of multipolar spindles.
FIGURE 6:
Kif2a morphant phenotypes caused by lagging chromosomes and failure of cytokinesis. (A) Embryos were injected at two-cell stage with GAP43 RFP:RNA and H2B:GFP RNA (Control) and coinjected with Kif2a morpholino (Kif2aMO), MO plus human 2a RNA (Rescue 2a), or MO plus human 2b RNA (Rescue 2b). The animal caps were microdissected at stage 9 and placed in an imaging chamber for confocal microscopy. A time-lapse movie was made on a Zeiss 780 Confocal Microscope with the 25× objective and a framing rate of 30 s. Still frames of these movies are shown from the indicated time postfertilization. The morphant embryos display lagging chromosomes and failure of cytokinesis (indicated by white or yellow arrows). Scale bar, 20 μm. (B) From time-lapse movies taken from stages 9–12 (similar to those described earlier), quantification of divisions with lagging chromosomes was made for control embryos, Kif2a morphants at stage 9.5, Kif2a morphants at stage 10.5, and morphants rescued with either human Kif2a or Kif2b. Bars represent the percentage of divisions imaged with lagging chromosomes. n = 20 for each category of embryo. Control embryos displayed no divisions with lagging chromosomes. Error bars, SEM. (C) Percentage of divisions imaged with failed cytokinesis. n = 20 for the categories of embryos in B. Error bars, SEM.
To corroborate our findings that Kif2a-depleted embryos failed cytokinesis, we quantified the number of centrosomes and the ploidy in Kif2a-depleted cells. Kif2a-depleted embryos were fixed at stages 10 and 10.5, and mitotic spindles in animal caps were assessed for centrosome numbers using antibodies to centriomatrix protein Cep57 (Andersen et al., 2003; Emanuele and Stukenberg, 2007). Kif2a MO–injected multipolar cells had significantly more centrosomes than control cells (Figure 7). Control cells usually had one centrosome for each half-spindle (Figure 7). In contrast, Kif2a-depleted embryos had a large number of cells with more than three centrosomes. Usually, each pole had its own Cep57 foci in the multipolar spindles generated by Kif2a depletion. Thus it is likely that the multipolar spindles that arise after Kif2a depletion are caused by multiple centrosomes.
A second marker of failed cytokinesis is increased ploidy. We measured the ploidy of cells depleted of Kif2a by counting the paired kinetochores, since there is a single kinetochore per sister chromatid in mitosis. Kinetochore numbers were quantified for 50 mitotic figures from 10 separate animal caps in both the control and MO-injected embryos, using antibodies to Ndc80 as a marker. Control caps had 36 kinetochore pairs, as expected for X. laevis, which has 36 chromosomes. Almost all mitotic cells after Kif2a depletion had >36 chromosomes, with the number usually increasing in steps of N (Figure 7B). The multipolar spindles generally had double the wild-type ploidy, suggesting that cytokinesis failure preceded multipolarity. Thus depletion of Kif2a generated cells with extra centrosomes and increased ploidy. Therefore we conclude that Kif2a is required for proper chromosome segregation and cytokinesis within Xenopus embryos.
