Abstract
In a recent project, we collected the transcriptional profiles of Bacillus subtilis 168 after treatment with a large set of diverse antibacterial agents. One result of the data analysis was the identification of marker genes that are indicative of certain compounds or compound classes. We cloned these promoter regions in front of a luciferase reporter gene and reintroduced the constructs individually into the B. subtilis chromosome. Strains were analyzed for their responsiveness after treatment with a set of 37 antibacterials. Twelve functional reporter strains were generated that were selectively and significantly upregulated by the compounds. The selectivity of the reporter strains ranged from generic pathways like protein biosynthesis, cell wall biosynthesis, and fatty acid biosynthesis to compound classes (quinolones and glycopeptides) and individual compounds (rifampin, cycloserine, and clindamycin). Five of the strains are amenable for high-throughput applications, e.g., pathway-specific screening. In summary, we successfully generated B. subtilis reporter strains that are indicative of the mechanisms of action of various classes of antibacterials. The set of reporter strains presented herein can be used for mode-of-action analyses and for whole-cell screening of compound libraries in a mode-of-action-specific manner.
Many strategies to discover novel antibacterial entities make use of recent developments in genomics and postgenomics technologies (15). These approaches are of increasing importance in the context of the numerous reports on antimicrobial resistance, often of the multiresistance type. Pathogens that were once susceptible are becoming more and more accustomed to currently used drugs, and the outcome of this battle cannot be foreseen with the arsenal of antibiotics in use (24), and hence novel drugs are urgently needed.
Resistance is often accompanied by the upregulation of resistance genes, e.g., the VanA type of resistance to vancomycin in Enterococcus spp. (3). The promoter of this inducible resistance operon of Enterococcus faecium has been transferred to Bacillus subtilis in front of a lacZ reporter gene, and it has been shown that induction of lacZ in this strain is conferred by antibiotics that target the cell wall (32). This strain may be used as a tool to discover novel compounds that inhibit similar cellular functions. A similar approach uses the inducible β-lactamase of Citrobacter freundii in the heterologous host Escherichia coli (31). This elegant approach quantifies directly the gene product which elicits resistance, since it can be measured spectrophotometrically. Other recent advances in this direction are the utilization of genetically tailored strains that are more sensitive for certain compounds (29) or strains that generally facilitate the penetration of the compounds through the outer membrane (31). Also, several genes that have been described in the literature were reinvestigated for their potential use as marker genes, e.g., heat shock or cold shock genes as indicators of H-type and C-type protein biosynthesis inhibitors (6, 33) or the extracytoplasmic sigma factor σE as an indicator of compounds that damage the outer membrane or interfere with peptidoglycan biosynthesis (6, 10).
All the examples mentioned above require precise biological knowledge about a given biological pathway. By applying a genomewide analysis of the transcriptional response of B. subtilis to the inhibition of a broad range of essential biological processes (14), we generated a comprehensive data set of expression profiles that enabled the identification of potential marker genes independent of precise knowledge of the signaling events within each pathway. In the study presented here, we demonstrate that such marker genes can be successfully used to generate reporter strains. Such reporter strains may help in the search for novel antibacterial entities. They may be used as mode-of-action-specific whole-cell screening assays or as tools to assign a mode of action to uncharacterized whole-cell-active compounds.
MATERIALS AND METHODS
Expression profiling and data processing.
In a previous study, we collected expression profiles of B. subtilis 168 after treatment with more than 40 different antibacterial agents of various classes (14). The main goal of this project was the generation of a database of expression profiles that enabled the prediction of the mode of action of novel uncharacterized chemical entities. As a result, this database enabled the identification of marker genes which are indicative of certain compounds or compound classes.
Processing of expression profiling data has been described earlier (14, 22). Data were stored in CodeBase, an in-house-developed gene expression database.
Data analysis.
Gene expression profiles were analyzed in order to identify genes that are specifically upregulated by given classes of compounds and hence may be used as marker genes. At the time the project was started, CodeBase contained the expression profiling data for B. subtils 168 after treatment with 16 antibacterial agents (cefoxitin, cycloserine, oxacillin, vancomycin, ciprofloxacin, moxifloxacin, novobiocin, cerulenin, triclosan, trimethoprim, chloramphenicol, clindamycin, erythromycin, neomycin, spectinomycin, and rifampin). The gene expression profiling data obtained following treatment with these 16 compounds represent the basis for the selection of the marker genes presented herein.
As a general approach, a filter was applied according to which genes had to fulfill four criteria: (i) the gene is upregulated at least fivefold with respect to the corresponding control sample; (ii) upregulation is significant at a 5% significance level (t test); (iii) the gene is upregulated at least fivefold with respect to all other compounds not belonging to the same class; and (iv) the normalized expression level is larger than 0.5 (the average expression level over all genes was normalized to 1). However, for some compounds and compound classes, no genes that passed this filter were identified, and therefore some concessions had to be made. Details are described in the Results section.
Wherever possible and necessary, the apparent operon structures were taken into account. For example, if several potential reporter genes were identified, preference was given to genes which are part of an operon in which all genes are upregulated or genes in which the potential promoter region does not overlap adjacent genes. For each compound class or individual compound, the most promising genes were selected, and the genomic sequence was analyzed for presumable promoter regions.
