Abstract
The sul3 gene recently described in Escherichia coli was found in 22 of 512 (4.3%) German Salmonella isolates from different regions and sources and of different serotypes, antimicrobial resistance phenotypes, and genomic groups. This is the first report on the prevalence of sul3 among Salmonella strains, and the findings support the strong potential of this determinant to spread within bacterial populations.
Sulfonamides represent the oldest group of antimicrobial agents and have been available since the 1930s. They competitively inhibit the bacterial enzyme dihydropteroic acid synthetase (DHPS). Resistance to sulfonamides appeared quite soon after their introduction into clinical practice. Resistance can result from mutations in the chromosomal DHPS or by acquisition of DHPS drug resistance genes (sul genes), whose products have lower affinities for sulfonamides (2, 7, 8, 13, 16-19). For many years, only two DHPS drug resistance genes, sul1 and sul2, which share 57% nucleotide sequence (DNA) identity, were detected. While sul1 is part of the 3′ conserved segment of class 1 integrons, sul2 often appears to be associated with genes that confer resistance to streptomycin (strA and strB). Both genes have been found on plasmids and chromosomes (2, 7, 8, 13, 16-19). The sul1 and sul2 genes seem to be equally distributed among sulfonamide-resistant Escherichia coli isolates of clinical origin (6-8, 17, 18). In some studies, only 70% of sulfonamide resistance could be explained by these genes (10; unpublished data).
Recently, Perreten and Boerlin (13) described a new DHPS sulfonamide resistance gene, designated sul3 (GenBank accession number AJ459418), which has 50.4% amino acid identity to Salmonella enterica plasmid pHCM1, 40.6% amino acid identity to sul2 from E. coli plasmid RSF1010, and 40.9% amino acid identity to sul1 from E. coli plasmid R388 (GenBank accession number X12869). This sul3 gene was found in Swiss E. coli strains from pigs. For Salmonella, a gene named sul3 can be found in GenBank (accession number AY047357); however, this gene seems to be a defective sul1 gene. The objective of this work was to ascertain the presence and spread of the sul3 gene described for the Swiss E. coli isolates in non-typhoid Salmonella strains.
The study included 512 epidemiologically unrelated German sulfonamide-resistant Salmonella strains from the German National Salmonella Reference Laboratory (NRL-Salm; Berlin, Germany) strain collection. They were isolated in 2001 from livestock (100, 87, and 75 isolates from poultry, cattle, and swine, respectively), food products (236 isolates), and feed (14 isolates) in different laboratories in the 16 German states (Länder).
Several steps were used in the study. (i) Dot blotting was used to screen the isolates for the sul3 gene. For dot blotting, 5 μl (about 100 ng) of boiled DNA (11) was spotted onto nylon membranes (Roche Applied Sciences, Mannheim, Germany), cross-linked for 2 min, and hybridized by a nonradioactive method (Roche Applied Sciences) with a sul3-specific probe obtained from plasmid pVP440 (13). (ii) PCR amplification was then used to confirm the presence of sul3 in the suspected positive isolates. PCR was carried out as described previously (4) with primers sul3-F (GAGCAAGATTTTTGGAATCG) and sul3-B (CATCTGCAGCTAACCTAGGGCTTTGGA) and the conditions described elsewhere (13). (iii) The sequence of the sul3 gene found in one Salmonella-positive strain was then analyzed. First, only the PCR product obtained with the sul3-specific primers was sequenced, as described previously (4). Second, the 5′ flanking region was also sequenced by using internal primers specific for the orf and sul3 genes (GenBank accession number AJ459418). (iv) Restriction fragment length polymorphism (RFLP) analysis of all the sul3 PCR products was then performed as described previously (4) with 5 U of HindIII or SphI endonuclease (Amersham Biosciences, Freiburg, Germany). (v) Molecular typing of the strains was performed by plasmid analysis (9) and pulsed-field gel electrophoresis (PFGE) with the XbaI endonuclease (11) and the run conditions recommended by the Salm-Net project (12). (vi) Finally, the location of the sul3 gene was determined by Southern blot hybridization (14) of the plasmid patterns with the sul3-specific probe.
Twenty-two of the 512 (4.3%) sulfonamide-resistant Salmonella strains (Table 1) hybridized with the sul3-specific probe. By PCR, all 22 strains gave amplification products of about 789 bp. The sequence of the PCR product generated by Salmonella strain NRL-01-02571 showed 100% identity with the sequence of the sul3 gene found in E. coli deposited in GenBank (accession number AJ459418). Restriction of all sul3 PCR products with the HindIII or the SphI endonuclease generated the same RFLP patterns generated for the control (fragments of 591 and 198 bp and fragments of 700 and 89 bp, respectively).