Kif2a has a role in pole coalescence
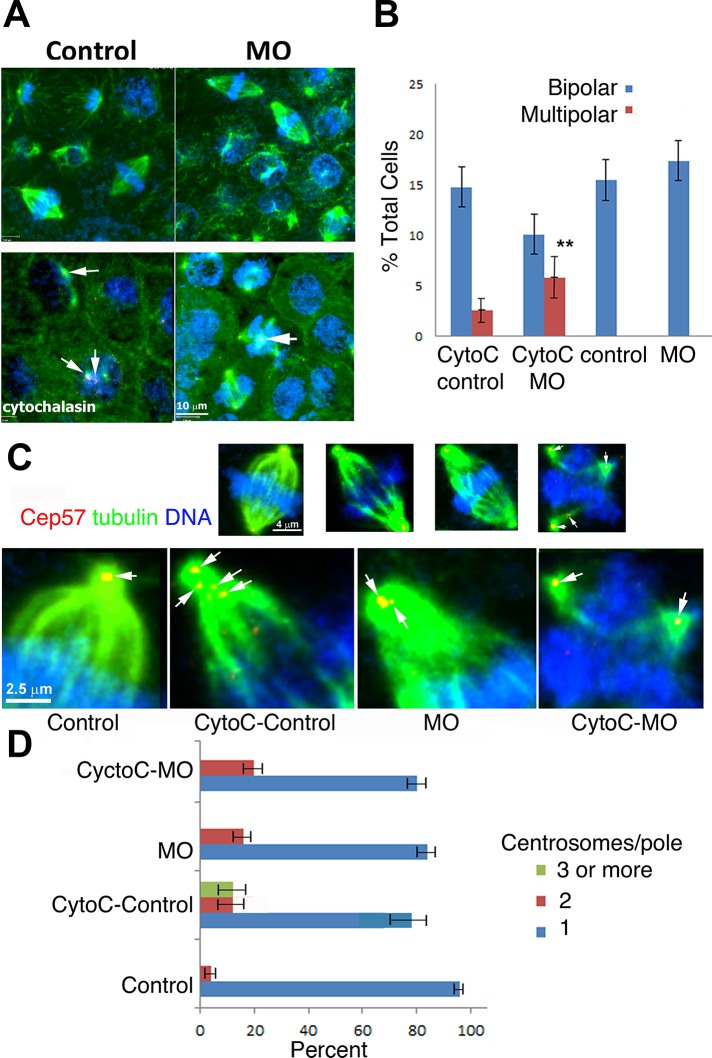
Normally, during stage 11, there are few mitotic figures, but Kif2a-depleted embryos had a high percentage of multipolar spindles. This suggests that Kif2a depletion causes multipolar spindles that cannot be resolved, generating a prolonged mitotic arrest at stage 11. Human cells with extra centrosomes first generate multipolar spindles early in mitosis and then have mechanisms to coalesce the extra poles to form a bipolar spindle (Kwon et al., 2008). Pole coalescence is required to fulfill the spindle checkpoint and segregate chromosomes (Kwon et al., 2008). We hypothesized that Kif2a-depleted cells were deficient in their ability to coalesce poles. Alternatively, it was possible that cells by stage 11 have failed several rounds of cytokinesis, generating multipolar structures that the pole-coalescing machinery cannot resolve. To distinguish between these two models, we generated multipolar spindles in stage 9 embryos (before cells normally fail cytokinesis) and measured the ability of Kif2a-depleted cells to coalesce poles in the first mitosis after cytokinesis failure. Stage 8/9 MO-injected and control embryos were treated with cytochalasin B (80 μM) for 1 h, and then the drug was removed by extensive washes into 0.3× MMR (see Materials and Methods) for 45 min. Animal caps were dissected at stage 10 and immunostained with antibodies against Cep57 and α-tubulin. As expected, disrupting F-actin blocks cytokinesis in embryos and leads to the precocious appearance of the multipolar spindles within both the control and MO-injected animal caps (Figure 8, A and B). However, multipolar spindles were more abundant within the combined cytochalasin B and MO–injected caps compared with those only treated with cytochalasin B (Figure 8B). We counted the centrosomes per pole to measure coalescence. In control cells treated with actin inhibitors, we could measure poles with more than three centrosomes, demonstrating that Xenopus embryos can coalesce poles. In contrast, we did not detect any poles with more than three centrosomes in Kif2a-depleted embryos combined with the brief cytochalasin treatment. This correlates with the increased number of multipolar spindles in Kif2a-depleted embryos. We conclude that Kif2a is required for efficient pole coalescence but note that poles with two centrosomes can be measured after Kif2a depletion, so Kif2a is not absolutely required for the process.
FIGURE 8:
Kif 2a-depleted cells poorly coalesce centrosomes. (A) Embryos were treated with 80 μM cytochalasin B at stages 8/9 and washed out of the drug after 1 h. At stage 10 embryos were fixed and imaged. Caps were immunostained for α-tubulin (green) and Cep57 (red) and stained for DNA (blue). Arrows (cytochalasin group) indicate centrosomes and multicentrosomes within cells. (B) Inhibition of cytokinesis generates multipolar spindles within stage 10 embryonic caps at a stage before their normal appearance in untreated MO-injected caps. Both cytochalasin-treated groups had multipolar spindles, but they were more abundant within the MO-injected group treated with DMSO instead of cytochalasin (control), depleted of Kif2a and DMSO (MO), control MO treated with cytochalasin B (CytoC control), or Kif2a-depleted embyros treated with cytochalasin (CytoC-MO). (C) Measuring pole coalescence after cytochalasin treatment by quantifying centrosomes at poles. Top, whole-cell micrographs; bottom, confocal micrographs, enlarged half-spindles of the same micrographs. Arrows indicate centrosomes within the spindles. (D) Quantification of C. For each group, 25 spindles from 10 caps were assessed.
