Abstract
The 24-alkylated sterols have been shown previously to be absent in membranes of amphotericin B (AmB)-resistant Leishmania donovani promastigotes, suggesting that the S- adenosyl-l-methionine:C-24-Δ-sterol-methyltransferase (SCMT or ERG6) was not functional or not expressed in AmB-resistant (AmB-R) parasites. From an L. donovani wild-type clone, we cloned two cDNAs with an identical open reading frame encoding a putative SCMT, the enzyme responsible for a first sterol methylation at the C-24 position. The two cDNAs differed by their 3′-untranslated region (3′-UTR) and 5′-UTR sequences. One transcript (A) had a normal structure with a spliced leader and was highly expressed in normal cells but absent in AmB-R cells. The other (B), which did not possess the spliced leader sequence, was weakly expressed in normal cells but strongly expressed in AmB-R cells. As a functional test, ERG6 null mutant Saccharomyces cerevisiae yeasts were transformed using the pYES2.1 TOPO TA expression vector containing the candidate SCMT1/ERG6 coding sequence cloned from L. donovani. The transformed yeasts exhibited C-24 alkylated sterol expression, mainly ergosterol, within their membranes, proving that the isolated cDNA encodes on a SCMT responsible for sterol methylation. In AmB-R L. donovani promastigotes, the absence of the normal transcript (A) and the expression of an abnormal species (B) devoid of a spliced leader could explain the absence of sterol methylation in these cells. Further studies using a homologous system will allow us to draw conclusions about the relationship between SCMT expression and AmB resistance in Leishmania.
Leishmania donovani is a protozoan parasite that is responsible for fatal visceral leishmaniasis in the absence of treatment. Usually, pentavalent antimonials and amphotericin B (AmB) are the first-line treatments. However, their irregular effectiveness of these treatments is ascribed to an increase in the number of cases that exhibit primary resistance (antimonials) or relapse following antimonials or AmB therapy (5, 16). Several studies in India have demonstrated that acquired resistance to pentavalent antimonials is already affecting the treatment of visceral leishmaniasis. In the major focus of visceral leishmaniais in Bihar, India, 30 to 60% cases do not respond to drug treatment (14, 19). The antifungal agent AmB has long been recognized as a powerful antileishmanial drug. Its activity results from the specific target of AmB at the level of sterols, mainly ergosterol, which is found in the membrane of Leishmania genus and fungi. Largely because of the decline in the effectiveness of antimonials in India, AmB has been rediscovered as an effective treatment in visceral leishmaniasis despite its toxicity. To overcome toxicity. AmB formulations have been set up, and lipid formulations of AmB have been proven to increase the efficacy and to limit the toxicity of conventional AmB (4). In developing countries, the cost of lipid formulations of AmB precludes their use, and so AmB desoxycholate is used but is more toxic. Considering the widespread use of AmB in the treatment of visceral leishmaniasis, the emergence of AmB resistance is at risk. To anticipate clinical AmB resistance, we described some biochemical characteristics from an AmB-resistant (AmB-R) L. donovani promastigote line selected by stepwise drug pressure (15). From the specific affinity of AmB for ergosterol, we identified and quantified the sterol composition of the wild-type and AmB-R parasite membrane and found that C-24 alkylated sterols were absent from AmB-R parasites, indicating that the enzyme system responsible for this reaction could therefore be defective in AmB-R parasites. Thus, this total absence of C-24 alkylated sterols could result from the nonexpression of S-adenosyl-l-methionine:C-24-Δ-sterol-methyltransferase (SCMT orERG6; EC 2.1.1.41) or expression of inactive mutant SCMT. This paper describes the cloning of SCMT from L. donovani promastigotes.
MATERIALS AND METHODS
Chemical compounds.
AmB and bis(trimethylsilyl)trifluoroacetamide (BSTFA) were purchased from Sigma Chemicals (Saint-Quentin Fallavier, France). RPMI 1640 medium, fetal calf serum, HEPES, and l-glutamine were purchased from Life Technologies, Cergy-Pontoise, France.
Biological materials.
