Abstract
Malignant rhabdoid tumor (MRT), a highly aggressive cancer of young children, displays inactivation or loss of the hSNF5/INI1/SMARCB1 gene, a core subunit of the SWI/SNF chromatin-remodeling complex, in primary tumors and cell lines. We have previously reported that reexpression of hSNF5 in some MRT cell lines causes a G1 arrest via p21CIP1/WAF1 (p21) mRNA induction in a p53-independent manner. However, the mechanism(s) by which hSNF5 reexpression activates gene transcription remains unclear. We initially searched for other hSNF5 target genes by asking whether hSNF5 loss altered regulation of other consensus p53 target genes. Our studies show that hSNF5 regulates only a subset of p53 target genes, including p21 and NOXA, in MRT cell lines. We also show that hSNF5 reexpression modulates SWI/SNF complex levels at the transcription start site (TSS) at both loci and leads to activation of transcription initiation through recruitment of RNA polymerase II (RNAPII) accompanied by H3K4 and H3K36 modifications. Furthermore, our results show lower NOXA expression in MRT cell lines compared with other human tumor cell lines, suggesting that hSNF5 loss may alter the expression of this important apoptotic gene. Thus, one mechanism for MRT development after hSNF5 loss may rely on reduced chromatin-remodeling activity of the SWI/SNF complex at the TSS of critical gene promoters. Furthermore, because we observe growth inhibition after NOXA expression in MRT cells, the NOXA pathway may provide a novel target with clinical relevancy for treatment of this aggressive disease.
Introduction
Malignant rhabdoid tumor (MRT) is a rare and extremely aggressive childhood cancer, originally described as an unfavorable histologic variant of the pediatric renal Wilms’ tumor (1). The most common locations are in the kidney and central nervous system, although MRTs may arise in almost any site (2, 3). Despite significant advances in treatment, for MRTs diagnosed before the age of 6 months, patient survival at 4 years drops to approximately 8.8% (4). Therefore, improved patient outcome requires a better understanding of MRT and the development of novel therapeutic strategies. The common genetic alteration in MRTs is inactivation of hSNF5 (also known as SMARCB1, INI1, and BAF47), located in chromosome band 22q11.2 (5), which implicates the loss of hSNF5 function as the primary cause of these tumors (6). Therefore, the elucidation of hSNF5 function should lead to the identification of the key molecular steps necessary for MRT tumorigenesis.
hSNF5 is a component of the SWI/SNF chromatin-remodeling complex. SWI/SNF complexes are ATP-dependent chromatin-remodeling complexes that regulate gene transcription by causing conformational changes in chromatin structure (7). SWI/SNF subunits can be subclassified into 3 categories: (i) ATPase subunit (either BRG1 or BRM), (ii) core subunits (hSNF5, BAF155, and BAF170), and (iii) accessory subunits (BAF53, BAF57, BAF180, and others; ref. 8). How loss of the core SNF5 subunit leads to the development of a rare pediatric cancer remains one of the most challenging questions in the cancer epigenetic field.
Our previous study has shown that while hSNF5 reexpression in MRT cells increases both p21CIP1/WAF1 and p16INK4A expression during the induction of G1 cell-cycle arrest, p21CIP1/WAF1 upregulation precedes p16INK4A. Furthermore, we have showed that p21CIP1/WAF1 transcription shows both p53-dependent and -independent mechanisms of induction after hSNF5 reexpression. hSNF5 was confirmed to bind to the p21CIP1/WAF1 promoter by chromatin immunoprecipitation (ChIP) analysis (9). However, little is known about how hSNF5 activates transcription at its target promoters.
Therefore, in this study, we initially sought additional hSNF5-regulated genes to characterize the effects of hSNF5 reexpression of its target genes. We focused on hSNF5 regulation of other p53 target genes because mutations that inactivate the p53 pathway rarely appear in MRTs (10, 11). We assessed whether hSNF5 regulates the transcription of representative p53 target genes by real-time quantitative reverse transcription PCR (qRT-PCR) and Western blotting. We then clarified how hSNF5 regulates its target genes by conducting ChIP assays for hSNF5, histone modifications, and SWI/SNF complexes. Our results show that hSNF5 can regulate its target genes either through its modulation of SWI/SNF complex activity or recruitment of complementary transcriptional factors. We also establish the NOXA/PMAIP1 gene as a downstream target of hSNF5 that may provide a new therapeutic target for treatment of MRTs.
Materials and Methods
Cell culture and adenovirus infection
MCF7, A204.1, G401.6, TTC642, TM87-16, and TTC549 cells were cultured in RPMI-1640 containing 10% FBS. The MCF7, A204, and G401 cell lines came from the American Type Tissue Collection and were used within 6 months of receipt or from frozen stocks within a similar time frame. The remaining 3 cell lines came directly from their originator, Dr. Timothy Triche (Children’s Hospital Los Angeles, Los Angeles, CA). Similar time limitations were also used to maintain the identities of these cell lines. The TTC642pLKO.1 and TTC642p53KD cell lines were culture in RPMI-1640 containing 10% FBS and 1 μg/mL puromycin (Cellgro) (9). The Ad/pAdEasyGFPINI-SV+ adenoviral vectors expressing hSNF5 and coexpressing the GFP (designated Ad-hSNF5) and the Ad/pAdEasyGFP-expressing GFP (designated Ad-GFP) were previously published (9, 12, 13). To achieve infection of more than 90% cells, we infected at a multiplicity of infection (MOI) of 10 for the G401.6TG cell line, 20 for the A204.1 and TTC549 cell line, 80 for the TM87-16 cell line, and 200 for the TTC642 cell line.