DISCUSSION
We characterized the loss-of-function phenotypes of the kinesin-13 microtubule depolymerase Kif2a in Xenopus embryos. Microtubule depolymerases are a major regulator of microtubule dynamics and are used for many cellular functions (Ems-McClung and Walczak, 2010). However, their requirements for tissue dynamics are poorly understood. Our work confirmed a number of in vitro results, including that Kif2a is a major regulator of spindle size and orientation and chromosome segregation (Wilbur and Heald, 2013). We identified new roles for Kif2a in cytokinesis and spindle pole coalescence, as well as in gastrulation movements. We showed that the early embryonic functions of Xenopus Kif2a can be rescued by both human Kif2a and Kif2b, suggesting that they contain overlapping functions for early embryogenesis. Finally, we demonstrated that Aurora kinase phosphorylation on T70 is required to localize Kif2a to both poles and centromeres.
Kif2a-deficient mitoses result in failure of epiboly
Our studies demonstrate that Kif2a is required to complete gastrulation of Xenopus embryos. We show that whole-embryo Kif2a depletion results in failure of epiboly of the animal region and failure of marginal zone/endodermal internalization and blastopore closure at the opposite, vegetal end of the embryo. Surprisingly, depletion of Kif2a in the animal cap results in the whole-embryo phenotype, whereas depletion in the marginal zone—the region that normally drives the vegetal internalization and blastopore closure—has no effect. This suggests a requirement for Kif2a in animal cap epiboly that is not found in the marginal zone and also suggests that the absence of epiboly in Kif2a morphants, both in whole-embryo and animal cap depletions, mechanically interferes with vegetal gastrulation movements. We confirmed this hypothesis by removing the offending animal cap region of whole-embryo Kif2a morphants, which rescues the vegetal internalization movements. The blastocoel roof cells of intact embryos divide to form three layers—a superficial epithelium, and two deep mesenchymal layers at stage 9. The superficial cells divide and flatten, whereas the two layers of deep cells intercalate radially (along the radii of the spherical embryo, i.e., normal to the surface of the embryo) to form a single, thinner layer of greater surface area (Keller, 1978, 1980). This spreading allows the marginal zone to move vegetally over the vegetal endoderm as it generates circumblastoporal convergence forces to internalize the yolk plug and close the blastopore. Keller and Jansa (1992) reported that the animal cap is not necessary for blastopore closure. This suggests that animal cap spreading is a rate-limiting step in blastopore closure.
Mechanical effects of failed epiboly on vegetal gastrulation movements are to be expected. Epiboly of the animal cap and emboly (decrease in area) of the vegetal region with involution of the marginal zone contribute to blastopore closure and convergence and extension (reviewed in Keller and Shook, 2004). Removal of all or part of the animal cap in normal embryos results in a consistently earlier onset and faster rates of several aspects of the internalization movements, including bottle cell formation, involution, and blastopore closure (Keller and Jansa, 1992). Epiboly does not push the marginal zone vegetally but occurs at a somewhat slower rate than the internalization movements, acting as a “brake” and supplying tissue to the internalization movements, which are slightly slower than is required, thereby preventing folding or buckling of the tissue, and smoothly integrating the spreading and internalization tissue movements. Radial intercalation drives epiboly, reducing the thickness of the animal cap, allowing for more cells on the surface to spread vegetally. The data presented here show that Kif2a depletion results in cytokinesis defects. Radial intercalation involves planarly polarized divisions and directed polarized neighbor exchange to establish and maintain fewer layers of cells of greater area (Keller, 1980; Marsden and DeSimone, 2001; Petridou et al., 2012; Woolner and Papalopulu, 2012). Both behaviors are sensitive to defects in cytokinesis. The multipolar, multicentrosomal cells with persistent connections due to failed cytokinesis and lagging chromosomes in Kif2a-depleted embryos appear incompatible with the polarized cell behavior and neighbor exchange required during radial intercalation and oriented division, and the result is a thick, multilayered animal cap of small area. Because the mutants of Kif2a at a mitotic phosphorylation (T70) site fail to rescue both chromosome segregation and gastrulation defects, we argue that the mitotic defects cause the gastrulation phenotypes. Here we show that lack of Kif2a results in failed epiboly, but we do not believe that the mechanism of this failure is primarily an effect on cell division because blocking cell division with hydroxyurea and aphidicolin to produce fewer, larger cells does not block gastrulation.