Construction of reporter plasmids.
pSUGAR (Fig. 1) was generated by digesting pSWEET-bgaB (5) with BamHI and HindIII and inserting a luciferase reporter gene amplified from pGEM-luc (Promega) with primers AAAAGGATCCTAAGTAGGTGACCGGTAAAGCGGCCGCAAGGAGGGCCCGCTAGCATGGAAGACGCCAAAAACATAAAGA and AAAAAAGCTTttacaatttggactttccgc, which were digested with the same restriction enzymes. Restriction sites which enable the cloning of test promoters were introduced via one of the primers during the amplification of luc. These restriction sites are located on both sides of a Shine-Dalgarno sequence (restriction sites are italic in the sequence shown above, and the Shine-Dalgarno sequence is indicated in bold).
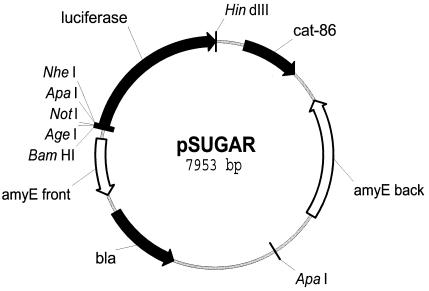
FIG. 1.
Map of plasmid pSUGAR. pSUGAR is a derivative of pSWEET-bgaB (5), which allows integration onto the B. subtilis chromosome at the amyE site. pSUGAR carries an origin of replication for E. coli but not B. subtilis. Restriction sites for cloning of the test promoters are located in front of the luciferase reporter gene.
Test promoters were amplified from B. subtilis 168 genomic DNA and introduced into pSUGAR via NotI and NheI with the primers listed in Table 1. All cloning work was done in Escherichia coli DH5α. Derivatives of pSWEET-bgaB and pSUGAR were selected on Luria broth (LB) containing ampicillin (50 μg/ml).
TABLE 1.
Primers used for amplification of the test promoters
| Gene | Orientationa | Primerb |
|---|---|---|
| dinB | F (Not) | AAAAGCGGCCGCTAGTTTACCCCGCTAAACTTTA |
| R (Nhe) | AAAAGCTAGCATTCCCCCTTTCGTGTGTATAG | |
| yneA | F (Not) | AAAAGCGGCCGCTCAAAACGTCGATTTTAAGAAG |
| R (Nhe) | AAAAGCTAGCAACCTCCAACAGGAATGTTTGT | |
| yorB | F (Not) | AAAAGCGGCCGCTTAGAGGAAATGAAATTATGTT |
| R (Nhe) | AAAAGCTAGCATCCCCTGTTTTGAAATTTTTG | |
| fabHB | F (Not) | AAAAGCGGCCGCTCATAGATTCCTATCTACACTT |
| R (Nhe) | AAAAGCTAGCCACTCCTTATGGTCAGATTATA | |
| glpD | F (Not) | AAAAGCGGCCGCAGTAATACTATGGTATAATGGT |
| R (Nhe) | AAAAGCTAGCTCCTCCTTGTTGTCACGGTAAA | |
| ytrA | F (Not) | AAAAGCGGCCGCGATTGACTTTGTGAGTCAAAGT |
| R (Nhe) | AAAAGCTAGCCCCTACTTTCTATACGATCTGA | |
| ywoB | F (Not) | AAAAGCGGCCGCTCATGTAAGATTTCCTGACATG |
| R (Nhe) | AAAAGCTAGCCCCTCAGTGTATTATTTGATGT | |
| yrzI | F (Not) | AAAAGCGGCCGCAGATGTTTACAAAATGGAATTT |
| R (Nhe) | AAAAGCTAGCCACCCCCTTTCAAAGTCCGCAT | |
| ypbG | F (Not) | AAAAGCGGCCGCAGCCCGGAGCCTCAGCTTATAC |
| R (Nhe) | AAAAGCTAGCCTCTCCATTCTTTTTAGAACTT | |
| ydeK | F (Not) | AAAAGCGGCCGCCGTTGTTCTCCTAACTGGTATG |
| R (Nhe) | AAAAGCTAGCCACTCCACATATCTTTCTTGTT | |
| yvgS | F (Not) | AAAAGCGGCCGCAACCGATTTCGAAGTGAAATCG |
| R (Nhe) | AAAAGCTAGCCACCTCCAGAAAATAGTTGACA | |
| expZ | F (Not) | AAAAGCGGCCGCAAAATGAGAGCAGGAGTTTTTT |
| R (Nhe) | AAAAGCTAGCCCCTCGCTTTAAAGGGAGAATA |
F, forward primer; R, reverse primer. Restriction sites that were introduced via the oligonucleotides are indicated.
Restriction sites are underlined for the forward primers (NotI) and the reverse primers (NheI).
Generation of reporter strains.
Reporter plasmids that were generated as described above were transformed into B. subtilis 168 and selected on LB containing chloramphenicol (5 μg/ml). Colonies were picked and analyzed by PCR for a double crossover with appropriate primer combinations.
Reporter assays.