Colony-forming assay
Approximately, 1 × 106 TTC642 cells were then transfected with 6 μg plasmid DNA (pcDNA3, pcDNA3-fSNF5, and pcDNA-wt-hNOXA) using Fugene 6 Reagent (Promega). At 24 hours posttransfection, each 10-cm plate was split into 8 cm × 10 cm plates and placed on selective media containing 300 μg/mL neomycin (Gibco). Two weeks after transfection, the plates were fixed with 95% ethanol and stained with Coomassie blue (50% methanol, 10% glacial acetic acid, and 1 g/L Coomasie blue). Colonies with greater than 1,000 cells were counted by visual inspection.
Protein extracts and Western blotting
Western blotting was carried out as described previously(13). Western blot analyses of proteins were conducted by using anti-NOXA (OP180; Calbiochem), anti-hSNF5 (612110, BD-Transduction Laboratories), anti-actin (A2066; Sigma), and horseradish peroxidase–conjugated anti-rabbit or anti-mouse immunoglobulin G (IgG; GE Healthcare).
RNA extraction and qRT-PCR analysis
RNA was extracted using the RNeasy Mini Kit (Qiagen) and 1 μg was used for cDNA synthesis primed with Random Primers (Invitrogen). cDNA was analyzed using TaqMan (Applied Biosystems) real-time qRT-PCR analysis with β-actin as the reference gene in each reaction. Reactions were carried out on an ABI 7900 HT sequence detection system (Applied Biosystems) and relative quantification was determined using the 2−ΔΔCt method (14, 15). The primers used the TaqMan gene expression assay primer/probe set in this study, which is described in Table 1.
Table 1.
TaqMan gene expression assay primer/probes
| Gene symbol | Gene name | Assay ID |
|---|---|---|
| ACTB | Actin, β | Hs99999903_m1 |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | Hs00355782_m1 |
| TP53 | Tumor protein p53 | Hs01034249_m1 |
| BBC3 | BCL2-binding component 3 (PUMA) | Hs00248075_m1 |
| BAX | BCL2-associated X protein | Hs00180269_m1 |
| PMAIP1 | Phorbol-12-myristate-13-acetate-induced protein 1(NOXA) | Hs00560402_m1 |
| SFN | Strati3n (14-3-3σ) | Hs00968567_s1 |
| MDM2 | Mdm2 p53-binding protein homolog | Hs01066930_m1 |
| GADD45B | Growth arrest and DNA-damage-inducible, β | Hs00169587_m1 |
Chromatin immunoprecipitation
ChIP was carried out as described by Donner and colleagues (15). Immunoprecipitation was conducted with an antibody specific to hSNF (Dr. Tony Imbalzano, University of Massachusetts School of Medicine, Amherst, MA), histone H3 trimethylation of lysine 4 (H3K4 me3; 39159; Active Motif), BRG-1 (J1; Dr. Weidong Wang, National Institute of Aging, Baltimore, MD), BAF155 (sc-10756; Santa Cruz Biotechnology), RNA polymerase II (RNAPII; 8WG16; Covance), histone H3 trimethylation of lysine 4 (H3K36 me3; ab9050; Abcam), normal rabbit IgG (sc-2027; Santa Cruz Biotechnology), normal mouse IgG (sc-2025; Santa Cruz Biotechnology), histone H3 C-terminal (39163; Active Motif), or p53 (DO-1; Calbiochem). DNA present in each immunoprecipitation was quantified by real-time qRT-PCR using gene-specific primers on an ABI 7000 sequence detection system. All expression values were normalized against input DNA or histone H3. PCR primer sequences are shown in Table 2 (15).
Table 2.
PCR primer sequences for ChIP Assays
| Gene | Site | Forward primer | Reverse primer |
|---|---|---|---|
| NOXA | −4,578 bp | GGT TGG TGT GAT TGC TTG GCC G | AGG GCT GCC TGG GAG AGC AA |
| −769 bp | ACT CAT GGC CTC GCC AAA CAT T | AGG GCT GAG CTA CCT GGG AAC G | |
| −158 bp | GCG GGT CGG GAG CGT GTC | AGA CGG CGT TAT GGG AGC GGA | |
| 43 bp | CGG GCC GGG CGT CTA GTT TC | CGC GCC AGA GAC CAC GCT TT | |
| 104 bp | CCC TGC CTG CAG GAC TGT TCG | CCC GGG AAC CTC AGC CTC CA | |
| 874 bp | AGT TTT CAG GCC AGC GCC CC | GGC CCA CAC AGA CTT CGG GC | |
| 2,573 bp | AGA GCT GGA AGT CGA GTG TGC T | TGC CGG AAG TTC AGT TTG TCT CCA | |
| *p21CIP1/WAF1 | −3,000 bp | CCGGCCAGTATATATTTTTAATTGAGA | AGTGGTTAGTAATTTTCAGTTTGCTCAT |
| −2,283 bp | AGCAGGCTGTGGCTCTGATT | CAAAATAGCCACCAGCCTCTTCT | |
| −1,391 bp | CTGTCCTCCCCGAGGTCA | ACATCTCAGGCTGCTCAGAGTCT | |
| −20 bp | TATATCAGGGCCGCGCTG | GGCTCCACAAGGAACTGACTTC | |
| 182 bp | CGTGTTCGCGGGTGTGT | CATTCACCTGCCGCAGAAA | |
| 507 bp | CCAGGAAGGGCGAGGAAA | GGGACCGATCCTAGACGAACTT | |
| 2,786 bp | GCACCATCCTGGACTCAAGTAGT | CGGTTACTTGGGAGGCTGAA | |
| 4,001 bp | AGTCACTCAGCCCTGGAGTCAA | GGAGAGTGAGTTTGCCCATGA |
NOTE:
p21 primers were based on ref. (15).