Studies from Kif2a−/− mice suggest that Kif2a plays a role in directional movement of growth cones during neuron development (Harris and Hartenstein, 1991; Homma et al., 2003). However, Kif2a−/− mice gastrulate well. Our ability to rescue the gastrulation phenotypes using RNA encoding either human Kif2a or Kif2b argues strongly that these homologues share redundant functions for the mitotic roles, which we confirmed directly. We suggest that the mouse phenotypes are caused by nonoverlapping roles of the homologues. We did not follow brain development or neuronal growth cones of our embryos rescued with either human Kif2a or Kif2b to determine whether they both retained brain development functions. However, such an approach could be used to determine whether frogs could be used as a disease model for retardation.
Kif2a has multiple roles in animal cap mitoses
The first observable cellular phenotype of Kif2a injection was increased spindle length in stage 10 embryos. Average metaphase spindle lengths increased ∼1.4 times at both stages 10.5 and 11. Recently Aurora kinase regulation of Kif2a was shown to regulate the length of midzone microtubules (Uehara et al., 2013). Kif2a also controls spindle size during Xenopus development (Wilbur and Heald, 2013). It was argued that Kif2a was inhibited by increases in RanGTP activity in Xenopus eggs to generate larger spindles. Our finding that spindles are larger after loss of Kif2a in embryos is consistent with these observations. The fact that we see larger spindles in embryos depleted of Kif2a thus supports an emerging theme that mitotic spindle size is largely controlled by regulating microtubule dynamics through microtubule depolymerases and severing proteins (Levy and Heald, 2012; Whitehead et al., 2013; Wilbur and Heald, 2013).
The most obvious phenotype of Kif2a depletion in embryos is multipolar spindles, which first appear at stage 10.5 and increase at stage 11. Two possible “mechanisms” can promote multipolar spindle generation. One such method is centrosome fragmentation, by which spindles initially assemble with two poles but then split to form supernumerary poles by fragmentation of these original poles (Ehrhardt and Sluder, 2005). Second, cells with extra centrosomes generate a pole from each centrosome, and cells then have mechanisms to coalesce these poles to form bipolar spindles with multiple centrosomes in each pole (Meraldi et al., 2002; Kwon et al., 2008). To distinguish these models, we followed mitotic events by live-cell imaging. Kif2a-depleted cells often had anaphase-lagging chromosomes that became entrapped by the cytokinetic furrow. Although sometimes the cells were able to resolve these defects, they resulted in eventual regression of the cytokinetic furrow ∼50% of the time.
Kif2a had not been implicated in cytokinesis before, but this role may not have been observed because tissue culture cells arrest with monopolar spindles before cytokinesis can occur. Of interest, the few cells that escape the arrest after depletion of Kif2b fail in cytokinesis (Manning et al., 2007).
Cells in culture have robust methods to coalesce poles to transform the multipolar spindles generated by extra centrosomes to bipolar spindles (Kwon et al., 2008). Cell proliferation decreases significantly at stage 11, and thus control embryos have only 7% of cells in mitosis. However, Kif2a-depleted embryos had >25% of cells in mitosis, and most of those had multipolar spindles. This suggested that either Kif2a had an additional role in centrosome clustering or clustering is deficient in embryos compared with tissue culture systems. To distinguish these models, we generated multipolar spindles in control and Kif2a at stage 9 before Kif2a depletion caused multipolarity or cytokinesis failure at stage 10. After cytochalasin treatment, we could detect multipolar spindles and extra centrosomes in both control and Kif2a-depleted embryos, demonstrating that cytokinesis failure had occurred. However, the number of multipolar spindles was higher in the Kif2a-depleted embryos, and no pole structures contained more than two centrosomes, indicating a role for Kif2a in pole coalescence.