An overnight culture of the reporter strain was cultivated in basal limitation medium (30) at 37°C and 200 rpm. A 10-ml culture was inoculated from the overnight culture to an A600 of 0.05 in fresh basal limitation medium and grown to an A600 of 0.5 under the conditions described above. Aliquots of 25 μl of this suspension were transferred into the wells of a 384-well microplate (white, sterile with clear bottom; Nunc), each already containing 25 μl of basal limitation medium plus the test compounds at various concentrations. Plates were incubated at 37°C for a period depending on the induction kinetics of the reporter strain (details below). After this incubation step, 25 μl of each well was transferred to a fresh 384-well microplate with a Quadra 384 model 230 workstation (Tomtec). Each well of this second microplate was prefilled with 25 μl of luciferase assay reagent (Labsystems). All experiments were run at least in duplicate.
Z′ factors were calculated as a measure of the applicability of the system for use in high-throughput applications (35); 120 replicates of both a positive and a negative control sample were measured in parallel in the system described above.
As a basis for the concentrations to test, we used copt, the maximum subinhibitory concentration of each compound, as determined in culture flasks for expression profiling experiments (14). The concentration range tested was the 0.125-fold to 8-fold this reference concentration for most of the compounds and included the MIC as determined in 96-well microtiter plates. The copt and MIC, respectively, in basal limitation medium for the compounds used in this study were (all values in micrograms per milliliter) 0.5 and 1 for cefoxitin (FOX), 16 and 32 for cycloserine (CYC), 0.25 and 0.5 for oxacillin (OXA), 0.5 and 0.5 for ristocetin (RIS), 0.25 and 0.25 for vancomycin (VAN), 0.25 and 1 for actinomycin D (AMY), 1 and 2 for ethidium bromide (EBR), 0.5 and 0.5 for ciprofloxacin (CIP), 0.25 and 0.25 for moxifloxacin (MXF), 8 and 8 for nalidixic acid (NAL), 0.5 and 2 for norfloxacin (NOR), 1 and 1 for coumermycin A1 (COU), 0.25 and 2 for novobiocin (NOV), 4 and 8 for cerulenin (CER), 0.03 and 0.015 for hexachlorophene (HCP), 1 and 2 for triclosan (TCL), 0.25 and 0.06 for 5-fluoruracil (5FU), 64 and 16 for sulfacetamide (SUA), 1 and 0.5 for trimethoprim (TMP), 0.03 and 1 for gramicidin A (GRA), 0.125 and 0.5 for monensin (MON), 0.008 and 1 for nigericin (NIG), 2 and 4 for nitrofurantoin (NIT), 64 and 128 for polymyxin B sulfate (PMY), 64 and 128 for Triton X-114 (TRX), 2 and 2 for azaserine (AZA), 32 and 4 for actinonin (ACT), 4 and 4 for chloramphenicol (CHL), 0.25 and 1 for clarithromycin (CLR), 2 and 2 for clindamycin (CLI), 4 and 0.25 for erythromycin (ERY), 0.06 and 0.25 for fusidic acid (FUS), 1 and 0.125 for neomycin (NEO), 64 and 64 for puromycin (PUR), 128 and 64 for spectinomycin (SPT), 0.5 and 2 for tetracycline (TET), and 0.008 and 0.125 for rifampin (RIF). Dimethyl sulfoxide was used as a negative control.
RESULTS
Outline of the approach.
The approach of the work shown here consists of four steps. First, a subset of the gene expression data (see Materials and Methods) of a preceding study (14) was used to identify genes which are upregulated in B. subtilis after treatment with certain compounds or a compound class (Table 2). Second, we identified the promoter regions of the selected genes by inspection of the individual DNA sequences, and the predicted regions were cloned in front of the luciferase reporter gene of pSUGAR with NotI and NheI. Third, we transformed B. subtilis 168 cells with the plasmid constructs and selected for integration into the chromosome via a double crossover. Finally, all reporter strains were tested in the system described below.
TABLE 2.
Overall performance of functional reporter strains
| Type | Class | Gene | Reference compounda | Maximal induction (min)
|
Induction factorb (fold)
|
RLUc
|
Concn (μg/ml)
|
Z′d | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arraye | Reporter | Array | Reporter | Basal | Induced | MIC | Inductionf | |||||
| Generic pathways | Fatty acid biosynthesis | fabHB | Cerulenin | All | 200 | 28.3 | 3.7 | 419 | 1,628 | 8 | 1-64 | 0.36 |
| glpDg | Cerulenin | All | 180 | 12.6 | 2.0 | 74 | 218 | 8 | 1-8 | <0 | ||
| Protein biosynthesis | yrzIh | Clindamycin | 80 | 360 | 104.1 | 2.5 | 195 | 610 | 2 | 0.25-8 | <0 | |
| Cell wall biosynthesis | ypbGi | Vancomycin | 40, 80 | 80 | 11.4 | 2.0 | 216 | 882 | 0.25 | 0.125-0.25 | <0 | |
| Compound classes | Quinolonesj | dinBk | Ciprofloxacin | 80 | 240 | 14.8 | 15.4 | 1,272 | 16,928 | 0.5 | 0.06-4 | 0.43 |
| yneAl | Ciprofloxacin | 80 | 240 | 19.0 | 16.1 | 218 | 4,487 | 0.5 | 0.125-4 | 0.54 | ||
| yorBl | Ciprofloxacin | 80 | 200 | 14.1 | 28.1 | 616 | 18,181 | 0.5 | 0.06-4 | 0.51 | ||
| Glycopeptides | ytrA | Vancomycin | 10 | 40 | 42.9 | 3.4 | 228 | 1,550 | 0.25 | 0.125-2 | 0.26 | |
| ywoB | Vancomycin | 10 | 60 | 62.1 | 1.9 | 45 | 101 | 0.25 | 0.125-2 | <0 | ||
| Individual compounds | Cycloserine | ydeK | Cycloserine | 40 | 160 | 56.2 | 2.2 | 92 | 382 | 32 | 8-64 | 0.02 |
| Rifampicin | yvgS | Rifampicin | 80 | 80 | 46.2 | 3.7 | 86 | 322 | 0.125 | 0.008-0.125 | <0 | |
| Clindamycin | expZ | Clindamycin | All | 300 | 76.3 | 5.1 | 116 | 644 | 2 | 0.25-2 | 0.03 | |
Compound for which the highest level of upregulation was observed in expression profiling experiments. This compound was also used to determine Z′.
