Abstract
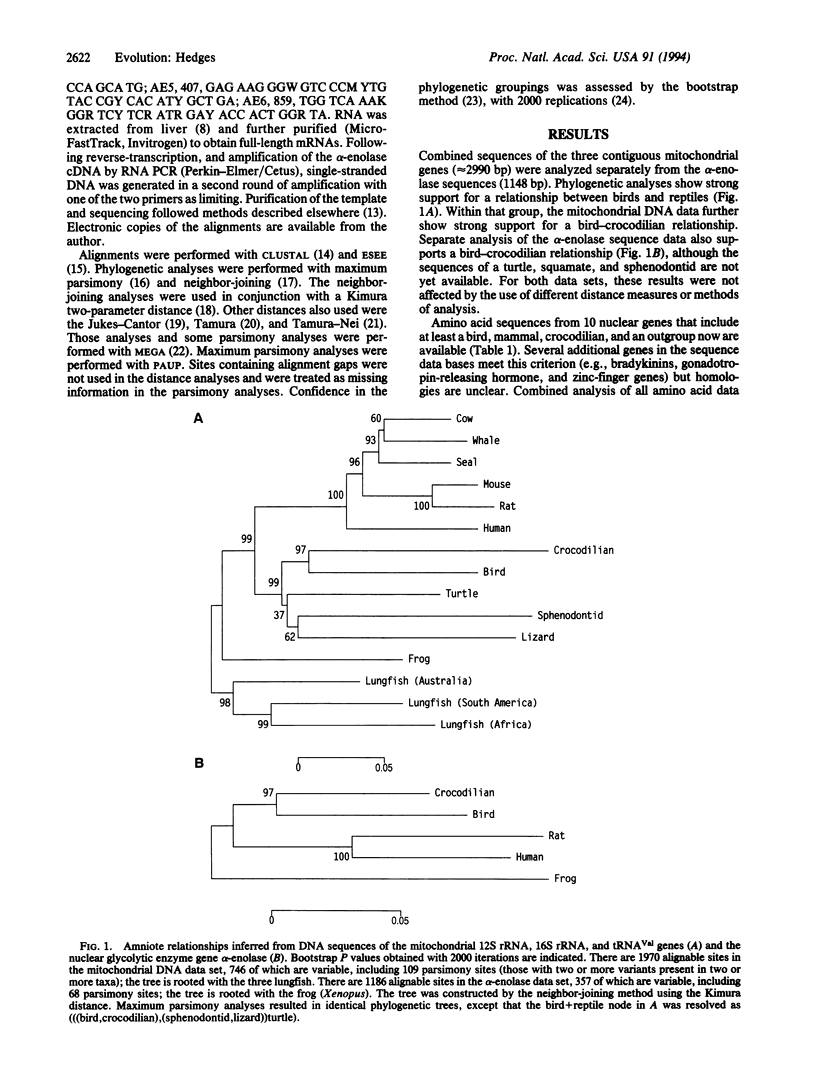
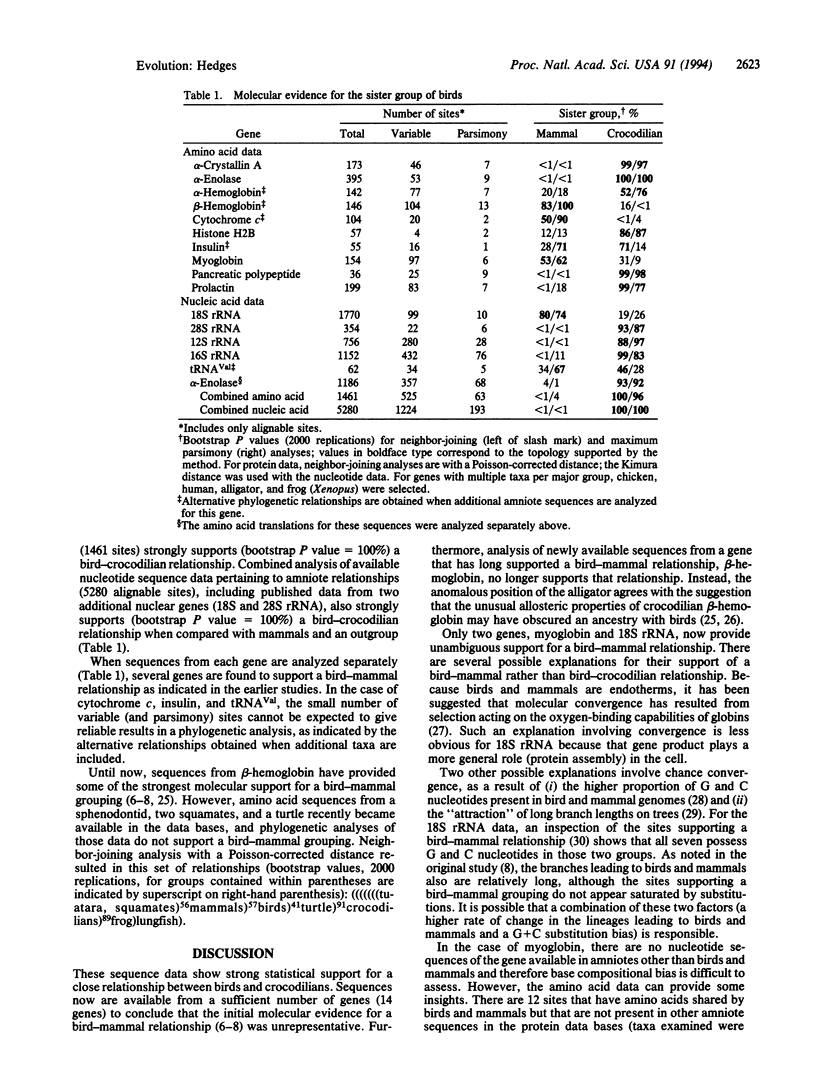
The major groups of amniote vertebrates appeared during a relatively short time span at the end of the Paleozoic Era, a fact that has caused difficulty in estimating their relationships. The fossil record suggests that crocodilians are the closest living relatives of birds. However, morphological characters and molecular sequence data from living amniotes have repeatedly challenged this hypothesis by indicating a bird-mammal relationship. DNA sequences from four slow-evolving genes (mitochondrial 12S and 16S rRNA, tRNAVal, and nuclear alpha-enolase) now provide strong statistical support for a bird-crocodilian relationship.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Olofsson B., Filipski J., Zerial M., Salinas J., Cuny G., Meunier-Rotival M., Rodier F. The mosaic genome of warm-blooded vertebrates. Science. 1985 May 24;228(4702):953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Cabot E. L., Beckenbach A. T. Simultaneous editing of multiple nucleic acid and protein sequences with ESEE. Comput Appl Biosci. 1989 Jul;5(3):233–234. doi: 10.1093/bioinformatics/5.3.233. [DOI] [PubMed] [Google Scholar]
- Dene H., Sazy J., Goodman M., Romero-Herrera A. E. The amino acid sequence of alligator (Alligator mississippiensis) myoglobin. Phylogenetic implications. Biochim Biophys Acta. 1980 Aug 21;624(2):397–408. doi: 10.1016/0005-2795(80)90081-1. [DOI] [PubMed] [Google Scholar]
