Abstract
The Drosophila lymph gland is a hematopoietic organ in which progenitor cells, which are most akin to the common myeloid progenitor or CMP in mammals, proliferate and differentiate into three types of mature cells – plasmatocytes, crystal cells and lamellocytes – the functions of which are reminiscent of mammalian myeloid cells1. During the first and early second instars of larval development, the lymph gland contains only progenitors, whereas in the third instar, a medial region of the primary lobe of the lymph gland called the medullary zone (MZ) contains these progenitors2, while maturing blood cells are found juxtaposed in a peripheral region designated the cortical zone (CZ)2. A third group of cells referred to as the posterior signaling center (PSC) functions as a hematopoietic niche3,4. Similar to mammalian myeloid cells, Drosophila blood cells respond to multiple stresses including hypoxia, infection, and oxidative stress5–7. However, how systemic signals are sensed by myeloid progenitors to regulate cell fate determination has not been well described. Here, we show that the hematopoietic progenitors of Drosophila are direct targets of systemic (insulin) and nutritional (essential amino acid) signals, and that these systemic signals maintain the progenitors by promoting Wingless (WNT in mammals) signaling. We expect that this study will promote investigation of such possible direct signal sensing mechanisms by mammalian myeloid progenitors.
Keywords: Insulin signaling, dTOR, slimfast, amino acid sensing, myeloid lineage, metabolic control, inflammation, metabolic disorder, hematopoiesis
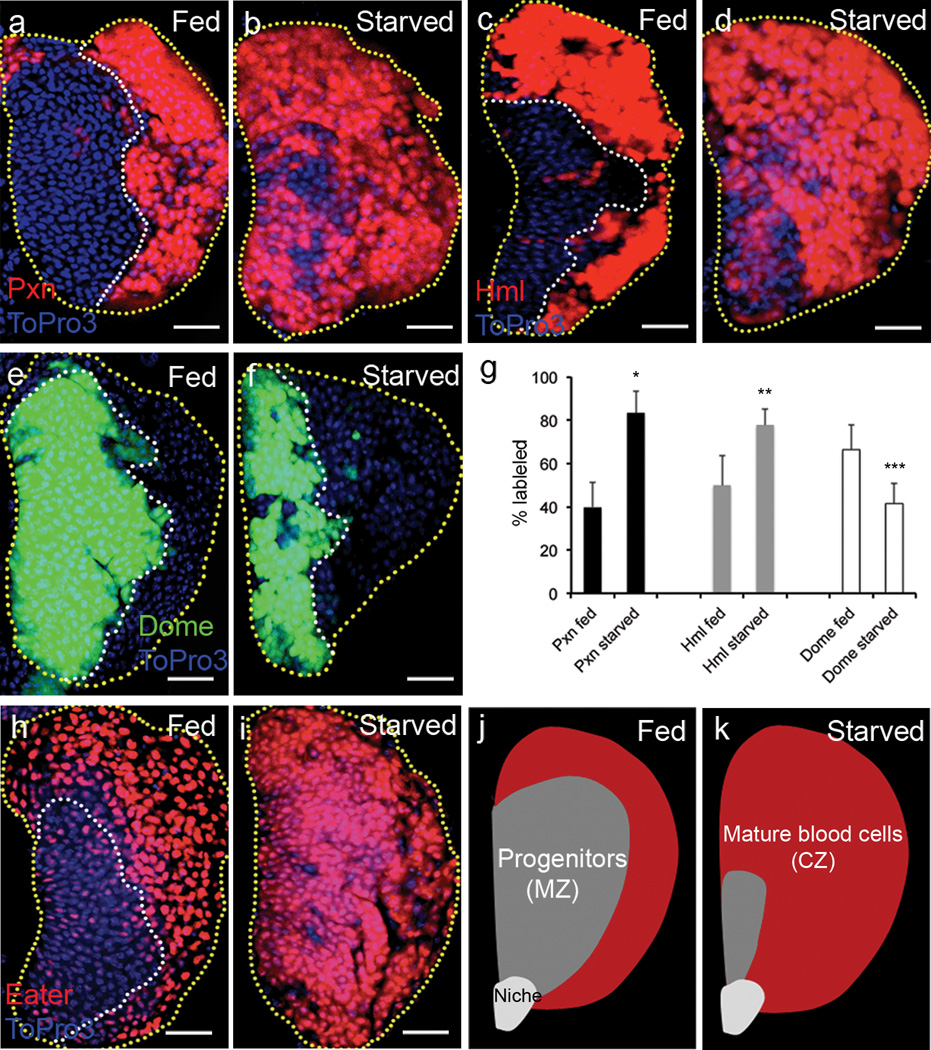
It is known that metabolic dysfunction in mammals causes abnormal inflammatory responses in the blood system. However, how metabolic stresses impinge on hematopoiesis is still unclear. Here, we found that starvation of Drosophila larvae leads to blood cell phenotypes. The most striking effect is acceleration of blood cell differentiation both in time and number of cells affected in the lymph gland. Following 24 hours of starvation, cells occupying the position of the MZ begin to express differentiation markers such as Peroxidasin (Pxn)8 and Hemolectin (Hml)9 normally restricted to the CZ (Fig 1a–d, g). Corresponding to this increase, a substantial reduction of Domeless (Dome) marking the progenitor population is also evident (Fig 1e–g). The protein Eater10, normally expressed at very low levels in the progenitors and at high levels in differentiated cells, is expressed at high levels in all cells upon starvation (Fig 1h–i). These data are schematically summarized in Fig 1j–k.
Figure 1. Starvation induces abnormal differentiation in the lymph gland.