Induction factors for the reporter strains were calculated from a set of experiments designed to evaluate the reporter strains in a high-throughput mode.
RLU, mean relative light units, as measured in a set of experiments designed to evaluate the functionality of the reporter strains (see Results). Indicated are the values measured without compound treatment (basal) and after treatment with the reference compounds (induced) at the optimal time points and concentrations.
The Z′ factor is a measure of assay robustness in high-throughput applications (35).
Gene expression profiles were collected after 10, 40, and 80 min of compound treatment.
Concentration window in which significant upregulation was observed.
glpD was selected as a specific marker gene for triclosan, but the reporter strain was responsive to cerulenin as well.
Two false-negative (puromycin and actinonin) and three compounds that elicited unexpected positive responses (5-fluoruracil, nitrofurantoin, and nalidixic acid) were detected with the yrzI reporter strain.
One false-negative (ristocetin) and one compound that elicited an unexpected positive response (polymyxin B) were detected with the ypbG reporter strain.
Genes were selected as marker genes for topoisomerases, but not all strains elicited a signal with the coumarins.
Two compounds that elicited unexpected positive responses were detected with the dinB reporter strain, azaserine and 5 fluoruracil.
One compound that elicited an unexpected positive response was detected with the yneA and the yorB reporter strains, azaserine.
Using the filter described in the Materials and Methods section, we were able to identify marker genes for six compounds or compound classes. Most of the selected genes could be successfully used for the generation of functional reporter strains (see below and Table 2). In order to cover all important classes of antimicrobials, we allowed for less stringent criteria for those classes for which we could not identify marker genes. In particular, it was difficult to identify marker genes for the classes of protein biosynthesis and cell wall biosynthesis. yrzI was the best marker gene we could identify for inhibition of protein biosynthesis. Similarly, ypbG and ypuA were chosen as marker genes for inhibition of cell wall biosynthesis. Although these genes did not fulfill the criteria set above, they were the best we could identify.
Initial characterization and kinetics of upregulation.
First we investigated for each reporter strain whether the expected response occurred within a similar time frame as in the expression profiling analysis. This is an essential prerequisite to further characterize the individual reporter strains thereafter. Furthermore, by treating the reporter strains with the model antimicrobials of the classes, these experiments indicate whether the individual reporter strains are functional at all.
Altogether, 12 functional reporter strains were generated (see below and Table 2). For all reporter strains, we observed a good correlation to the kinetics of transcript production in expression profiling experiments (Table 2). However, the time required to trigger the highest level of induction was delayed compared to the profiling results.
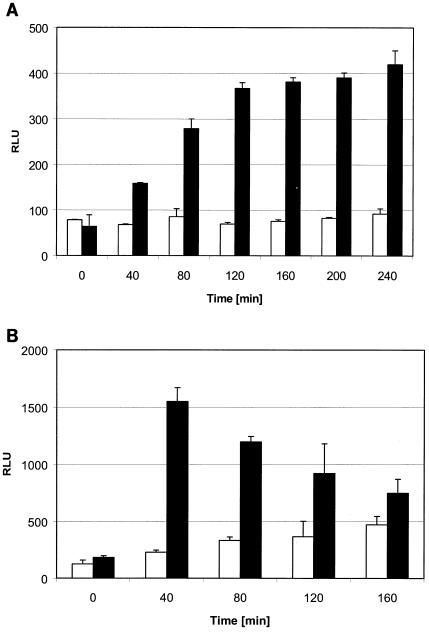
Figure 2A shows a reporter strain indicative of cycloserine. Its signal increased continuously for 2 h. In contrast, Fig. 2B is an example of a strain indicative of glycopeptides. This strain already showed the highest level of upregulation at the earliest time point measured, and thereafter the signal decreased steadily.
FIG. 2.
Kinetics of upregulation of a cycloserine (A, ydeK) and a glycopeptide (B, ytrA) reporter strain. Activities are indicated as relative light units (RLU). Solid bars show values for reporter strains treated with cycloserine at 16 μg/ml (A) and vancomycin at 0.25 μg/ml (B). Open bars show values for the untreated control strains.
In summary, the compound-induced upregulation of most genes was confirmed by the reporter strain approach. However, the time needed to obtain a maximum readout had to be optimized for each strain individually.
Sensitivity and specificity of reporter strains.
After the incubation time had been optimized for each functional reporter strain, we next evaluated each strain in terms of sensitivity and specificity. To do so, the response of all 12 functional reporter strains to a panel of 37 antibacterial agents at a wide range of concentrations was tested.