TABLE 1.
Features of the Salmonella strains carrying the sul3 gene
| NRL reference strain no. | Serotype (phage type) | German region of isolation | Origin | Resistance patterna | PP | PFP | Amplification by PCRb
|
||
|---|---|---|---|---|---|---|---|---|---|
| sul3 | sul1 | sul2 | |||||||
| 01-00835 | Agona | Brandenburg | Turkey | AMP-CHL-STR-SPT-SUL-TET | 14 | X13 | + | − | − |
| 01-01887 | Agona | Nordrhein-Westfalen | Turkey | AMP-CHL-KAN-NEO-STR-SPT-SUL-TET | 9 | X13 | + | − | − |
| 01-01055 | Agona | Brandenburg | Turkey | CHL-STR-SPT-SUL | 8 | X14 | + | − | − |
| 01-01647 | Anatum | Nordrhein-Westfalen | Turkey | CHL-STR-SPT-SUL | 13 | X15 | + | − | − |
| 01-00463 | Brandenburg | Brandenburg | Turkey | CHL-STR-SPT-SUL | 4 | X11 | + | − | − |
| 01-00799 | Brandenburg | Bayern | Chicken | AMP-CHL-KAN-NEO-STR-SPT-SUL-TET | 6 | X12 | + | − | − |
| 01-00461 | Heidelberg | Brandenburg | Turkey | CHL-STR-SPT-SUL-TET | 3 | X1 | + | − | − |
| 01-00564 | Heidelberg | Brandenburg | Turkey | CHL-STR-SPT-SUL | 3 | X1 | + | − | − |
| 01-00867 | Heidelberg | Brandenburg | Turkey | AMP-CHL-STR-SPT-SUL | 3 | X1 | + | − | − |
| 01-00830 | Heidelberg | Brandenburg | Turkey | AMP-CHL-STR-SPT-SUL-TET | 3 | X2 | + | − | − |
| 01-01645 | Heidelberg | Thüringen | Turkey | CHL-KAN-NEO-STR-SPT-SUL | 3 | X1 | + | − | − |
| 01-01744 | Heidelberg | Rheinland-Pfalz | Poultry meat | CHL-STR-SPT-SUL | 3 | X1 | + | − | − |
| 01-00466 | Heidelberg | Brandenburg | Turkey | AMP-CHL-KAN-NEO-STR-SPT-SUL-TET | 5 | X4 | + | − | − |
| 01-00828 | Heidelberg | Brandenburg | Turkey | CHL-STR-SPT-SUL | 11 | X5 | + | − | − |
| 01-00973 | Heidelberg | Brandenburg | Turkey | AMP-CHL-KAN-NEO-STR-SPT-SUL-TET | 7 | X3 | + | − | − |
| 01-01743 | Heidelberg | Rheinland-Pfalz | Turkey meat | CHL-STR-SPT-SUL | 12 | X1 | + | − | − |
| 01-00617 | Subspecies I, roughc | Brandenburg | Turkey | CHL-KAN-NEO-STR-SPT-SUL | 3 | X1 | + | − | − |
| 01-00614 | Subspecies I, rough | Brandenburg | Turkey | CHL-STR-SPT-SUL | 3 | X10 | + | − | − |
| 01-00055 | Subspecies I, rough | Bayern | Chicken | CHL-KAN-NEO-SPT-SUL-TET | 2 | X8 | + | − | − |
| 01-00398 | Subspecies I, rough | Bremen | Swine meat | AMP-CHL-STR-SPT-SUL-TET-TMP-SXT | 10 | X9 | + | − | − |
| 01-03713 | Monophasic [4,5,12:i:−] (U302) | Sachsen | Swine meat | AMP-CHL-GEN-STR-SPT-SUL-TET-TMP-SXT | 15 | X7 | + | − | + |
| 01-02571 | Typhimurium (DT104A) | Sachsen-Anhalt | Swine | AMP-CHL-FFN-STR-SPT-SUL-TET-TMP-SXT | 1 | X6 | + | − | − |
The MICs of 17 antimicrobial agents were assessed by the NCCLS broth microdilution method as described elsewhere (6). AMP, ampicillin; CHL, chloramphenicol; FFN, florfenicol; GEN, gentamicin; KAN, kanamycin; NEO, neomycin; STR, streptomycin; SPT, spectinomycin; SUL, sulfamethoxazole; TET, tetracycline; TMP, trimethoprim; SXT, trimethoprim-sulfamethoxazole.
The strains did not agglutinate with the specific sera used.
Seventeen of the sul3 carrier strains were from poultry livestock, one was from swine livestock, and four were from food products (two from poultry products and two from swine products) (Table 1). The poultry isolates were predominately isolated from turkeys or turkey products (at least 16 isolates). The sul3 gene could not be detected among isolates originating from cattle or feed.