Results
The effects of reexpression of hSNF5 on p53 target genes in MRT cell lines
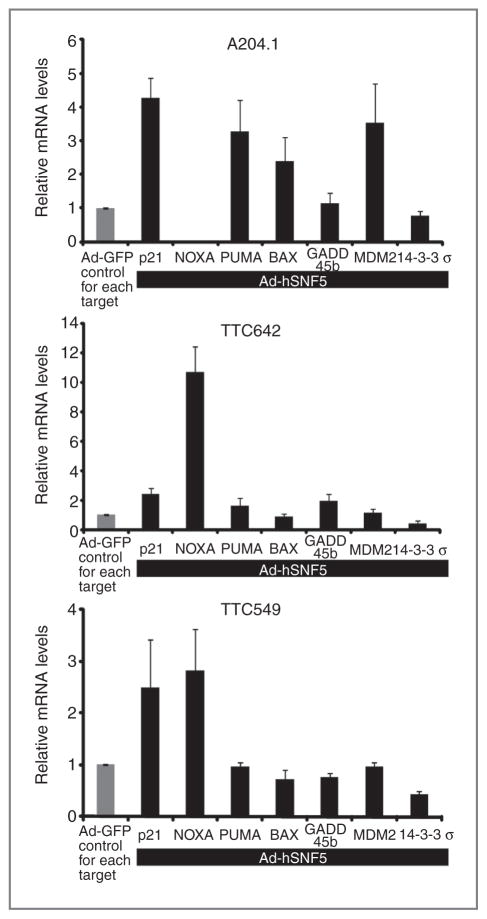
To determine whether hSNF5 regulates additional p53 target genes, we analyzed the change of mRNA levels using real-time qRT-PCR in 3 MRT cell lines, A204.1, TTC642, and TTC549, at 24 hours after infection with Ad-hSNF5 and Ad-GFP (negative control) adenoviruses [Kuwahara and colleagues (9)]. Reexpression of hSNF5 differentially modified expression of subsets of p53 target genes among the MRT cell lines (Fig. 1). All 3 cell lines showed modest to robust induction of p21CIP1/WAF1 expression and decreased expression of 14-3-3σ. However, expression of proapoptotic genes varied. While the A204.1 cell line showed induction of both PUMA and BAX expression, the other 2 cell lines showed only NOXA expression (Fig. 1). The expression of the remainder of the genes also diverged among the lines with increased MDM2 expression in A204.1 and higher GADD45b expression in TTC642. These results suggest that inactivation of hSNF5 does not recapitulate p53 loss in MRT cell lines.
Figure 1.
hSNF5-induced p53 target genes’ expression. RNA was extracted at 24 hours after infection with Ad-hSNF5 and Ad-GFP. The mRNA levels were measured for each gene by real-time qRT-PCR and normalized for β-actin expression and relative to the Ad-GFP for each genes. Values are the mean of 3 independent experiments; bars, ± SD.
hSNF5-induced NOXA mRNA and protein expression in MRT cell lines
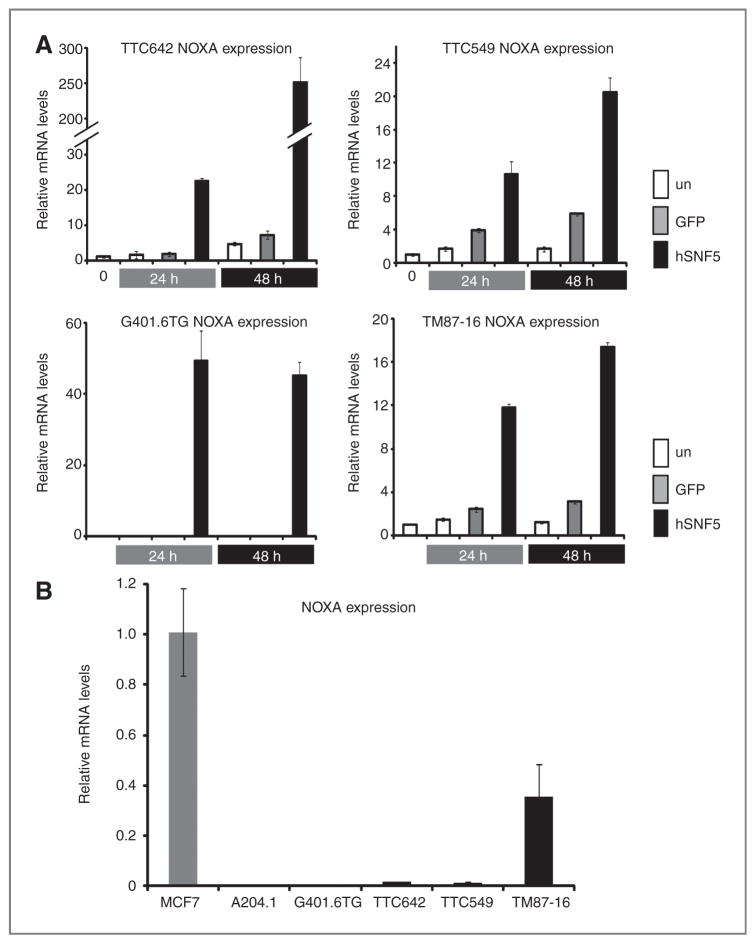
Intriguingly, our real-time qRT-PCR analysis showed that hSNF5 reexpression caused an increase in NOXA mRNA in the TTC642 and TTC549 cell lines. This result suggested that NOXA might be a common downstream target of the hSNF5 protein in MRT cell lines. To test this notion, we examined NOXA mRNA levels by real-time qRT-PCR in 2 additional MRT cell lines (TM87-16 and G401.6TG) after hSNF5 reexpression. We found the level of NOXA mRNA increased 24 hours after Ad-hSNF5 infection in comparison with Ad-GFP infection in the TM87-16 MRT cell line followed by a marked increase at 48 hours after infection. In the G401.6TG cell line, although basal expression on NOXA mRNA is not detected, reexpression of hSNF5 induced NOXA mRNA within 24 hours (Fig. 2A). We next evaluated the basal NOXA mRNA expression level in all MRT cell lines by real-time qRT-PCR. We used 6 MRT cell lines as well as the MCF7 human breast carcinoma cell line. We found significantly lower levels of NOXA mRNA in the MRT cell lines compared with MCF7 except for the TM87-16 cell line with no NOXA mRNA expression in the A204.1 and G401.6TG cell lines (Fig. 2B). Furthermore, we tested whether NOXA protein levels increased after Ad-hSNF5 infection in comparison with Ad-GFP infection. We observed that the level of NOXA protein increased at 24 hours after Ad-hSNF5 infection in comparison with Ad-GFP infection in 2 MRT cell lines (TTC642 and TTC549), followed by a marked increase at 48 hours after infection (Fig. 3A). Finally, we asked whether NOXA expression might contribute to the potent growth arrest induced by hSNF5. Using a colony-forming assay, we found that both hSNF5 and NOXA expression in the TTC642 cell line gave similar levels of growth inhibition (Fig. 3B).