(i) Parasites. A strain of L. donovani (MHOM/IN/80/DD8) promastigotes from the World Health Organization collection at the London School of Hygiene and Tropical Medicine (London, United Kingdom) was used for selection of resistance to AmB by an in vitro stepwise selection process as previously described (15). Both wild-type (WT) and AmB-resistant (AmB-R) strains were cloned on semisolid RPMI 1640 medium containing 1% Bacto Agar (Difco) and 10% fetal calf serum as described by Iovannisci and Ullman (9). Colonies were picked up and transferred separately into liquid RPMI 1640 medium. The obtained clones were called L. donovani WT1 and AmB-R1. The susceptibilities of the WT1 and AmB-R1 clones to AmB were determined by calculation of the 50% inhibitory concentrations, which were found to be 0.09 ± 0.01 μM and 2.01 ± 0.14 μM, respectively. The resistance index was therefore estimated at 22.3. This value is similar to those obtained with WT and AmB-R parent lines.
(ii) Yeast.
The Saccharomyces cerevisiae WT strain, and S. cerevisiae ERG6 deletion strain BKY48-5C (α leu2-3 ura3-52 erg6Δ:LEU2) were kindly given by M. Bard, Biology Department, Indiana University. This last strain was used as the recipient strain for transformation with the SCMT1 cDNA.
In vitro promastigote culture.
Promastigote forms of L. donovani WT1 and AmB-R1 were cultivated in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf scrum, 25 mM HEPES, 2 mM l-glutamine, and gentamicin (50 μg/ml). Flasks were placed in an orbital incubator (Gallenkamp) under continuous shaking (150 rpm) at 27°C.
Yeast culture.
S. cerevisiae WT was grown at 30°C in liquid yeast-peptone-dextrose (YPD) complete medium containing 1% yeast extract (Difco), 1% Bacto Peptone (Difco), and 2% glucose.
Cloning of L. donovani SCMT.
From the L. major genome database (www.ebi.ac.uk/parasites/parasite-genome.html), we selected a genomic sequence with homology to sequences encoding SCMT from different organisms. Primers were designed in the open reading frame of the putative SCMT gene from L. major to amplify the equivalent cDNA from L. donovani and design internal primers. The total sequences of SCMT cDNAs were obtained by rapid amplification of cDNA ends (RACE) using a kit purchased from Clontech (Palo Alto, Calif.). The protocol accompanying the kit was used without modification, except that the 39-bp spliced leader sequence (5′-AACTAACGCTATATAAGTATCAGTTTCTGTACTTTATTG-3′) was used as the template for the sense primers in 5′-RACE. PCR products were cloned into the pCR-TOPO TA cloning vector (Invitrogen, Inc.) for sequencing.
Parasite RNA extraction and Northern blot analysis.
When parasite cultures reached a density of 15 × 106 to 20 × 106/ml (logarithmic phase), promastigotes were harvested by centrifugation and washed three times with large volumes of cold phosphate-buffered saline (pH 7.5). Total RNA was extracted by the method of Chomczynski and Sacchi (3). For Northern blot analysis, total RNA (20 μg) was denaturated and electrophoresed in a 1% agarose gel containing formaldehyde. Denatured RNAs were transferred by diffusion blotting onto a nylon membrane (Stratagene, La Jolla, Calif.) using 20× SSC (1× SSC is 0.15 M NaCl plus 15 mM sodium citrate). The membrane was first incubated for 4 h at 65°C in 0.25 M sodium phosphate buffer (pH 6.8) containing 1 mM EDTA, 7% sodium dodecyl sulfate, and 10 mg of bovine serum albumin per ml and then hybridized overnight at 65°C with the heat-denatured probe cDNA. The cDNA probes were α-32P labeled by random-priming extension. The membrane was washed twice for 20 min in 2× SSC-0.1% SDS at 65°C and once for 30 min in 0.1× SSC-0.1% SDS at 65°C and then autoradiographed at −80°C using Hyperfilm MP (Amersham Pharmacia Biotech); the SCMT cDNA probes used were prepared by reverse transcription-PCR (RT- PCR) using total RNA from L. donovani. The sense and antisense primers, designed on the basis of the 3′- untranslated region (3′-UTR) of SCMT1 cDNA A (GenBank accession number AY488058) were 5′- CATCTTCCCTCCCTTTCCTC-3′ and 5′-CCGCATGAACAACAGAGAGA-3′, respectively. The sense and antisense primers designed on the basis of the 3′-UTR of SCMT1 cDNA B (GenBank accession number AY488059) were 5′-TGTCCAGTAGTCCCGGAAAC-3′ and 5′-CGGCGATGATAGTGATGTTG-3′, respectively.
RT-PCR of the L. donovani RNA.