All reporter strains elicited a response at concentrations below the MIC, i.e., at concentrations that do not inhibit growth of the organism under investigation (Fig. 3, Table 2). Sublethal concentrations were also used in the expression profiling experiments, and the behavior of the reporter strains hence reflects the conditions used for the identification of the marker genes.
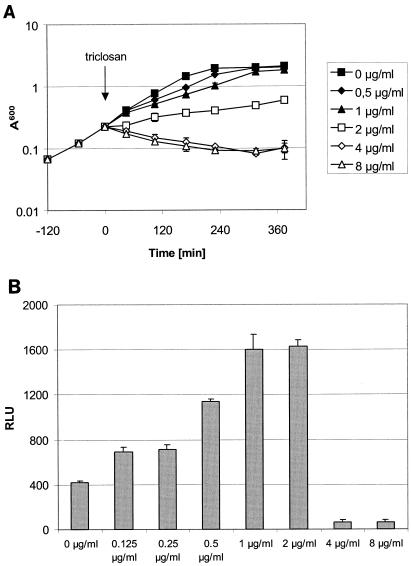
FIG. 3.
Concentration dependence of a fatty acid biosynthesis reporter strain. (A) A reporter strain carrying a luciferase reporter gene under the control of the fabHB promoter was grown in basal limitation medium (30) and treated with triclosan as indicated by the arrow. Growth was monitored by optical density measurements. (B) The same reporter strain was treated with triclosan in the system described herein. Luciferase activity was measured after 200 min of treatment at the concentrations indicated. Activities are given as relative light units (RLU).
Next, we determined the spectrum of antibacterial agents to which the individual reporter strains responded. As shown above, all strains exhibited luciferase activity after treatment with the reference compounds, and only two reporter strains did not elicit a signal with all compounds of the class (Table 2). The reporter strain for protein biosynthesis inhibitors did not show a signal with puromycin and actinonin, and the reporter strain for cell wall biosynthesis inhibitors was insensitive to ristocetin. As described above, these are exactly the two classes for which it was difficult to identify marker genes.
With the exception of the three reporter strains for the quinolones, all of which elicited a signal with azaserine, the other strains did not give rise to any unexpected positive responses. It is noteworthy that azaserine was also classified as an inhibitor of topoisomerase in our bioinformatic analysis of the gene expression data (14).
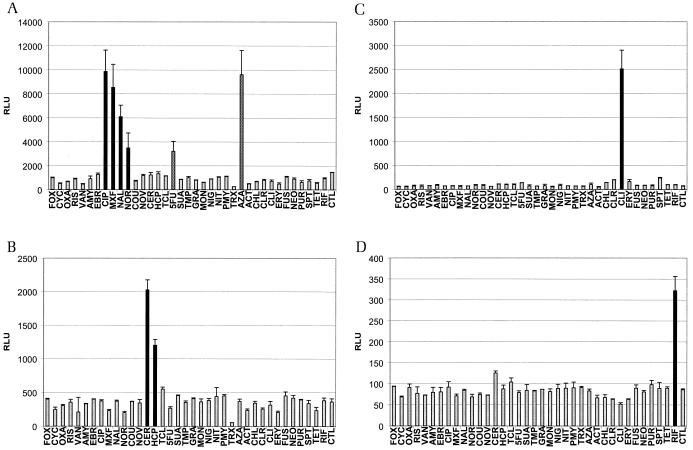
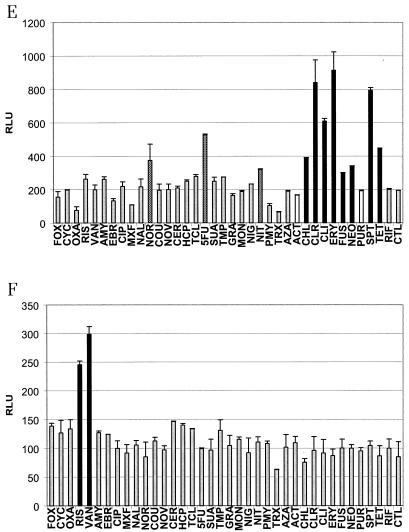
The results for a subset of reporter strains are shown in Fig. 4. Figure 4A and B show the functionality of quinolone and fatty acid biosynthesis reporter strains, respectively. Two unexpected positive responses, for 5-fluoruracil and azaserine, were identified for the quinolone reporter strain (Fig. 4A). The two reporter strains for the individual compounds clindamycin and rifampin (Fig. 4C and D) elicited strong signals and did not yield in any misclassifications. The same held true for a reporter strain for glycopeptides (Fig. 4F). Figure 4E shows the results with the reporter strain for inhibitors of protein biosynthesis. It was much more difficult, as described above, to use this strain for meaningful classification.
FIG. 4.
Response patterns of six reporter strains. Reporter strains were induced as follows: the quinolone reporter strain (dinB) for 240 min (A), the fatty acid reporter strain (fabHB) for 200 min (B), the clindamycin reporter strain (expZ) for 300 min (C), the rifampin reporter strain (yvgS) for 80 min (D), the protein biosynthesis reporter strain (yrzI) for 360 min (E), and the glycopeptide reporter strain (ytrA) for 40 min (F). Compounds were added at concentrations just low enough not to inhibit growth of the organism (see Materials and Methods). Black bars show values for compounds that correctly elicited a signal. Grey bars show values for compounds that, as expected, did not show a signal significantly above that of the control sample. White bars indicate false-negatives, and checkered bars show values for compounds that elicited unexpected positive responses. Activities are given as relative light units (RLU). See text for abbreviations of drug names.