Figure 2.
hSNF5-induced NOXA mRNA expression. A, RNA was extracted at the indicated times after infection with Ad-hSNF5 and Ad-GFP. The mRNA levels were measured for each gene by real-time qRT-PCR and normalized for β-actin expression. Values are the mean of 3 independent experiments; bars, ±SD; un, uninfected control. B, NOXA mRNA expression in 6 MRT cell lines (A204.1, TTC642, G401.6TG, TTC549, TM87-16, and TTC549) by real-time qRT-PCR. The MCF7 cell line was used as a control. The NOXA mRNA levels were measured for each gene by real-time qRT-PCR and normalized for β-actin expression. Values are the mean of 3 independent experiments; bars, ±SD.
Figure 3.
hSNF5-induced NOXA protein expression. A, cells were harvested at the indicated times after infection with Ad-hSNF5 and Ad-GFP. Total cell protein (30 μg) were separated on a 4% to 20% SDS-PAGE and probed with either anti-SNF5, anti-β-actin, or anti-NOXA. un; uninfected control. B, cells were transfected with the designated plasmids and selected in neomycin for 2 weeks. The results represent the colony numbers from 3 independent experiments; bars, ±SD.
Recruitment of hSNF5 on NOXA locus correlates with NOXA transcription in MRT cell lines
We analyzed the chromatin status at NOXA promoter in both TTC642 and TTC549 cell at 24 hours after Ad-hSNF5 infection to clarify the mechanism of NOXA activation by hSNF5. To analyze the recruitment of hSNF5 and other factors associated with gene transcription, we made sets of primers for the NOXA promoter regions from −4,758 to +2,573 (Fig. 4A). Our ChIP data confirmed that hSNF5 bound within 1 kb of the transcription start site (TSS) with maximal enrichment at the TSS in both cell lines (Fig. 4B). Furthermore, a modest increase of BRG1 and BAF155 (~2×) also appeared across the promoter region after hSNF5 induction of NOXA expression in the TTC642 cell line (Fig. 4C). We next determined the effect of hSNF5 reexpression on the H3K4me3, a chromatin mark associated with gene activation. H3K4me3 increased after hSNF5 reexpression at TSS with the maximal peak near the TSS in both the TTC642 (Fig. 4C). These results showed that lower levels of NOXA mRNA expression in MRT cell lines correlates with the absence of hSNF5 expression. This finding suggests that loss of hSNF5 expression might lead to an epigenetic modification at the NOXA promoter altering transcriptional activity.
Figure 4.
Recruitment of hSNF5, SWI/SNF complexes, and RNAPII to the NOXA locus and histone modification on NOXA locus after hSNF5 reexpression. A, schematic of the NOXA locus indicating the 1 p53-binding sites and overall gene structure. Primers used in real-time qRT-PCR of ChIP-enriched DNA are named according to their relative distance (bp) to the TSS. B–E, at 24 hours after infection with Ad-hSNF5 and Ad-GFP, protein was extracted for ChIP assays. ChIP assays were conducted using antibodies directed against hSNF5 (B), BRG-1 (C), BAF155 (C), H3K4me3 (C), H3K36me3 (C), RNAPII (D), and p53 (E) on indicated site of NOXA promoter. Values are the mean of duplicate or triplicate; bars, ±SD. F, RNA was extracted at 24 hours after infection with Ad-hSNF5 and Ad-GFP. The mRNA levels were measured for each gene by real-time qRT-PCR and normalized for β-actin expression. Values are the mean of 3 independent experiments; bars, ±SD; un; uninfected control; IP, immunoprecipitation.
Reexpression of hSNF5 induces p21CIP1/WAF1 and NOXA transcription accompanied with SWI/SNF complex recruitment
In our previous report, we showed that hSNF5 directly controlled p21CIP1/WAF1 transcription activity. However, the mechanism of p21CIP1/WAF1 and NOXA transcription activity induced by hSNF5 has not yet been clarified. To investigate the occupancy of reexpressed hSNF5 on the p21CIP1/WAF1 locus, we first conducted a ChIP assay for hSNF5, BAF155, and BRG-1 at 24 hours after infection of Ad-hSNF5 compared with Ad-GFP infection. We made sets of primers for the p21CIP1/WAF1 promoter regions from −3,000 to +4,001 as previously described (ref. 15; Fig. 5A). We found that reexpressed hSNF5 binds within 1 kb of TSS with maximal enrichment site at TSS in both A204.1 and TTC642 cell lines (Fig. 5B). In contrast, BRG-1 and BAF155 levels increased on p21CIP1/WAF1 locus throughout the whole region (Fig. 5B). These results obtained from p21CIP1/WAF1 locus were congruent with those obtained from the NOXA locus (Fig. 4). These observations suggest that hSNF5 might bind near the TSS and either recruit other SWI/SNF complex contents such as BRG-1 and BAF155 to the target gene locus or re-activate an existing SWI/SNF complex to initiate gene transcription.