Total RNA (2 μg) was treated with 15 U of ThermoScript reverse transcriptase (Invitrogen, Inc.) in 20 μl of RT-PCR buffer for 90 min at 60°C as specified by the manufacturer. The control was run without the reverse transcriptase. The SCMT cDNA was amplified by 30 cycles (95°C for 5 s, 60°C for 10 s, and; 72°C for 1 min) in a GeneAmp 2400 thermal cycler (Perkin-Elmer) with 50 μl of prewarmed PCR buffer containing 1 μl of Advantage 2 polymerase Mix (Clontech), 250 nM each sense and antisense oligonucleotide primer, and 200 μM each deoxynucleoside triphosphate. The sense PCR primers for SCMT cDNA were designed on the basis of the L. major genomic sequence and were 5′- CCACACCTACACGCCGGAAC-3′ and 5′-ATGTCTGCCGGTGGCCGTG-3′, and the antisense primer for SCMT cDNA was designed on the basis of the L. major genomic sequence was 5′-CTAGGCCTGCTTGGACGGCTT-3′. The amplified product were expected to contain 1,084 bp and 1,061 bp respectively.
SCMT expression in the yeast.
The SCMT1 coding sequence was amplified by PCR using 5′-ATGTCTGCCGGTGGCCGTG-3′ and 5′-CTAGGCCTGCTTGGACGGCTT-3′ as sense and antisense primers, respectively. The PCR product was cloned in the pYES2.1/V5-His-TOPO expression vector from the pYES2.1 TOPO TA expression kit (Invitrogen, Inc.). The clones were purified and sequenced with the BigDyeTerminator cycle-sequencing method on an automated ABI Prism 310 sequencer (Perkin-Elmer Applied Biosystems). The transformation of the SCMT (ERG6) null mutant yeast strain was performed by the method of Ito et al. (10). Cultures of ERG6 null mutant yeast strain were grown to mid-log phase, diluted back to 2 × 106 to 3 × 106 cells per ml in YPD, and grown to 6 × 106 to 7 × 106 cells per ml on an orbital shaker at 30°C. A 50-ml sample of culture was harvested, washed twice in 0.1 M lithium acetate-10 mM Tris (pH 8.0)-1 mM EDTA, and suspended in a final volume of 200 μl of 0.1 M lithium acetate-10 mM Tris (pH 8.0)-1 mM EDTA. Immediately afterward, 20 μg of plasmid DNA and 100 μg of sheared calf thymus DNA were added, and the cell suspension was incubated on a roller at 30°C for 1.5 h. Aliquots (100 μl) of cell suspension were then transferred to microcentrifuge tubes, to which 0.7 ml of 36% polyethylene glycol-0.1 M lithium acetate-1× Tris-EDTA was added. The cells were vortexed thoroughly, incubated at room temperature for 1 h, and heat shocked at 42°C for 2 min before being plated out on selective medium (yeast nitrogen base plus amino acids without uracil).
Sterol determination.
S. cerevisiae WT, S. cerevisiae ERG6 null mutant, and ERG6 transformed cells were studied for ergosterol expression, this sterol being the end compound resulting from the C-24 sterol methylation. Yeast cells were grown as described above in a total volume of 500 ml of culture medium. The cells were harvested, washed three times in phosphate- buffered saline (pH 7.5), and pooled, and the pellet was suspended in 20 ml of dichoromethane- methanol (2:1, vol/vol) for about 24 h at 4°C. After centrifugation (11,000 × g for 1 h at 4°C), the extract was evaporated under vacuum. The residue was saponified with 30% KOH in methanol at 80°C for 2 h. Nonsaponifiable sterols were extracted from stationary-phase yeast cells into n-heptane. The sterol residue was dissolved in dichloromethane. An aliquot of clear yellow sterol solution was added to 2 volumes of BSTFA, and the sealed tubes were heated at 80°C for 1 h. The trimethylsilyl ethers of sterols were subjected to gas chromatography/mass spectrometry analysis. Three experiments were performed, and the results are expressed as mean of percentages relative to total sterols ± standard deviations.
RESULTS
Cloning of two SCMT1 cDNAs from L. donovani WT1 promastigotes.
A BLAST analysis of the L. major genome database (www.ebi.ac.uk/parasites/parasite-genome.html), using the S. cerevisiae SCMT Erg6 sequence (Swiss-Prot accession number P25087) as a query evidenced two genomic sequences from L major Friedlin strain V1. The largest was a 3,529,852-bp fragment (GenBank accession number AL499624), corresponding to L. major assembled shotgun reads from chromosome 36. It comprised a 1,062 bp open reading frame which encodes a 353-amino-acid predicted protein exhibiting 42% identity to the G23-A381 portion of the S. cerevisiae SCMT. The second was a 569-bp fragment (GenBank accession number AQ843968) corresponding to the region of this ORF from positions 484 to 1047.