In summary, we successfully generated and evaluated reporter strains corresponding to the following compound classes: protein biosynthesis inhibitors, fatty acid biosynthesis inhibitors, cell wall biosynthesis inhibitors, quinolones, and glycopeptides. In addition, functional reporter strains were constructed for the individual compounds rifampin, cycloserine, and clindamycin. For most of the strains, the level of upregulation was lower than observed in the expression profiling experiments (Table 2). We assume that this is due to the different detection methods used, the dissimilar induction kinetics, and the accumulation of active luciferase in the reporter strains. However, the induction levels were adequate for the applications discussed herein. Interestingly, the reporter genes that are indicative of quinolones (dinB, yneA, and yorB) show the same magnitude of upregulation as observed during array-based gene expression analysis (Table 2).
Amenability for high-throughput applications.
The data described above allow the use of the indicated reporter strains for mode-of-action analysis and for mode-of-action-specific whole-cell compound screening on a laboratory scale. We next aimed at evaluating the reporter strains for their suitability for high-throughput applications.
To do so, we calculated the Z′ factor, a screening window coefficient which reflects both the signal difference between a positive and a negative control sample and the signal variance associated with the measurements (35). Z′ factors above zero indicate that a screen is amenable to high-throughput applications. One hundred twenty replicates of both a negative and a positive control sample were assayed with a Quadra workstation. The results are summarized in Table 2.
The levels of upregulation were smaller for some of the strains in this large-scale approach compared to the smaller-scale experiments described above. The best Z′ factors were obtained with the three reporter strains for the quinolones. Also amenable for high-throughput applications are the reporter strains for fatty acid biosynthesis inhibitors (fabHB) and glycopeptides (ytrA). The latter two strains showed only a relatively low level of upregulation (3.7- and 3.4-fold, respectively), but the coefficient of variation of the expression levels was very small in these strains, giving rise to high Z′ factors. The other strains revealed Z′ factors around or below zero, and therefore their use is only reasonable in smaller-scale applications.
DISCUSSION
We generated a panel of luciferase reporter strains which are indicative of various compounds or compound classes. The genes used for construction of these strains were selected by analyzing a first subset of gene expression data collected in a preceding project (14). The plenitude of data enabled the selection of genes that are highly indicative of most of the compound classes that we were interested in. Most but not all of the genes identified as reporter genes are of unknown function. dinB, a marker gene for the quinolones, is known to be DNA damage inducible (8, 21), and fabHB, encoding a β-ketoacyl-acyl carrier protein synthase III, is involved in fatty acid biosynthesis (9). glpD, a marker gene for fatty acid biosynthesis inhibitors, is involved in glycerol metabolism (4, 13), and expZ, a marker gene for clindamycin, encodes an ATP-binding transport protein (25). expZ is not upregulated by other compounds that inhibit protein biosynthesis at the peptidyl transferase site (e.g., chloramphenicol or macrolide antibiotics). In contrast to two other compounds, the binding side of clindamycin overlaps both the A site and the P site of the ribosome (28), and this distinct feature might be the basis for the specific upregulation of expZ.
In this context, it should be noted that not all of the reporter strains that we generated were functional. These nonfunctional reporter strains carried upstream sequences of the genes dppA and ykfA (fatty acid biosynthesis), yheH (protein biosynthesis), ypuA (cell wall biosynthesis), yumD (trimethoprim), racE (chloramphenicol), hrcA (neomycin), and veg (coumarines). A reason for the nonperformance of these strains may be selection of incorrect promoter fragments or the need for adjacent regulatory sequences that have been taken out of context in the reporter strains.
As described, the time required to induce the maximum readout for the reporter strains was delayed compared to the expression profiling results. We assume that this delay is due to the need for translation of the mRNA and to the accumulation of functional reporter protein, leading to a steadily increasing signal. However, strains which induced gene expression early in the microarray study were also the first to produce functional luciferase in the reporter strains.
Interestingly, the reporter strains which are indicative of the quinolones did not trigger a signal with any of the coumarins. This is surprising because both compound classes act on the same molecules, type II topoisomerases. These two compound classes, however, act on different subunits of the topoisomerase enzyme. Topoisomerase II introduces negative supercoils into DNA, utilizing energy derived from the hydrolysis of ATP. A key step in this supercoiling reaction is the gyrase-mediated cleavage of DNA. Quinolones act on the alpha subunit of topoisomerase, interrupting this cleavage and resealing reaction (11). This results in DNA damage and the induction of DNA repair. In contrast, the coumarins bind to the ATP binding site located on the beta subunit of the enzyme, inhibiting the enzymatic activity of the enzyme but leaving the DNA largely intact (20). The reporter strains discussed herein are obviously able to distinguish between these two activities.
Furthermore, all reporter strains indicative of the quinolones also elicited a signal with azaserine. Several papers have been published on the possible mode of action of azaserine. Inhibition of glutamine synthase (16) and purine biosynthesis (18) have both been suggested to be the main target of this drug. Other reports, however, point to DNA damage as the crucial effect of this compound (34). Azaserine may act as a carboxymethylating agent, and the onset of DNA repair has been demonstrated after azaserine treatment (19). This effect may explain the response observed with the quinolone reporter strains.