Regulation of kinesin-13 localization by Aurora kinases
Kinesin-13s are tightly regulated by multiple mechanisms, including Ran-regulated importin binding, interaction with the ICIS protein and EB1, and phosphorylation by polo and Aurora mitotic kinases. The N-terminus of kinesin-13 plays a primary role in targeting these kinesins to subcellular structures (Zhang et al., 2007). Phosphorylation by Aurora kinase on the N-terminus of MCAK has been shown to have a complex role in targeting MCAK to chromatin and centromeres. Phosphorylation promotes binding to chromosome arms, whereas dephosphorylation promotes centromere targeting (Zhang et al., 2007). In this study, we characterized a related site on Kif2a. The importance of this phosphorylation event was demonstrated by the fact that injection of Kif2aT70A was unable to rescue any of the observed phenotypes. Moreover, at least one of the functions of the phosphorylation became apparent by the fact that Kif2aT70A was not properly localized to either centromeres or spindle poles. It is likely that Aurora A kinase phosphorylates T70 to localize Kif2a to spindle poles, whereas Aurora B localizes to Kif2a to centromeres, since this corresponds to the predominant pool of both kinases. We previously showed that Aurora kinases can regulate microtubule depolymerase activity in both MCAK and Kif2a and localization of MCAK. Thus Aurora regulation of both activity and localization appears to be a common regulatory theme of the kinesin-13 family.
Xenopus animal caps as a system in which to study mitotic events
The study of mitosis has matured to a point at which the critical in vivo experiments are knockdown and replacements with mutants. Although Xenopus extracts are outstanding for biochemistry and generating assays that isolate complex reactions (such as self-organization of microtubules into a bipolar spindle structure), their robustness in generation of whole spindles can limit one's ability to detect subtle phenotypes. We feel that the Xenopus embryo is developing into an outstanding complement to tissue culture studies, as one can visualize subtle phenotypes in vertebrate tissues in nontransformed cells that would develop in a normal vertebrate animal (Kieserman et al., 2008; Kieserman and Wallingford, 2009; Woolner et al., 2008). The two-layer thick animal cap can be easily dissected from above the blastocoel of a Xenopus early embryo. This explant will live in a simple salt solution for >2 wk, using endogenous yolk as an energy source, and, in the absence of external factors, will develop into skin. Animal caps provide high-resolution imaging of cell division events in normal euploid cells. Moreover, they are in a tissue context with normal cell–cell interactions. Another advantage of animal caps is that the cells are rapidly dividing, with a 90- to 180-min cell cycle time (Howe et al., 1995). Animal caps will normally develop into skin, but they are pluripotent, and years of embryological experiments have determined methods to convert animal caps into a large number of tissue types. A long-term goal of this system is to exploit this pluripotency to dissect cell cycle events in different tissue types. Finally, animal caps allow us to use the set of Xenopus antibodies that we have assembled to move seamlessly between biochemistry and antibody depletions in Xenopus extracts and in vivo knockdown and rescue experiments in animal caps.
Kif2a as a cancer target
Kinesin-13s are druggable and emerging as exciting potential cancer targets. Both MCAK and Kif2a have been shown to contribute to Taxol resistance, presumably because increasing the depolymerase activity can restore more normal dynamics to Taxol-treated cells (Hedrick et al., 2008; Rizk et al., 2009). Therefore depleting kinesin-13 activity may increase the efficacy of taxanes. Our finding that Kif2a has roles in pole coalescence provides another mechanism by which Kif2a inhibitors could specifically kill cancer cells. Multiple centrosomes are often found in the cells in human tumors, making drugs that block pole coalescence an exciting new avenue for cancer therapy (Kwon et al., 2008; Godinho et al., 2009).
MATERIALS AND METHODS
Chemicals are from Sigma-Aldrich (St. Louis, MO) unless specified.
Extract and embryo preparation
Oocytes, eggs, and embryos were obtained from X. laevis females, which were injected with 800 U of human chorionic gonadatropin into the dorsal lymph sac 18 h before use. Eggs were laid into 0.1× MMR (10 mM NaCl, 0.2 mM KCl, 0.1 mM MgCl2, and 0.5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), pH 7.4, and fertilized by adding macerated testes. At 30 min after fertilization, embryos were dejellied in 2% cysteine (in 0.1× MMR, pH 8.0) and rinsed several times with 0.1× MMR. Embryos were maintained in 0.1× MMR at 14.8oC until microinjection. IE and ME were generated as described (Lan et al., 2004). MOE is a high-speed supernatant of an egg extract that preserves the mitotic state. Briefly, dejelled eggs were lysed in an equal volume of ice-cold extraction buffer, EB (80 mM b-glycerol phosphate, 20 mM EGTA, 10 mM MgCl2) + 1 µM microcystin, 1 mM ATP, and 10 mM DTT. Lysate was clarified at 256,000 × g in a T-70 rotor, and the clear fraction between the lipid and membranes was used.