- Giallongo A., Feo S., Moore R., Croce C. M., Showe L. C. Molecular cloning and nucleotide sequence of a full-length cDNA for human alpha enolase. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6741–6745. doi: 10.1073/pnas.83.18.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges S. B., Bezy R. L., Maxson L. R. Phylogenetic relationships and biogeography of xantusiid lizards, inferred from mitochondrial DNA sequences. Mol Biol Evol. 1991 Nov;8(6):767–780. doi: 10.1093/oxfordjournals.molbev.a040689. [DOI] [PubMed] [Google Scholar]
- Hedges S. B., Hass C. A., Maxson L. R. Relations of fish and tetrapods. Nature. 1993 Jun 10;363(6429):501–502. doi: 10.1038/363501b0. [DOI] [PubMed] [Google Scholar]
- Hedges S. B., Maxson L. R. Pancreatic polypeptide and the sister group of birds. Mol Biol Evol. 1991 Nov;8(6):888–891. doi: 10.1093/oxfordjournals.molbev.a040694. [DOI] [PubMed] [Google Scholar]
- Hedges S. B., Moberg K. D., Maxson L. R. Tetrapod phylogeny inferred from 18S and 28S ribosomal RNA sequences and a review of the evidence for amniote relationships. Mol Biol Evol. 1990 Nov;7(6):607–633. doi: 10.1093/oxfordjournals.molbev.a040628. [DOI] [PubMed] [Google Scholar]
- Hedges S. B. The number of replications needed for accurate estimation of the bootstrap P value in phylogenetic studies. Mol Biol Evol. 1992 Mar;9(2):366–369. doi: 10.1093/oxfordjournals.molbev.a040725. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Perutz M. F., Bauer C., Gros G., Leclercq F., Vandecasserie C., Schnek A. G., Braunitzer G., Friday A. E., Joysey K. A. Allosteric regulation of crocodilian haemoglobin. Nature. 1981 Jun 25;291(5817):682–684. doi: 10.1038/291682a0. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol. 1992 Jul;9(4):678–687. doi: 10.1093/oxfordjournals.molbev.a040752. [DOI] [PubMed] [Google Scholar]
- Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993 May;10(3):512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]