The lowest sensitivity and specificity were observed with the reporter strains for protein biosynthesis and cell wall biosynthesis. These are the two most diverse pathways among all the reporter strains generated. Inhibition of cell wall biosynthesis may occur at various different stages. β-Lactams of the penicillin or the cephalosporin class act on a variety of penicillin-binding proteins (12, 23), whereas other compounds target specific enzymes or intermediates during peptidoglycan biosynthesis (26, 27). Likewise, inhibitors of protein biosynthesis act on various steps and at different locations on the ribosome (17, 28). In contrast, actinonin inhibits the removal of N-terminal formyl groups from newly synthesized proteins and exerts its action at a completely different step of protein biosynthesis (1, 2). It is hence not too surprising that reporter strains for these two classes show less sensitivity and specificity than the other strains.
The complete set of reporter strains covers most classes of currently used antibacterial agents. It therefore represents a novel and convenient tool to categorize novel chemical entities with antibacterial activity. We assume that it is possible to generate reporter strains for most compounds or compound classes of interest. In fact, we expanded our data set of expression profiles to almost 40 compounds (14). Reanalysis of the responses elicited with this further set of compounds confirmed the specificity of the selected reporter strains. Of note, azaserine, which triggered a response with all quinolone reporter strains as describes above, leads to upregulation of all genes selected as markers for this compound class (data not shown; see http://www.gpc-biotech.com/supplementary_material.htm).
One advantage of the approach described herein lies in the fact that expression profiling allows quantification of the expression level of each individual transcript. Selection of marker genes is therefore not restricted to the limited number of genes that have previously been studied in detail (6, 29, 32). This is illustrated by some of our reporter strains which carry the promoter sequences of previously uncharacterized genes, which would not have been chosen as marker genes by a traditional approach.
Since the incubation times differ for the individual reporter strains, we handled them individually or in smaller groups of strains with similar kinetics. Ultimately, however, a system that allows the parallel assessment of all strains would be preferable. One system that detects the growth of microorganisms in preconfigured microplates has been described (7). Each well carries a different growth supplement or inhibitory compound. Statistical tools that link the phenotypic observations, i.e., growth or no growth, with a given genetic predisposition or the effect a drug exerts onto the organism are then employed. A similar technological setup might be conceivable for the approach described here, although another reporter system that enables kinetic measurements will have to be introduced.
Another application of the reporter strains is their use for pathway-based drug-screening applications. We found that five of the strains resulted in Z′ factors suitable for high-throughput applications. As mentioned above, the performance of the other strains is still good enough to utilize them on a laboratory scale.
In conclusion, we generated reporter strains for antimicrobial agents that act by several modes of action. Selection of marker genes was achieved solely by analyzing the data generated in a large-scale gene expression profiling project. We have shown that this technique produces reliable results that can be exploited further. The reporter strains generated here are novel tools that can be used for the mode-of-action prediction of antibacterials or in pathway-based drug-screening applications.
Acknowledgments
We thank Eric Brown for the kind gift of pSWEET-bgaB and Nicola Saunders and Ruairi Friel for critical reading of the manuscript.
REFERENCES
- 1.Apfel, C. M., S. Evers, C. Hubschwerlen, W. Pirson, M. G. Page, and W. Keck. 2001. Peptide deformylase as an antibacterial drug target: assays for detection of its inhibition in Escherichia coli cell homogenates and intact cells. Antimicrob. Agents Chemother. 45:1053-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apfel, C. M., H. Locher, S. Evers, B. Takacs, C. Hubschwerlen, W. Pirson, M. G. Page, and W. Keck. 2001. Peptide deformylase as an antibacterial drug target: target validation and resistance development. Antimicrob. Agents Chemother. 45:1058-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur, M., and R. Quintiliani, Jr. 2001. Regulation of VanA- and VanB-type glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 45:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beijer, L., and L. Rutberg. 1992. Utilisation of glycerol and glycerol 3-phosphate is differently affected by the phosphotransferase system in Bacillus subtilis. FEMS Microbiol. Lett. 79:217-220. [DOI] [PubMed] [Google Scholar]