From these sequences, primers were designed in two conserved domains of SCMTs to amplify the equivalent regions by using L. donovani WT1 cDNAs. Sequencing of the amplification product allowed the design of internal primers for 3′ and 5′-RACE.
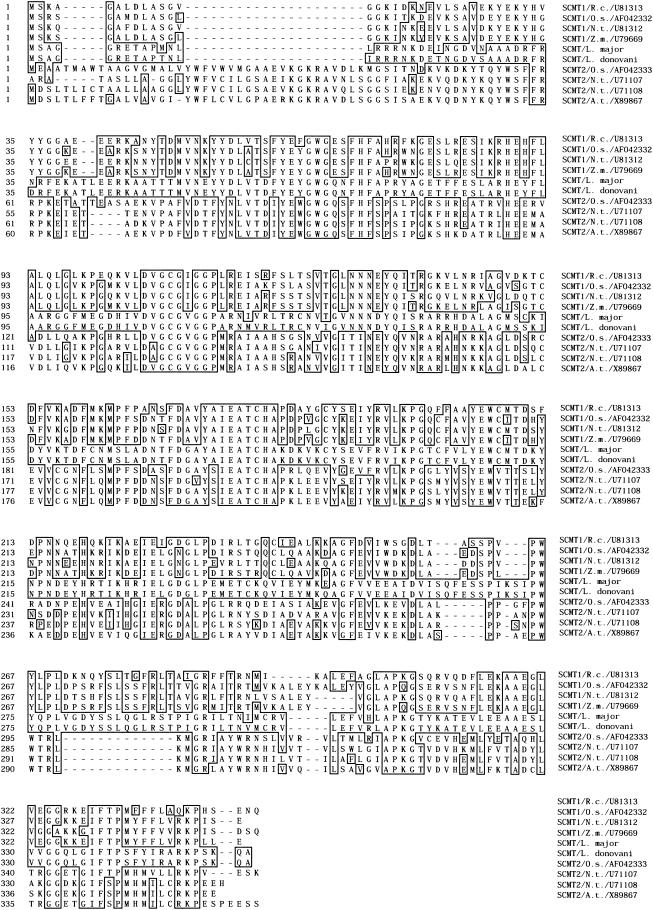
Two cDNAs (A and B) were cloned. Both contained the same 1,062-bp ORF which encodes a 353-amino-acid protein which is 96% identical to the predicted L. major SCMT. The sequences of the putative SCMT from L. major and L. donovani are highly similar to plant type 1 and 2 SCMTs (Fig. 1). However, the sequence of L. donovani SCMT is closer to that of the type 1 (41 to 45% identity) than the type 2 (32 to 34% identity) family of plant SCMTs. The putative SCMT cloned from L. donovani should therefore catalyze a first methylation of the sterol at the C-24 position rather than a second sterol methylation.
FIG. 1.
Multiple amino acid sequence alignment of L. major, L. donovani, and eight plant SCMTs. The L. major sequence is predicted from the genomic sequence. L. donovani SCMT is predicted by conceptual translation of the ORF of the cloned cDNAs. Plant SCMT sequences were predicted from their cDNA, whose GenBank accession number is indicated on the right. Residues that exactly match the L. donovani SCMT sequence are shown in boxes. Plant sequences from family 1 and 2 SCMTs are represented above and below the two Leishmania members, respectively. Plant sequences were from Ricinus communis (R.c.), Oryza sativa (O.s.), Zea mays endosperm (Z.m.), Nicotiana tabacum (N.t.), and Arabidopsis thaliana (A.t.). The alignment was prepared with Megalign program using the Clustal method.
The protein was also 36, 38, and 41% identical to the SCMT from Neurospora crassa, candida albicans, and S. cerevisae (ERG6), respectively. It consists of a predicted 39.9-kDa protein which comprises no signal peptide and is probably cytoplasmic, according to its average hydrophobicity, although its C-terminal (L329-A346) region contains a hydrophobic tail.
Expression of SCMT transcripts A and B in WT1 and AmB-R1 cells.