Figure 5.
Recruitment of hSNF5, SWI/SNF complexes, RNAPII, and p53 to the NOXA locus and histone modification on NOXA locus after hSNF5 reexpression. A, schematic of the p21 locus indicating the 2 p53-binding sites (p53-HABS: high-affinity p53-binding site and p53-LABS: low-affinity p53-binding site) and overall gene structure. Primers used in real-time qRT-PCR of ChIP-enriched DNA are named according to their relative distance (bp) to the TSS. B and C, at 24 hours after infection with Ad-hSNF5 and Ad-GFP, protein was extracted for ChIP assays. ChIP assays were conducted using antibodies directed against hSNF5 (B), BRG-1 (B), BAF155 (B), H3K4me3 (C), H3K36me3 (C), and RNAPII (C) on indicated site of p21 promoter. Values are the mean of duplicate or triplicate; bars, ±SD.
Reexpression of hSNF5 induces p21CIP1/WAF1 and NOXA transcription accompanied with RNAPII recruitment and histone modification
To assess the effects of reexpressed hSNF5 on known steps of transcriptional activation, we conducted ChIP assays for RNAPII, H3K4me3, and H3K36me3 on the p21CIP1/WAF1 and NOXA loci at 24 hours after infection of Ad-hSNF5 compared with Ad-GFP infection. Our ChIP results for RNAPII showed an increase at the TSS on both p21CIP1/WAF1 and NOXA locus after hSNF5 reexpression with maximal enrichment site at TSS (Figs. 4D and 5C). The RNAPII occupancy pattern was similar to the hSNF5 occupancy pattern. Moreover, the H3K4me3-binding pattern on p21CIP1/WAF1 locus after hSNF5 reexpression showed an increase in binding at the TSS with a maximal peak near the TSS (Fig. 5C). These results support the H3K4me3 ChIP results of obtained from the NOXA experiments (Fig. 4C). Because increased H3K4me3 binding correlate with promoter activation (16), our results strongly suggest that hSNF5 reexpression leads to the transcriptional activation by inducing the initiation step with recruitment of RNAPII. Furthermore, H3K36me3 levels also increased downstream of the TSS in the p21CIP1/WAF1 and NOXA promoters (Figs. 4C and 5C). These results imply that reexpression of hSNF5 presumably precedes the transcription elongation step (17).
p53 is not required for hSNF5-induced transcriptional activity on the NOXA and p21CIP1/WAF1 promoters
We next determined whether hSNF5 recruitment to the NOXA promoter affected p53 binding. We conducted a ChIP assay for p53 binding at these promoters in the TTC642 and TTC549 cell lines. We assessed p53-binding site on the NOXA promoter at −158 bp, the consensus p53-binding site (18), and did not observe binding after hSNF5 reexpression (Fig. 4E). In our previous study, we determined that upregulation of p21CIP1/WAF1 transcription by hSNF5 operated through a p53-dependent mechanism in A204.1 cells but through a p53-independent mechanism in TTC642 cells (9). To determine p53 dependency for NOXA transcription, we used 2 previously characterized p53 stable knockdown MRT cell lines from TTC642 cells and a negative control cell line (pLKO.1; ref. 9). Infection of the pLKO.1 and p53KD cells with Ad-GFP did not alter levels of NOXA mRNA 24 hours after infection compared with the uninfected parental cell lines (Fig. 4F). However, we found an increase of NOXA mRNA after hSNF5 reexpression that was not significantly different among the TTC642, TTC642 pLKO.1, and TTC642 p53KD cells (Fig. 4F). These observations suggest that hSNF5 binding could initiate transcription on the p21CIP1/WAF1 and NOXA promoters in the TTC642 cell line without additional p53 recruitment.
Discussion
The objective of this study was to identify additional genes, besides p21WAF/CIP1, regulated by both hSNF5 and p53 in MRT cells and to determine the mechanism of hSNF5 regulation of their transcriptional activity. While hSNF5 could regulate expression of varying p53 target genes among the MRT cell lines, NOXA was the only common target gene in 5 of 6 cell lines. We then used ChIP analyses of the promoter regions of the p21WAF/CIP1 and NOXA genes to understand how hSNF5 regulates their expression. Our results provide major insights into the control of hSNF5 target genes and how hSNF5 and the SWI/SNF complex functions in the regulation of their transcription.
Recently, Lee and colleagues showed hSNF5 is the only gene recurrently mutated at a high frequency in MRTs (19). This finding implicates MRTs as true “epigenetically driven” cancers and indicates that downstream target genes of hSNF5 must drive tumorigenesis for this cancer. Because the patterns of epigenetic changes, especially those associated with chromatin landscape, can vary from cell to cell, it does not seem surprising that we observed a slightly different pattern of gene expression after restoration of hSNF5 expression in each MRT cell line. Our results also indicate that hSNF5 may regulate a subset of p53 target genes such as p21CIP1/WAF1 and NOXA in MRT cell lines but generally through a p53-independent mechanism.
Intriguingly, we observed an increase in NOXA mRNA and protein levels after reexpression of hSNF5 in most MRT cell lines. Our ChIP assays also showed that reexpression of hSNF5 increased NOXA transcriptional activity through the recruitment of BRG1 and RNAPII to the NOXA promoter. This is the first report that hSNF5 regulates NOXA transcriptional activity and establishes NOXA as a clinically relevant hSNF5 target gene. NOXA was initially identified as a phorbol ester-responsive gene (20). The NOXA protein, containing a Bcl2 homology domain 3 (BH-3), has been previously implicated in apoptosis associated with DNA damage, hypoxia, or exposure to inhibitors of the proteasome (21). Apoptosis-associated NOXA activation is primarily achieved through transcriptional upregulation through a number of transcription factors including p53 and Myc (20). Because NOXA binds with high affinity to the antiapoptotic Bcl-2 family members Mcl-1, it seems to act as a mediator of apoptosis in cells showing a dependency on Mcl-1 expression (22, 23). Furthermore, some reports showed that NOXA and Mcl-1 control sensitivity to some chemotherapeutics agents, such as Taxol, vincristine, and platinum-based drugs (24, 25).