Antisense morpholino, RNA preparation, and embryo microinjection
Antisense morpholino oligonucleotides (MO; Gene Tools) were generated based on Xenopus Kif2a genes [CTTCTGCTCCCCCTCCCTGTGGCTT]. A dilution to 4 ng/nl at 10-nl injection gave Kif 2a depletion and was used in all experiments. The 40 ng was injected into single- or two-celled embryos. For MO + RNA rescue experiments, 2–4 ng of in vitro–transcribed RNA (mMessage mMachine Kit; Ambion) encoding hKif2a RNA was coinjected with the Kif2a-MO mixture.
Embryos were injected at the one- and two-cell stage(s). On most occasions, controls were microinjected with MilliQ water (the morpholino diluent), but their survival was no worse than that of noninjected controls; therefore sometimes controls were not injected.
Immunofluorescence
The Kif2a, Ndc80, and Cep57 antibodies were previously described (McCleland et al., 2003; Emanuele and Stukenberg, 2007; Knowlton et al., 2009). At stage 9, control and MO-injected embryos were assessed for viability. At stages 10, 10.5, and 11, the epithelial animal cap of these embryos were dissected and fixed overnight at 4°C in MEMFA (100 mM MOPS, 2 mM EGTA, 1 mM magnesium sulfate, 4% formaldehyde) or Dent's solution (80% methanol/20% DMSO) and rotated on a nutator. They were postfixed in methanol overnight at −20°C. After postfixing, the caps were hydrated, bleached, washed several times in Tris-buffered saline (TBS; pH 7.4) with 1% SDS detergent (TBS and Tween 20 [TBST]). Next they were blocked in 10% fetal serum/5% dimethyl sulfoxide in TBS. The caps were then incubated in the first antibody (DM1α, Kif2a; 1:500 dilution) for 48 h at 4°C. After incubation in the primary antibodies, epithelial caps were washed several times in TBST, incubated in blocking solution, and then immersed overnight at 4°C with secondary (anti-mouse and anti-rabbit) antibodies. Again after many rinses in TBST, the caps were subjected to a DNA stain, dehydrated, and then cleared in Murray's solution. The caps were mounted on slides and observed and photographed under a confocal microscope. Immunofluorescence in cytostatic factor–arrested egg extracts and S3 cells were performed as previously described (Lan et al., 2004; Knowlton et al., 2009).
Embryo lysates and immunoblotting
Embryo lysates were prepared from control and Kif2a MO–injected embryos. Embryos were collected at stages 9, 10, and 10.5 for both groups. Staged embryos were homogenized in a lysis buffer (50 mM NaCl, 50 mM Tris, pH 8, 5 mM EDTA, 0.5% Triton-X, 0.5% NP-40, and protease inhibitors). They were first rinsed in lysis buffer, and then 150 μl of buffer was added to 15–20 embryos and homogenized over ice using a 100-ml pipette tip. The middle layer was carefully drawn off, avoiding the top membrane layer. This lysate was stored at −80°C until used for immunoblotting. Samples were separated by 10% SDS–PAGE, transferred to nitrocellulose, and analyzed by Western blot according to standard protocols using anti–α-tubulin (1:2500 dilution) as a standard.
Cytochalasin B treatment
Single-celled embryos were injected with Kif2a MO (4 ng/nl), and controls were not injected. For comparison purposes, Kif2a MO–injected and uninjected control embryos were not incubated in cytochalasin B. Embryos were allowed to develop to stage 8/9 and then incubated for 1 h at room temperature in 80 μM cytochalasin B in 0.3× MMR. They were washed in 0.3× MMR for 45 min, and then (stage 10) animal caps were immediately dissected, fixed, and stained for Cep57 and α-tubulin. Control animals were used to ensure that fixation occurred at stage 10.
Quantification and statistical analysis
To quantify spindle phenotypes in animal caps, we tabulated the number of bipolar metaphase spindles, multipolar spindles, and cell counts using 40× confocal micrographs of α-tubulin (DMIα antibodies) staining; >10 animal caps were analyzed for each sample. To test for a statistically significant difference between samples, we performed unpaired Student's t tests.
Supplementary Material
Acknowledgments
We thank Dan Burke for critical reading of the manuscript. The work was supported primarily by National Institutes of Health Grants GM099108, GM063045, and HD069352.