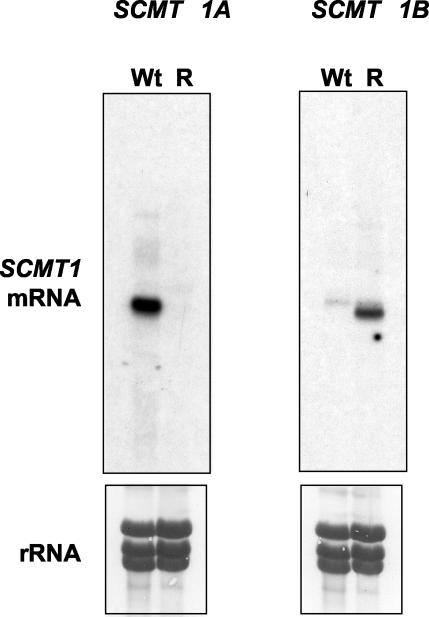
Both transcripts (A and B) encoded the same protein but exhibited different 3′-UTR sequences. The 3′-UTR of transcript A was longer (1,768 nucleotides [nt]) than that of transcript B (1,109 nt), although the first 182 nt downstream of the stop codon were identical in the two transcripts. Differences in the 3′-UTR sequences allowed us to analyze the expression of each species in WT and AmB-R cells by Northern blot analysis Figure 2 shows that transcript A was highly expressed in WT1 parasites but undetectable in resistant cells. By contrast, the expression of transcript B was high in AmB-R cells but was not detected in WT cells (Fig. 2).
FIG. 2.
Expression profile of SCMT1A and SCMT1B mRNA in WT and resistant strains. A Northern blot is shown for 20 μg of total RNA from L. donovani WT and resistant strains that were hybridized with the α-32P-labeled 3′-UTR SCMT1A probe (left) or with the α-32P- labeled 3′-UTR SCMT1B probe (right). The lower panels show the rRNA bands detected by methylene blue staining.
Further characterization of SCMT1 A and B mRNAs.
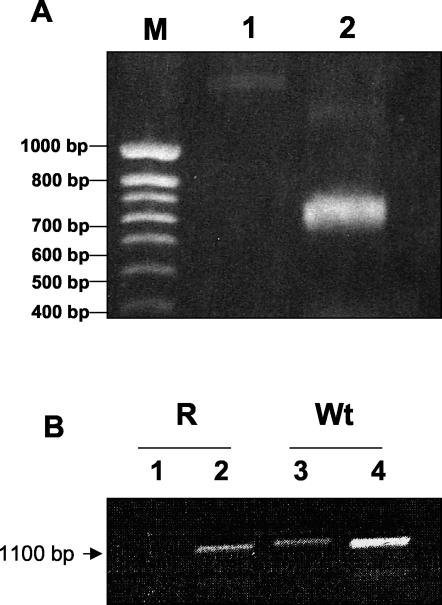
PCR experiments were performed to analyze the 5′-UTR sequences of transcripts A and B. With a forward primer matching on the spliced leader and a reverse primer in shared the ORF sequence of the two transcripts, only cDNA from the WT cells could be amplified, indicating that transcript B expressed in AmB-R cells had no spliced leader (Fig. 3A). In addition, PCR experiments were performed with forward primers matching just upstream of the start codon and reverse primer in the common ORF sequence. Under these conditions, no amplification product was obtained with the cDNAs from AmB-R (Fig. 3B, lane 1) whereas a product of the expected size was obtained with the forward primer matching just downstream of the start codon (lane 2). The expected amplification products were also obtained in control PCR experiments performed under identical conditions with cDNAs from WT cells, whatever the forward primer used (lanes 3 and 4). In conclusion, PCR experiments indicated that (i) transcript B was abnormal since it had no spliced leader, (ii) transcripts A and B had different 5′-UTR sequences, and (iii) transcript A was not expressed at all in resistant cells, as previously suggested by the Northern blot analyses (Fig. 2).
FIG. 3.