MRTs have a chemoresistance character such that chemotherapy does not prove an effective treatment for most patients (26). To resolve the chemoresistance character of MRTs, some candidate drugs such as EGF receptor (EGFR) kinase inhibitors, HER2 inhibitors, fenretinide, histone deacetylase (HDAC) inhibitors, and flavopilidol have been described (27–32). However, a promising effective therapy has not yet been established. Nocentini reported that perturbation of the p53 pathway and a reduced apoptotic response in MRT cell lines might contribute to their resistance to chemotherapies (33). They suggested that the lack of positive correlation between an increase in the Bax/Bcl2 ratio and cell death constitutes an abnormality in the control of apoptosis. In this study, we showed that NOXA expression in MRT cell lines inhibited growth as efficiently as hSNF5 reexpression. Although several reports have shown NOXA as a positive growth regulator, taken together, these data implicate NOXA expression as a contributor to the strong growth inhibition induced by hSNF5 reexpression (34, 35). Alternately, hSNF5 loss may result in a failure to regulate NOXA expression in the case of DNA damage by some chemotherapy agents such as doxorubicin. Additional studies will help elucidate the function of NOXA in MRT development.
Our results also showed the function of hSNF5 in the context of SWI/SNF complex. Some researchers have reported that reexpressed hSNF5 appeared at 1 or multiple sites within the promoters of several genes (13, 36–38). In our previous report, we also showed hSNF5 binds at a point close to the TSS on the p21CIP1/WAF1 and p16 promoters by ChIP assay (9). However, the pattern of hSNF5 occupancy of target gene promoters has remained unclear. Our results showed hSNF5 appeared on the promoter region with maximal enrichment site at the TSS. Kia and colleagues reported that on p16 promoter, reexpressed hSNF5 binds more at −0.3 kb and +85 bp site than other upstream and downstream sites (13, 36, 37). Our results with the p21CIP1/WAF1 and NOXA promoters provide strong evidence that hSNF5 is generally associated with the transcriptional activity at TSS on its target genes. In contrast, BRG-1 and BAF155 occupancy increased equally from downstream to upstream on p21CIP1/WAF1 and NOXA locus after reexpression of hSNF5. Therefore, our results indicate a discrepancy between hSNF5 occupancy and BRG-1 occupancy after hSNF5 reexpression. A more detailed analysis using ChIP-seq to identify the binding sites of the hSNF5 and the other SWI/SNF complex members will resolve this question.
Does this recruitment of the SWI/SNF complex lead to a change in the histone modification in their target genes? In our previous study, we found that H3K4me3 decreased at the p53-binding sites in the p21CIP1/WAF1 promoter (−2,283 and −1,391 kb) after hSNF5 reexpression, although p21CIP1/WAF1 transcription increased (9). Previous studies have concluded that nucleosomes with H3K4me3 are associated with actively transcribed genes in various eukaryotes (39–41). Guenther and colleagues also found H3K4me3 enriched within 1 kb of known TSS with maximal enrichment downstream of the TSS (42). Our results showed H3K4me3 increased at or near the TSS in the p21CIP1/WAF1 and NOXA promoters, similar to the results from Guenther and colleagues’ report. Because the transcriptional activity changes reflected the occupancy of H3K4me3, it is unclear whether the methylation of H3K4 is primary or secondary effects of hSNF5 reexpression. However, Lee and colleagues indicated that the C-terminal SET domain of H3K4 methyltransferase (MLL3 and MLL4) directly interacts with hSNF5 (43). The fact that H3K4me3 occupancy is likely linked to hSNF5 occupancy suggests that reexpressed hSNF5 potentially interacted with a H3K4 methyltransferase. In contrast, our H3K36me3 ChIP data showed that H3K36me3 occupancy was detected in transcribed regions of the p21CIP1/WAF1 and NOXA genes, peaking toward the 3′-end (39–41, 44). Our results concur with several earlier reports showing (39–41, 44) that H3K36me3 occupancy tracks within the body of transcriptionally active genes and associates with transcriptional elongation activity. These histone modifications after hSNF5 reexpression indicate that hSNF5 can regulate the transcription initiation followed by elongation activity in MRT cells.
We also showed that RNAPII occupancy increased after hSNF5 reexpression on p21CIP1/WAF1 and NOXA promoter. Guenther and colleagues found that most promoters (98%) occupied by RNAPII were also occupied by H3K4me3, whereas RNAPII occupied few genes (2%) that lack H3K4me3 (42, 45). Other researchers have shown that components of H3K4 methyltransferase complexes interact with the Ser5-phosphorylated form of RNAPII, indicating that transcription initiation coincides with H3K4me3 deposition (45). Similar studies showed that H3K4me3 modification occurs subsequent to RNAPII recruitment and Ser5 phosphorylation of RNAPII C-terminal domain (16, 40, 46). Indeed, we also showed that the occupancy of RNAPII follows a similar pattern for H3K4me3 occupancy. The increase of H3K4me3 occupancy in the p21CIP1/WAF1 and NOXA promoters may result from recruitment of H3K4 methyltransferase and RNAPII by hSNF5 reexpression.