Abbreviations used:
- CSF
cytostatic factor arrested Xenopus extract
- CytoC
cytochalasin B
- IE
interphase Xenopus extract
- MO
morpholino
- MOE
mature oocyte extract
- pT70
phosphorylated threonine-70 on Kif2a
- St
stage
- T70
Kif2a threonine-70
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-12-0721) on January 7, 2015.
*These authors contributed equally to the manuscript.
REFERENCES
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow JR. Aurora B regulates MCAK at the mitotic centromere. Dev Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Carmena M, Ruchaud S, Earnshaw WC. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr Opin Cell Biol. 2009;21:796–805. doi: 10.1016/j.ceb.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke J. Morphogenesis and regulation in spite of continued mitotic inhibition in Xenopus embryos. Nature. 1973;242:55–57. doi: 10.1038/242055a0. [DOI] [PubMed] [Google Scholar]
- Ehrhardt AG, Sluder G. Spindle pole fragmentation due to proteasome inhibition. J Cell Physiol. 2005;204:808–818. doi: 10.1002/jcp.20335. [DOI] [PubMed] [Google Scholar]
- Emanuele MJ, Stukenberg PT. Xenopus Cep57 is a novel kinetochore component involved in microtubule attachment. Cell. 2007;130:893–905. doi: 10.1016/j.cell.2007.07.023. [DOI] [PubMed] [Google Scholar]
- Ems-McClung SC, Walczak CE. Kinesin-13s in mitosis: Key players in the spatial and temporal organization of spindle microtubules. Semin Cell Dev Biol. 2010;21:276–282. doi: 10.1016/j.semcdb.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaetz J, Kapoor TM. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J Cell Biol. 2004;166:465–471. doi: 10.1083/jcb.200404015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Compton DA. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J Cell Biol. 2004;166:473–478. doi: 10.1083/jcb.200404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Upton K, Compton DA. Efficient mitosis in human cells lacking poleward microtubule flux. Curr Biol. 2005;15:1827–1832. doi: 10.1016/j.cub.2005.08.065. [DOI] [PubMed] [Google Scholar]
- Godinho SA, Kwon M, Pellman D. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 2009;28:85–98. doi: 10.1007/s10555-008-9163-6. [DOI] [PubMed] [Google Scholar]
- Harris WA, Hartenstein V. Neuronal determination without cell division in Xenopus embryos. Neuron. 1991;6:499–515. doi: 10.1016/0896-6273(91)90053-3. [DOI] [PubMed] [Google Scholar]
- Hedrick DG, Stout JR, Walczak CE. Effects of anti-microtubule agents on microtubule organization in cells lacking the kinesin-13 MCAK. Cell Cycle. 2008;7:2146–2156. doi: 10.4161/cc.7.14.6239. [DOI] [PubMed] [Google Scholar]
- Homma N, Takei Y, Tanaka Y, Nakata T, Terada S, Kikkawa M, Noda Y, Hirokawa N. Kinesin superfamily protein 2A (KIF2A) functions in suppression of collateral branch extension. Cell. 2003;114:229–239. doi: 10.1016/s0092-8674(03)00522-1. [DOI] [PubMed] [Google Scholar]
- Howe JA, Howell M, Hunt T, Newport JW. Identification of a developmental timer regulating the stability of embryonic cyclin A and a new somatic A-type cyclin at gastrulation. Genes Dev. 1995;9:1164–1176. doi: 10.1101/gad.9.10.1164. [DOI] [PubMed] [Google Scholar]
- Jaillard S, Andrieux J, Plessis G, Krepischi AC, Lucas J, David V, Le Brun M, Bertola DR, David A, Belaud-Rotureau MA, et al. 5q12.1 deletion: delineation of a phenotype including mental retardation and ocular defects. Am J Med Genet A. 2011;155A:725–731. doi: 10.1002/ajmg.a.33758. [DOI] [PubMed] [Google Scholar]
- Keller M, Shook D. Gastrulation in amphibians. In: Stern C, editor. Gastrulation. New York: Cold Spring Harbor Laboratory Press; 2004. pp. 171–203. [Google Scholar]
- Keller RE. Time-lapse cinemicrographic analysis of superficial cell behavior during and prior to gastrulation in Xenopus laevis. J. Morphol. 1978;157:223–248. doi: 10.1002/jmor.1051570209. [DOI] [PubMed] [Google Scholar]
- Keller RE. The cellular basis of epiboly: an SEM study of deep-cell rearrangement during gastrulation in Xenopus laevis. J Embryol Exp Morphol. 1980;60:201–234. [PubMed] [Google Scholar]
- Keller R, Jansa S. Xenopus gastrulation without a blastocoel roof. Dev Dyn. 1992;195:162–176. doi: 10.1002/aja.1001950303. [DOI] [PubMed] [Google Scholar]