- 5.Bhavsar, A. P., X. Zhao, and E. D. Brown. 2001. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl. Environ. Microbiol. 67:403-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianchi, A. A., and F. Baneyx. 1999. Stress responses as a tool to detect and characterize the mode of action of antibacterial agents. Appl. Environ. Microbiol. 65:5023-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bochner, B. R., P. Gadzinski, and E. Panomitros. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 11:1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheo, D. L., K. W. Bayles, and R. E. Yasbin. 1991. Cloning and characterization of DNA damage-inducible promoter regions from Bacillus subtilis. J. Bacteriol. 173:1696-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, K. H., R. J. Heath, and C. O. Rock. 2000. β-Ketoacyl-acyl carrier protein synthase III (FabH) is a determining factor in branched-chain fatty acid biosynthesis. J. Bacteriol. 182:365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dartigalongue, C., D. Missiakas, and S. Raina. 2001. Characterization of the Escherichia coli sigma E regulon. J. Biol. Chem. 276:20866-20875. [DOI] [PubMed] [Google Scholar]
- 11.Gmunder, H., K. Kuratli, and W. Keck. 1997. In the presence of subunit A inhibitors DNA gyrase cleaves DNA fragments as short as 20 bp at specific sites. Nucleic Acids Res. 25:604-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goffin, C., and J. M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmberg, C., L. Beijer, B. Rutberg, and L. Rutberg. 1990. Glycerol catabolism in Bacillus subtilis: nucleotide sequence of the genes encoding glycerol kinase (glpK) and glycerol-3-phosphate dehydrogenase (glpD). J. Gen. Microbiol. 136:2367-2375. [DOI] [PubMed] [Google Scholar]
- 14.Hutter, B., C. Schaab, S. Albrecht, M. Borgmann, N. A. Brunner, C. Freiberg, K. Ziegelbauer, C. O. Rock, I. Ivanov, and H. Loferer. Mechanism-of-action prediction of antibacterial compounds by gene expression profiling. Antimicrob. Agents Chemother., in press. [DOI] [PMC free article] [PubMed]
- 15.Ji, Y. 2002. The role of genomics in the discovery of novel targets for antibiotic therapy. Pharmacogenomics 3:315-323. [DOI] [PubMed] [Google Scholar]
- 16.Kim, C. H., and T. C. Hollocher. 1982. 13N isotope studies on the pathway of ammonia assimilation in Bacillus megaterium and Escherichia coli. J. Bacteriol. 151:358-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotra, L. P., J. Haddad, and S. Mobashery. 2000. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents Chemother. 44:3249-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubitschek, H. E., and R. J. Sepanski. 1982. Azaserine: survival and mutation in Escherichia coli. Mutat. Res. 94:31-38. [DOI] [PubMed] [Google Scholar]
- 19.Lawson, T. 1989. Nicotinamide and selenium stimulate the repair of DNA damage produced by N-nitrosobis (2-oxopropyl) amine. Anticancer Res. 9:483-486. [PubMed] [Google Scholar]
- 20.Lewis, R. J., O. M. Singh, C. V. Smith, T. Skarzynski, A. Maxwell, A. J. Wonacott, and D. B. Wigley. 1996. The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J. 15:1412-1420. [PMC free article] [PubMed] [Google Scholar]
- 21.Lovett, C. M. Jr., T. M. O'Gara, and J. N. Woodruff. 1994. Analysis of the SOS inducing signal in Bacillus subtilis using Escherichia coli LexA as a probe. J. Bacteriol. 176:4914-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machl, A. W., C. Schaab, and I. Ivanov. 2002. Improving DNA array data quality by minimising ′neighbourhood' effects. Nucleic Acids Res. 30:e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandell, G. L., and W. A. Petri. 1995. Antimicrobial agents. Penicillins, cephalosporins, and other β-lactam antibiotics, p. 1073-1101. In J. G. Hardman, L. E. Limbird, P. B. Molinoff, R. W. Ruddon, and. A. G. Gilman (ed.), The pharmacological basis of therapeutics, 9th ed. McGraw-Hill, New York, N.Y.
- 24.McManus, M. C. 1997. Mechanisms of bacterial resistance to antimicrobial agents. Am. J. Health Syst. Pharm. 54:1420-1433. [DOI] [PubMed] [Google Scholar]
- 25.Moszer, I., L. M. Jones, S. Moreira, C. Fabry, and A. Danchin. 2002. SubtiList: the reference database for the Bacillus subtilis genome. Nucleic Acids Res. 30:62-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rastogi, N., and H. L. David. 1993. Mode of action of antituberculous drugs and mechanisms of drug resistance in Mycobacterium tuberculosis. Res. Microbiol. 144:133-143. [DOI] [PubMed] [Google Scholar]
- 27.Reynolds, P. E. 1989. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 8:943-950. [DOI] [PubMed] [Google Scholar]
- 28.Schlünzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 29.Shapiro, E., and F. Baneyx. 2002. Stress-based identification and classification of antibacterial agents: second-generation Escherichia coli reporter strains and optimization of detection. Antimicrob. Agents Chemother. 46:2490-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stülke, J., R. Hanschke, and M. Hecker. 1993. Temporal activation of β-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 139:2041-2045. [DOI] [PubMed] [Google Scholar]
- 31.Sun, D., S. Cohen, N. Mani, C. Murphy, and D. M. Rothstein. 2002. A pathway-specific cell based screening system to detect bacterial cell wall inhibitors. J. Antibiot. (Tokyo) 55:279-287. [DOI] [PubMed] [Google Scholar]
- 32.Ulijasz, A. T., A. Grenader, and B. Weisblum. 1996. A vancomycin-inducible lacZ reporter system in Bacillus subtilis: induction by antibiotics that inhibit cell wall synthesis and by lysozyme. J. Bacteriol. 178:6305-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.VanBogelen, R. A., and F. C. Neidhardt. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:5589-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams-Hill, D. M., J. Olesen, C. Zucker, and H. E. Kubitschek. 1984. Azaserine: further evidence for DNA damage in Escherichia coli. Mutat. Res. 129:153-164. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, J. H., T. D. Chung, and K. R. Oldenburg. 1999. A simple statistical parameter for use in evaluation and validation of high-throughput screening assays. J. Biomol. Screen. 4:67-73. [DOI] [PubMed] [Google Scholar]