The strains carrying sul3 belonged to different serotypes, most of them to S. enterica serotype Heidelberg (10 strains), and originated from 8 of the 16 German states, although most of them came from the Brandenburg region (12 strains). The strains were very heterogeneous, showing nine different antimicrobial resistance patterns (RPs), 15 plasmid profiles (PPs), and 15 XbaI PFGE patterns (PFP-X) (Table 1; Fig. 1 and 2). The predominant genomic group was represented by five Salmonella serotype Heidelberg strains and one rough strain, which showed PP3 and PFP-X1. Only one strain carried a second sulfonamide resistance gene (sul2).
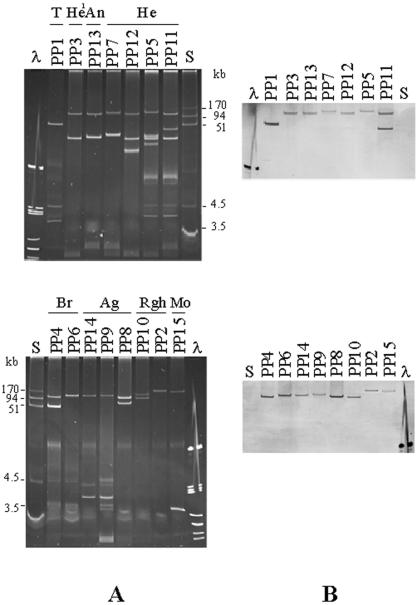
FIG. 1.
Plasmid analysis of the Salmonella strains carrying the sul3 gene. (A) plasmid patterns; (B) hybridization of the plasmid DNA in panel A with a sul3-specific probe. Lanes: λ, bacteriophage λ DNA digested with PstI; S, plasmids R27, R1, and V157 used as size standards (170 to 1.9 kb); T, Salmonella serotype Typhimurium; He, Salmonella serotype Heidelberg; An, Salmonella serotype Anatum; Br, Salmonella serotype Brandenburg; Ag, Salmonella serotype Agona; Rgh, rough; Mo, monophasic. 1, the same plasmid profiles were shown by rough strains.
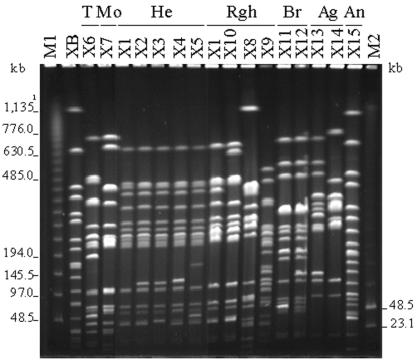
FIG. 2.
XbaI PFGE patterns of the Salmonella strains carrying the sul3 gene. Lanes: M1 and M2, Lambda Ladder and Low Range PFG-Markers (New England Biolabs, Schwalbach, Germany), respectively; XB, reference strain Salmonella serotype Braenderup H9812 (Centers for Disease Control and Prevention); T, Salmonella serotype Typhimurium; Mo, monophasic; He, Salmonella serotype Heidelberg; Rgh, rough; Br, Salmonella serotype Brandenburg; Ag, Salmonella serotype Agona; An, Salmonella serotype Anatum. 1, the size of the largest fragment of serotype Braenderup strain H9812.
The sul3 gene was located on large plasmids of different sizes, most of which were >90 kb (Fig. 1). One strain presented two copies of the gene on different plasmids.
The widespread resistance to sulfonamides is a good example of rapid adaptive evolution due to the horizontal transfer of resistance genes among mixed bacterial populations. Even though the use of sulfonamides in human medicine has decreased, selection pressures still exist in the veterinary, agriculture, and aquaculture fields in some countries (2, 7, 8, 18). Consequently, the genetic determinants for sulfonamide resistance are still very common in gram-negative bacterial plasmids. This persistence seems to be related to the incorporation of these determinants into very efficient vehicles for their spread: sul1 in class 1 integrons and sul2 in small multicopy plasmids or large transmissible multiresistance plasmids (1, 2, 5, 7, 17-19). The spread of the sul3 genes found in the Swiss E. coli isolates seems to be related to transposable elements (13; V. Perreten, personal communication).
The present work describes for the first time the presence of the sul3 gene in Salmonella strains. We have shown that sul3 can be found on different large plasmids and that it is not only spread among E. coli strains of different origins and from different countries (3, 6, 13). Sul3 can be detected in Salmonella strains of different origins, serotypes, and genomic groups. This study highlights the strong potential for the wide distribution of the sul3 resistance determinant in bacterial populations.