RT-PCR using total RNA isolated from L. donovani WT and resistant promastigotes. (A) 5′-RACE-PCR of cDNA. PCR was performed with a template consisting of single-stranded cDNA generated by reverse transcription of RNAs from WT and AmB-R L. donovani strains. Lanes: 1, cDNA template from resistant L. donovani strain amplified by PCR with sense and antisense oligonucleotide primers designed from the spliced leader sequence and from the coding sequence, respectively; 2, cDNA template from WT L. donovani amplified by PCR with sense and antisense oligonucleotide primers designed from the spliced leader sequence and from the coding sequence, respectively. The amplified products were analyzed by agarose gel electrophoresis and ethidium bromide staining. Size markers are shown on the left (lane M). (B) Identification of the 5′-UTR by RT-PCR analysis. Total RNA was reverse transcribed, as described in Materials and Methods. Lanes: 1, cDNA template from AmB-R L. donovani cells amplified by PCR with sense and antisense oligonucleotide primers designed from the 5′-UTR sequence of SCMT1A and from the coding sequence; 2, cDNA template from AmB-R L. donovani cells amplified by PCR with sense and antisense oligonucleotide primers designed from the coding sequence; 3, cDNA template from WT L. donovani amplified by PCR with sense and antisense oligonucleotide primers designed from the 5′-UTR sequence of SCMT1A and from the coding sequence; 4, cDNA template from WT L. donovani amplified by PCR with sense and antisense oligonucleotide primers designed from the coding sequence. The amplified products were analyzed by agarose gel electrophoresis and ethidium bromide staining.
Assessement of SCMT activity.
The putative SCMT activity of the SCMT1 gene product was estimated by the amount of ergosterol found within the yeast. The sterol analysis of total membranes showed that ergosterol represented 27.4% ± 3.1% of total sterol in the S. cerevisiae WT clone whereas it was totally absent from the S. cerevisiae SCMT null mutant clone. The yeast transformed with the SCMT1 candidate gene were able to induce ergosterol expression (31.1% ± 2.8% of the total sterols), whereas the same strain grown without an inducer was unable to express ergosterol. These results confirmed that the isolated gene was responsible for the C-24 methylation of sterols and therefore could be ascribed to SCMT1.
DISCUSSION
AmB is a polyene macrolide antibiotic derived from the actinomycete Streptomyces nodosus and is now a first-line treatment of visceral leishmaniasis refractory to antimonials. The toxicity of AmB is based on the affinity of this polyene antibiotic to bind to sterols and to form aqueous pores in the membranes of cells (1). The selective activity of AmB against fungi and Leishmania is due to the higher affinity of the drug for 24-substituted sterols, found in the plasma membrane of these eukaryotic microorganisms, than for cholesterol found in the plasma membranes of mammalian cells (1, 22). Thus, AmB binds strongly to ergosterol, the predominant plasma membrane sterol in fungi and Leishmania. This interaction leads to the formation of aqueous pores consisting of an annulus of eight amphotericin B molecules linked hydrophobically to the membrane ergosterol, which is thereby displaced from its normal phospholipid interactions. The consequence of this binding includes disruption of the osmotic integrity of the membrane, with leakage of intracellular potassium and magnesium, and disruption of oxidative enzymes in target cells, which leads to cell death (2).
Our previous study demonstrated that one of the most important characteristics of the AmB-R in L. donovani promastigotes was the absence of C-24-alkylated sterols in the membranes of AmB-resistant parasites, possibly explaining the lower affinity of AmB for AmB-R parasite membranes. The question was to identify the origin of the absence of the C- 24 methylation of sterols in the resistant parasites to study further the relationship between sterol alkylation and AmB-R. The most relevant hypothesis was to consider that the SCMT enzyme system was defective, leading to the following possibilities: (i) SCMT is not expressed in the AmB-R parasites, and the mRNA is absent; (ii) SCMT is expressed with a mutation(s) which could invalide its activity, and mRNA is present in the absence of ergosterol within the membranes. The aim of this study was therefore to determine the reason for the absence of ergosterol in AmB-R parasites at the molecular level.
We isolated from L. donovani WT cells two cDNAs corresponding to a normal transcript A with its spliced leader and to transcript B which exhibited different 3′- and 5′-UTRs and had no spliced leader. They had an identical ORF encoding the enzyme responsible for a first sterol methylation at the C-24 position. As a functional test, ERG6 null- mutant S. cerevisiae yeasts were transformed using a yeast expression vector containing the coding sequence of the putative SCMT (ERG6) from L. donovani. The transformed yeasts exhibited C-24-alkylated sterol expression, mainly ergosterol, within their membranes. Therefore, the isolated cDNA encoded the functional enzyme catalyzing the sterol methylation, which was thus identified as an SCMT. The SCMT from L. donovani was found to be close to those of plants and fungi.