However, H3K4me3 was still detected at the inactive NOXA promoter in the absence of hSNF5 with a low occupancy of RNAPII. Then, after reexpression of hSNF5, both RNAPII and H3K4me3 immediately increased on or near the TSS. Vastenhouw and colleagues indicated that many nonexpressed genes in embryonic stem cells also carry only H3K4me3 marks (44). They also showed most of these H3K4me3 domains are not associated with detectable levels of RNAPII, and H3K4me3 marks might be established in the absence of sequence-specific activators and without the stable association of RNAPII. These H3K4me3 domains might be paused genes for activation by creating a platform for the transcriptional machinery (12). Our data suggest that the inactive NOXA gene in MRT cells might be paused by loss of hSNF5 and reexpression of hSNF5 can release the pausing followed by recruitment of RNAPII. On the other hand, because transcriptional activity appears at the p21CIP1/WAF1 promoter, it has both H3K4me3 and RNAPII occupancy before reexpression of hSNF5. Taken together, hSNF5 can activate a transcription initiation step by recruitment of RNAPII accompanied with H3K4me3 nucleosome modification.
In conclusion, our results show that hSNF5 reexpression in MRT cells increases both p21CIP1/WAF1 and NOXA expression. Reexpression of hSNF5 leads to activation of transcription initiation by either recruitment of SWI/SNF complexes or activation of existing ones. Increased RNAPII binding at the TSS accompanied with H3K4 and H3K36 modifications follows. These results raise the exciting possibility that one mechanism for epigenetic changes caused by hSNF5 inactivation involves changes in promoter pausing. Furthermore, because MRT cells display repressed NOXA transcription activity due to hSNF5 loss, reexpression of the NOXA pathway might prove a promising new paradigm to treat MRT in the near future.
Acknowledgments
Grant Support
This work was supported by Public Health Service grants R01 CA91048 (to B.E. Weissman) and T32 ES007126 (to D. Wei) and a Grant-in-Aid for Research Activity start-up 22890157 by Japan Society for the Promotion of Science (to Y. Kuwahara).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
Conception and design: Y. Kuwahara, B.E. Weissman
Development of methodology: Y. Kuwahara, B.E. Weissman
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): Y. Kuwahara, D. Wei, J. Durand
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Y. Kuwahara, D. Wei, J. Durand, B.E. Weissman
Writing, review, and/or revision of the manuscript: Y. Kuwahara, D. Wei, J. Durand, B.E. Weissman
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): D. Wei, B.E. Weissman
Study supervision: B.E. Weissman
References
- 1.Beckwith JB, Palmer NF. Histopathology and prognosis of Wilms tumors: results from the First National Wilms’ Tumor Study. Cancer. 1978;41:1937–48. doi: 10.1002/1097-0142(197805)41:5<1937::aid-cncr2820410538>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 2.Biegel JA, Tan L, Zhang F, Wainwright L, Russo P, Rorke LB. Alterations of the hSNF5/INI1 gene in central nervous system atypical teratoid/rhabdoid tumors and renal and extrarenal rhabdoid tumors. Clin Cancer Res. 2002;8:3461–7. [PubMed] [Google Scholar]
- 3.Hoot AC, Russo P, Judkins AR, Perlman EJ, Biegel JA. Immunohistochemical analysis of hSNF5/INI1 distinguishes renal and extra-renal malignant rhabdoid tumors from other pediatric soft tissue tumors. Am J Surg Pathol. 2004;28:1485–91. doi: 10.1097/01.pas.0000141390.14548.34. [DOI] [PubMed] [Google Scholar]
- 4.Tomlinson GE, Breslow NE, Dome J, Guthrie KA, Norkool P, Li S, et al. Rhabdoid tumor of the kidney in the National Wilms’ Tumor Study: age at diagnosis as a prognostic factor. J Clin Oncol. 2005;23:7641–5. doi: 10.1200/JCO.2004.00.8110. [DOI] [PubMed] [Google Scholar]
- 5.Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–6. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 6.Biegel JA, Kalpana G, Knudsen ES, Packer RJ, Roberts CW, Thiele CJ, et al. The role of INI1 and the SWI/SNF complex in the development of rhabdoid tumors: meeting summary from the workshop on childhood atypical teratoid/rhabdoid tumors. Cancer Res. 2002;62:323–8. [PubMed] [Google Scholar]
- 7.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–92. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 8.Weissman B, Knudsen KE. Hijacking the chromatin remodeling machinery: impact of SWI/SNF perturbations in cancer. Cancer Res. 2009;69:8223–30. doi: 10.1158/0008-5472.CAN-09-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuwahara Y, Charboneau A, Knudsen ES, Weissman BE. Reexpression of hSNF5 in malignant rhabdoid tumor cell lines causes cell cycle arrest through a p21(CIP1/WAF1)-dependent mechanism. Cancer Res. 2010;70:1854–65. doi: 10.1158/0008-5472.CAN-09-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosson GB, Hazen-Martin DJ, Biegel JA, Willingham MC, Garvin AJ, Oswald BW, et al. Establishment and molecular characterization of five cell lines derived from renal and extrarenal malignant rhabdoid tumors. Mod Pathol. 1998;11:1228–37. [PubMed] [Google Scholar]
- 11.Xu J, Erdreich-Epstein A, Gonzalez-Gomez I, Melendez EY, Smbatyan G, Moats RA, et al. Novel cell lines established from pediatric brain tumors. J Neurooncol. 2012;107:269–80. doi: 10.1007/s11060-011-0756-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reincke BS, Rosson GB, Oswald BW, Wright CF. INI1 expression induces cell cycle arrest and markers of senescence in malignant rhabdoid tumor cells. J Cell Physiol. 2003;194:303–13. doi: 10.1002/jcp.10201. [DOI] [PubMed] [Google Scholar]