- Kieserman EK, Glotzer M, Wallingford JB. Developmental regulation of central spindle assembly and cytokinesis during vertebrate embryogenesis. Curr Biol. 2008;18:116–123. doi: 10.1016/j.cub.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Kieserman EK, Wallingford JB. In vivo imaging reveals a role for Cdc42 in spindle positioning and planar orientation of cell divisions during vertebrate neural tube closure. J Cell Sci. 2009;122:2481–2490. doi: 10.1242/jcs.042135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton AL, Vorozhko VV, Lan W, Gorbsky GJ, Stukenberg PT. ICIS and Aurora B coregulate the microtubule depolymerase Kif2a. Curr Biol. 2009;19:758–763. doi: 10.1016/j.cub.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Zhang X, Kline-Smith SL, Rosasco SE, Barrett-Wilt GA, Shabanowitz J, Hunt DF, Walczak CE, Stukenberg PT. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- Levy DL, Heald R. Mechanisms of intracellular scaling. Annu Rev Cell Dev Biol. 2012;28:113–135. doi: 10.1146/annurev-cellbio-092910-154158. [DOI] [PubMed] [Google Scholar]
- Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden M, DeSimone DW. Regulation of cell polarity, radial intercalation and epiboly in Xenopus: novel roles for integrin and fibronectin. Development. 2001;128:3635–3647. doi: 10.1242/dev.128.18.3635. [DOI] [PubMed] [Google Scholar]
- McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, Stukenberg PT. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003;17:101–114. doi: 10.1101/gad.1040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53-/- cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R, Sapra T, Howard J, Mitchison TJ. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol Biol Cell. 2004;15:2895–2906. doi: 10.1091/mbc.E04-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petridou NI, Stylianou P, Christodoulou N, Rhoads D, Guan J-L, Skourides PA. Activation of endogenous FAK via expression of its amino terminal domain in Xenopus embryos. PLoS ONE. 2012;7:e42577. doi: 10.1371/journal.pone.0042577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk RS, Bohannon KP, Wetzel LA, Powers J, Shaw SL, Walczak CE. MCAK and paclitaxel have differential effects on spindle microtubule organization and dynamics. Mol Biol Cell. 2009;20:1639–1651. doi: 10.1091/mbc.E08-09-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosasco-Nitcher SE, Lan W, Khorasanizadeh S, Stukenberg PT. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 2008;319:469–472. doi: 10.1126/science.1148980. [DOI] [PubMed] [Google Scholar]
- Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Sanhaji M, Friel CT, Wordeman L, Louwen F, Yuan J. Mitotic centromere-associated kinesin (MCAK): a potential cancer drug target. Oncotarget. 2011;2:935–947. doi: 10.18632/oncotarget.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R, Tsukada Y, Kamasaki T, Poser I, Yoda K, Gerlich DW, Goshima G. Aurora B and Kif2A control microtubule length for assembly of a functional central spindle during anaphase. J Cell Biol. 2013;202:623–636. doi: 10.1083/jcb.201302123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead E, Heald R, Wilbur JD. N-terminal phosphorylation of p60 katanin directly regulates microtubule severing. J Mol Biol. 2013;425:214–221. doi: 10.1016/j.jmb.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur JD, Heald R. Mitotic spindle scaling during Xenopus development by kif2a and importin alpha. Elife. 2013;2:e00290. doi: 10.7554/eLife.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolner S, O'Brien LL, Wiese C, Bement WM. Myosin-10 and actin filaments are essential for mitotic spindle function. J Cell Biol. 2008;182:77–88. doi: 10.1083/jcb.200804062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolner S, Papalopulu N. Spindle position in symmetric cell divisions during epiboly is controlled by opposing and dynamic apicobasal forces. Dev Cell. 2012;22:775–787. doi: 10.1016/j.devcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lan W, Ems-McClung SC, Stukenberg PT, Walczak CE. Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Mol Biol Cell. 2007;18:3264–3276. doi: 10.1091/mbc.E07-01-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.