Nucleotide sequence accession number.
The sul3 gene found in Salmonella strains has been submitted to GenBank and can be found under accession number AY316203.
Acknowledgments
We thank V. Perreten (Institute of Veterinary Bacteriology, University of Bern, Bern, Switzerland) for plasmid pVP440 and B. Hoog, K. Pries, G. Berendonk, and especially S. Ceccaroni for helpful assistance. We also thank A. Schroeter, C. Dorn, and A. Miko (NRL-Salm) for support.
This work was supported by grants from the German Ministry of Consumer Protection and Agriculture (BMVEL; grant AZ:1000-WK-17/00) and the Federal Institute for Risk Assessment (BfR [formerly BgVV]; grant F501-28/1322-136).
REFERENCES
- 1.Chu, C., D. H. Chiu, W. Y. Wu, C. H. Chu, T. P. Liu, and J. T. Ou. 2001. Large drug resistance virulence plasmids of clinical isolates of Salmonella enterica serovar Choleraesuis. Antimicrob. Agents Chemother. 45:2299-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enne, V. I., D. M. Livermore, P. Stephens, and L. M. Hall. 2001. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357:1325-1328. (Erratum, 357:1890.) [DOI] [PubMed] [Google Scholar]
- 3.Grape, M., L. Sundstrom, and G. Kronvall. 2003. Sulphonamide resistance gene sul3 found in Escherichia coli isolates from human sources. J. Antimicrob. Chemother. 29:1022-1024. [DOI] [PubMed]
- 4.Guerra, B., S. Soto, S. Cal, and M. C. Mendoza. 2000. Antimicrobial resistance and spread of class 1 integrons among Salmonella serotypes. Antimicrob. Agents Chemother. 44:2166-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guerra, B., S. Soto, R. Helmuth, and M. C. Mendoza. 2002. Characterization of a self-transferable plasmid from Salmonella enterica serotype Typhimurium clinical isolates carrying two integron-borne gene cassettes together with virulence and drug resistance genes. Antimicrob. Agents Chemother. 46:2977-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guerra, B., E. Junker, A. Schroeter, B. Malorny, S. Lehmann, and R. Helmuth. 2003. Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine and poultry. J. Antimicrob. Chemother. 52:489-492. [DOI] [PubMed] [Google Scholar]
- 7.Huovinen, P., L. Sundström, G. Swedberg, and O. Sköld. 1995. Trimethoprim and sulfonamide resistance. Antimicrob. Agents Chemother. 39:279-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huovinen, P. 2001. Resistance to trimethoprim-sulfamethoxazole. Clin. Infect. Dis. 32:1608-1614. [DOI] [PubMed] [Google Scholar]
- 9.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lanz, R., P. Kuhnert, and P. Boerlin. 2003. Antimicrobial resistance and resistance gene determinants in clinical Escherichia coli from different animal species in Switzerland. Vet. Microbiol. 91:73-84. [DOI] [PubMed] [Google Scholar]
- 11.Malorny, B., A. Schroeter, C. Bunge, B. Hoog, A. Steinbeck, and R. Helmuth. 2001. Evaluation of molecular typing methods for Salmonella enterica serovar Typhimurium DT104 isolated in Germany from healthy pigs. Vet. Res. 32:119-129. [DOI] [PubMed] [Google Scholar]
- 12.Peters, T. M., C. Maguire, E. J. Threlfall, I. S. Fisher, N. Gill, A. J. Gatto, and the Salm-Gene Project. 2003. The Salm-gene project—a European collaboration for DNA fingerprinting for Euro Surveill. Euro. Surveill. 8:46-50. [DOI] [PubMed] [Google Scholar]
- 13.Perreten, V., and P. Boerlin. 2003. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 47:1169-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1997. Characterization of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 157:177-181. [DOI] [PubMed] [Google Scholar]
- 16.Schwarz, S., and E. Chaslus-Dancla. 2000. Use of antimicrobials in veterinary medicine and mechanisms of resistance. Vet. Res. 32:201-225. [DOI] [PubMed] [Google Scholar]
- 17.Sköld, O. 2000. Sulfonamide resistance: mechanisms and trends. Drug Resist. Update 3:155-160. [DOI] [PubMed] [Google Scholar]
- 18.Sköld, O. 2001. Resistance to trimethoprim and sulfonamides. Vet. Res. 32:261-273. [DOI] [PubMed] [Google Scholar]
- 19.Sorum, H., and T. M. L'Abee-Lund. 2002. Antibiotic resistance in food-related bacteria—a result of interfering with the global web of bacterial genetics. Int. J. Food Microbiol. 78:43-56. [DOI] [PubMed] [Google Scholar]