The normal transcript A was completely absent from AmB-R cells. By contrast, these cells overexpressed the transcript B, but the absence of spliced leader probably prevented its translation and could be the cause of the absence of sterol methylation. Both transcripts probably arise from differential splicing in their UTR sequences but could also originate from two different genes. Further studies are necessary to characterize in more detail the 5′-UTR of transcript B and the genetic defect affecting the SCMT gene(s) in AmB-R parasites. In a previous study, DNA amplification was observed in two AmB-R mutants of L. tarentolae (17). The amplicons were extrachromosomal circles and were derived from different chromosomes. In one mutant, the circle was unusually stable, since it remained within the cell despite numerous passages free of drug. These authors observed a circumstantial link between the copy number of the amplicons and the resistance level. They conclude that several mutations could act together to lead to amphotericin B resistance (17).
Despite the widespread use of AmB against fungal infections and leishmaniasis, resistance to this polyene antifungal agent tends to be species dependent and emerges uncommonly and slowly in isolates from patients treated with AmB (7), whereas no clinical resistance has yet been found in Leishmania. Thus, Candida is a fungal genus developing AmB-R (7). The principal characteristic of the mechanism of resistance to AmB is a decrease in the amount of ergosterol in the fungal cell membrane (7), whereas a total absence of ergosterol was shown for AmB-R in Leishmania (15). Thus, a potential molecular mechanism of polyene resistance in Leishmania includes the absence of ergosterol.
The emergence of polyene-resistant fungi as a result of both qualitative and quantitative changes in the sterols of the cell membrane has been reported (20). In AmB-R mutant Aspergillus fennelliae strains, sterols other than ergosterol were seen in the membranes (13). Kelly et al. (12) studied two Cryptococcus neoformans isolates from a patient with AIDS before and after failing to respond to treatment with amphotericin B, and they reported that in the resistant strain there had been a depletion of ergosterol in favor of other sterols, due to a defect in the sterol isomerase enzyme. In a series of polyene-resistant C. albicans isolates, a 74 to 85% reduction in the ergosterol content was recorded (6). Published reports of the mechanisms of polyene resistance in fungi predominantly implicate lesions in ergosterol biosynthesis as the cause of such resistance (11, 12, 18). Lesions in enzymes involved in sterol biosynthesis, which result in the formation of functional sterols other than ergosterol and which give rise to polyene resistance, include defects in Δ-8-7-isomerase (12) and Δ-5-6-desaturase (11, 12). Such strains exhibiting these phenotypes accumulate Δ-8-sterols (ergosta-8-en-3β-ol and ergosta-8,22-dien-3β-ol) and delta-7-sterols (ergosta-7-en-3β-ol and ergosta-7,22-dien-3β-ol), respectively. A lesion in the latter also provides cross-resistance to azole antifungals (20). The mechanisms of resistance to AmB indicate fundamental differences between Leishmania and fungi. Thus, SCMT has not been linked to AmB resistance in fungi as it is in Leishmania, and the sterol composition of AmB-R Leishmania does not favor the involvement of Δ-8-7-isomerase and Δ-5-6-desaturase as observed in fungi. The present study hypothesizes the relationship between SCMT expression and AmB resistance in Leishmania. To obtain the final proof, we need to complete this investigation by using a homologous system. Therefore, further studies will be focused on the deletion of the SCMT gene in WT Leishmania spp. and its transfection in AmB-R parasites to verify the sensitivity to AmB in order to confirm the links between SCMT and AmB resistance.
Acknowledgments
This project was supported by a BQR grant (Bonus Qualité Recherche of the University Paris XI). Stanislas Morand is the recipient of a fellowship from the Ministère de l'Education Nationale de la Recherche et de la Technologie (MENRT).
M. Bard from Indiana University is acknowledged for supplying wild-type and ERG6 null mutant S. cerevisiae strains. P. Bouvier-Nave from the Institut de Botanique de Strasbourg (France) and F. Gamarro from the CSIC of Granada (Spain) are also acknowledged for fruitful discussions.