- 13.Chai J, Charboneau AL, Betz BL, Weissman BE. Loss of the hSNF5 gene concomitantly inactivates p21CIP/WAF1 and p16INK4a activity associated with replicative senescence in A204 rhabdoid tumor cells. Cancer Res. 2005;65:10192–8. doi: 10.1158/0008-5472.CAN-05-1896. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell. 2007;27:121–33. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–11. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 17.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol. 2002;22:1298–306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikiforov MA, Riblett M, Tang WH, Gratchouck V, Zhuang D, Fernandez Y, et al. Tumor cell-selective regulation of NOXA by c-MYC in response to proteasome inhibition. Proc Natl Acad Sci U S A. 2007;104:19488–93. doi: 10.1073/pnas.0708380104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K, Sougnez C, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest. 2012;122:2983–8. doi: 10.1172/JCI64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ploner C, Kofler R, Villunger A. Noxa: at the tip of the balance between life and death. Oncogene. 2008;27(Suppl 1):S84–92. doi: 10.1038/onc.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–8. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 22.Alves NL, Derks IA, Berk E, Spijker R, van Lier RA, Eldering E. The Noxa/Mcl-1 axis regulates susceptibility to apoptosis under glucose limitation in dividing T cells. Immunity. 2006;24:703–16. doi: 10.1016/j.immuni.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 23.Chonghaile TN, Letai A. Mimicking the BH3 domain to kill cancer cells. Oncogene. 2008;27(Suppl 1):S149–57. doi: 10.1038/onc.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheridan C, Brumatti G, Elgendy M, Brunet M, Martin SJ. An ERK-dependent pathway to Noxa expression regulates apoptosis by platinum-based chemotherapeutic drugs. Oncogene. 2010;29:6428–41. doi: 10.1038/onc.2010.380. [DOI] [PubMed] [Google Scholar]
- 25.Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–4. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 26.Rosson GB, Vincent TS, Oswald BW, Wright CF. Drug resistance in malignant rhabdoid tumor cell lines. Cancer Chemother Pharmacol. 2002;49:142–8. doi: 10.1007/s00280-001-0398-y. [DOI] [PubMed] [Google Scholar]
- 27.Smith ME, Cimica V, Chinni S, Jana S, Koba W, Yang Z, et al. Therapeutically targeting cyclin D1 in primary tumors arising from loss of Ini1. Proc Natl Acad Sci U S A. 2011;108:319–24. doi: 10.1073/pnas.0913297108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuwahara Y, Hosoi H, Osone S, Kita M, Iehara T, Kuroda H, et al. Antitumor activity of gefitinib in malignant rhabdoid tumor cells in vitro and in vivo. Clin Cancer Res. 2004;10:5940–8. doi: 10.1158/1078-0432.CCR-04-0192. [DOI] [PubMed] [Google Scholar]
- 29.Katsumi Y, Kuwahara Y, Tamura S, Kikuchi K, Otabe O, Tsuchiya K, et al. Trastuzumab activates allogeneic or autologous antibody-dependent cellular cytotoxicity against malignant rhabdoid tumor cells and interleukin-2 augments the cytotoxicity. Clin Cancer Res. 2008;14:1192–9. doi: 10.1158/1078-0432.CCR-07-1661. [DOI] [PubMed] [Google Scholar]
- 30.Das BC, Smith ME, Kalpana GV. Design and synthesis of 4-HPR derivatives for rhabdoid tumors. Bioorg Med Chem Lett. 2008;18:3805–8. doi: 10.1016/j.bmcl.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe M, Adachi S, Matsubara H, Imai T, Yui Y, Mizushima Y, et al. Induction of autophagy in malignant rhabdoid tumor cells by the histone deacetylase inhibitor FK228 through AIF translocation. Int J Cancer. 2009;124:55–67. doi: 10.1002/ijc.23897. [DOI] [PubMed] [Google Scholar]
- 32.Smith ME, Cimica V, Chinni S, Challagulla K, Mani S, Kalpana GV. Rhabdoid tumor growth is inhibited by flavopiridol. Clin Cancer Res. 2008;14:523–32. doi: 10.1158/1078-0432.CCR-07-1347. [DOI] [PubMed] [Google Scholar]
- 33.Nocentini S. Apoptotic response of malignant rhabdoid tumor cells. Cancer Cell Int. 2003;3:11. doi: 10.1186/1475-2867-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowman XH, McDonnell MA, Kosloske A, Odumade OA, Jenness C, Karim CB, et al. The proapoptotic function of Noxa in human leukemia cells is regulated by the kinase Cdk5 and by glucose. Mol Cell. 2010;40:823–33. doi: 10.1016/j.molcel.2010.11.035. [DOI] [PubMed] [Google Scholar]
- 35.Liu W, Swetzig WM, Medisetty R, Das GM. Estrogen-mediated upregulation of Noxa is associated with cell cycle progression in estrogen receptor-positive breast cancer cells. PLoS ONE. 2011;6:e29466. doi: 10.1371/journal.pone.0029466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang ZK, Davies KP, Allen J, Zhu L, Pestell RG, Zagzag D, et al. Cell cycle arrest and repression of cyclin D1 transcription by INI1/hSNF5. Mol Cell Biol. 2002;22:5975–88. doi: 10.1128/MCB.22.16.5975-5988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol. 2008;28:3457–64. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna ES, Tamayo P, Cho YJ, Tillman EJ, Mora-Blanco EL, Sansam CG, et al. Epigenetic inactivation of the tumor suppressor BIN1 drives proliferation of SNF5-deficient tumors. Cell Cycle. 2012;11:1956–65. doi: 10.4161/cc.20280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A. 2002;99:8695–700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–27. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 41.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–7. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 42.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee S, Kim DH, Goo YH, Lee YC, Lee SK, Lee JW. Crucial roles for interactions between MLL3/4 and INI1 in nuclear receptor transactivation. Mol Endocrinol. 2009;23:610–9. doi: 10.1210/me.2008-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, et al. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:922–6. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, et al. Menin associates with a trithorax family histone methyl-transferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–97. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 46.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–19. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]