REFERENCES
- 1.Brajtburg, J., and J. Bolard. 1996. Carrier effects on biological activity of amphotericin B. Clin. Microbiol. Rev. 9:512-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brajtburg, J., W. G. Powderley, G. S. Kobayashi, and G. Medoff. 1990. Amphotericin B: delivery systems. Antimicrob. Agents Chemother. 34:183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 4.Croft, S. L., and V. Yardley. 2002. Chemotherapy of leishmaniasis. Curr. Pharm. Des. 8:319-342. [DOI] [PubMed] [Google Scholar]
- 5.Desjeux, P., B. Piot, K. O'Neil, and J. P. Meert. 2001. Co-infections of Leishmania/HIV in south Europe. Med. Trop. 61:187-193. [PubMed] [Google Scholar]
- 6.Dick, J. D., W. G. Merz, and R. Saral. 1980. Incidence of polyene-resistant yeasts recovered from clinical specimens. Antimicrob. Agents Chemother. 18:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis, D. 2002. Amphotericin B: spectrum and resistance J. Antimicrob. Chemother. 49:7-10. [DOI] [PubMed] [Google Scholar]
- 8.Hebeka, E. K., and M. Solotorovsky. 1965. Development of resistance to polyene antibiotics in Candida albicans. J. Bacteriol. 89:1533-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iovannisci, D. M., and B. Ullman. 1983. High efficiency plating method for Leishmania promastigotes in semidefined or completely-defined medium. J. Parasitol. 69:633-636. [PubMed] [Google Scholar]
- 10.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joseph-Horne, T., N. J. Manning, D. W. Hollomon, and S. L. Kelly. 1995. Defective sterol Δ5(6) desaturase as a cause of azole resistance in Ustilago maydis. FEMS Microbiol. Lett. 127:29-34. [DOI] [PubMed] [Google Scholar]
- 12.Kelly, S. L., D. C. Lamb, M. Taylor, A. J. Corran, B. C. Baldwin, and W. G. Powderly. 1994. Resistance to amphotericin B associated to defective sterol Δ8,7 isomerase in a Cryptococcus neoformans strain from an AIDS patient. FEMS Microbiol. Lett. 122:39-42. [DOI] [PubMed] [Google Scholar]
- 13.Kim, S. J., K. J. Kwon-Chung, G. W. Milne, and B. Prescott. 1974. Polyene-resistant mutants of Aspergillus fennelliae: identification of sterols. Antimicrob. Agents Chemother. 6:405-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lira, R., S. Sundar, A. Makharia, R. Kenney, A. Gam, E. Saraiva, and D. Sacks. 1999. Evidence that the high incidence of treatment failures in Indian kala-azar is due to the emergence of antimony-resistant strains of Leishmania donovani. J. Infect. Dis. 180:564-567. [DOI] [PubMed] [Google Scholar]
- 15.Mbongo, N., P. M. Loiseau, M. A. Billion, and M. Robert-Gero. 1998. Mechanism of amphotericin B resistance in Leishmania donovani promastigotes. Antimicrob. Agents Chemother. 42:352-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray, H. W. 2000. Treatment of visceral leishmaniasis (kala-azar): a decade of progress and future approaches. Int. J. Infect. Dis. 4:158-177. [DOI] [PubMed] [Google Scholar]
- 17.Singh, A. K., B. Papadopoulou, and M. Ouellette. 2001. Gene amplification in amphotericin B-resistant Leishmania tarentolae. Exp. Parasitol. 99:141-147. [DOI] [PubMed] [Google Scholar]
- 18.Taylor, F. R., R. J. Rodriguez, and L. W. Parks. 1983. Requirement for a second sterol biosynthetic mutation for viability of a sterol C-14 demethylation defect in Saccharomyces cerevisiae. J. Bacteriol. 155:64-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thakur, C. P., G. P. Sinha, A. K. Pandey, N. Kumar, P. Kumar, S. M. Hassan, and S. Narain. 1998. Do the diminishing efficacy and increasing toxicity of sodium stibogluconate in the treatment of visceral leishmaniasis in Bihar, India, justify its continued use as a first-line drug? An observational study of 80 cases. Ann. Trop. Med. Parasitol. 92:561-569. [DOI] [PubMed] [Google Scholar]
- 20.Vanden Bossche, H., H. P. Marichal, and F. C. Odds. 1994. Molecular mechanisms of drug resistance in fungi. Trends Microbiol. 2:393-400. [DOI] [PubMed] [Google Scholar]
- 21.Watson, P. F., M. E. Rose, S. W. Ellis, H. England, and S. L. Kelly. 1989. Defective sterol C5-6 desaturation and azole resistance: a new hypothesis for the mode of action of azole antifungals. Biochem. Biophys. Res. Commun. 164:1170-1175. [DOI] [PubMed] [Google Scholar]
- 22.Werbovetz, K. A. 2002. Promising therapeutic targets for antileishmanial drugs. Expert Opin. Ther. Targets 6:407-422. [DOI] [PubMed] [Google Scholar]