Yellow dotted line indicates boundary of the lymph gland; white, the progenitors. Each lymph gland section is a middle section of a Z stack. White scale bars: 25 µm. Yellow scale bars: 50 µm. Blue indicates DNA marker, ToPro3. The number of lobes examined is indicated as n. Maturing cell markers, Peroxidasin (Pxn) and Hemolectin (Hml) (Pxn-gal4 UAS-GFP and hmlΔ-gal4 UAS-2xeGFP) are shown in red for consistency.
a, Pxn (red) is expressed only at the periphery of the lymph gland in well-fed animals (n=20).
b, Starvation induces abnormal differentiation of the progenitors, marked by Pxn (red), towards the medial region of the lymph gland (n=20).
c–d, This differentiation is also apparent using another mature cell marker, Hml (red, n=16 for control, n=14 for starved animals).
e–f, The progenitors, marked by dome-gal4 UAS-2xEYFP (green, borders demarcated by white dotted line) occupy medial region of the lymph gland in well-fed animals (n=14) (e), while starvation reduces the expression of Dome (green) (n=10) (f).
g, Quantification of starvation-induced differentiation phenotype. Percentage changes in markers (black, Pxn; grey, Hml; white, Dome) are as indicated (n=10 for each sample; *p=4.2×10−10, **p=2.8×10−5, ***p=0.001). Error bars indicate s.d.
h–i, Mature cell marker, eater-GFP (shown in red, for consistency with other panels) is only expressed in the CZ in well-fed wild-type lymph gland (n=14) (h), whereas starvation induces expression of eater-GFP (red) in cells occupying the progenitor region of the lymph gland (n=16) (i).
j–k, Schematic representation of a late third instar lymph gland primary lobe under normally fed and starved conditions.
j, In well-fed animals, mature blood cells (red) are restricted to the peripheral Cortical Zone (CZ) and the progenitors (grey) are seen in the central Medullary Zone (MZ).
k, Similarly staged starved animals exhibit abnormally accelerated maturation of blood cells (red) with reduced progenitor population (grey). Starvation does not alter the overall size of the lymph gland, but increases the relative proportion of differentiated cells.
The starvation experiments were performed either on PBS-soaked Whatman paper11(Supplementary Fig 1a–c) or on 1% agar plate12 (Fig 1a–d,g; Supplementary Fig 1d–f). Aseptic conditions to control against indirect effects due to bacterial infection were also used (Supplementary Fig 1g–i). In all controlled experimental conditions, starvation reduced progenitor population and caused an increase in the number of differentiating cells (Fig 1j–k), without an obvious alteration in the size of the hematopoietic organ, or the apoptotic profile of its cells (Supplementary Fig 1j–l).
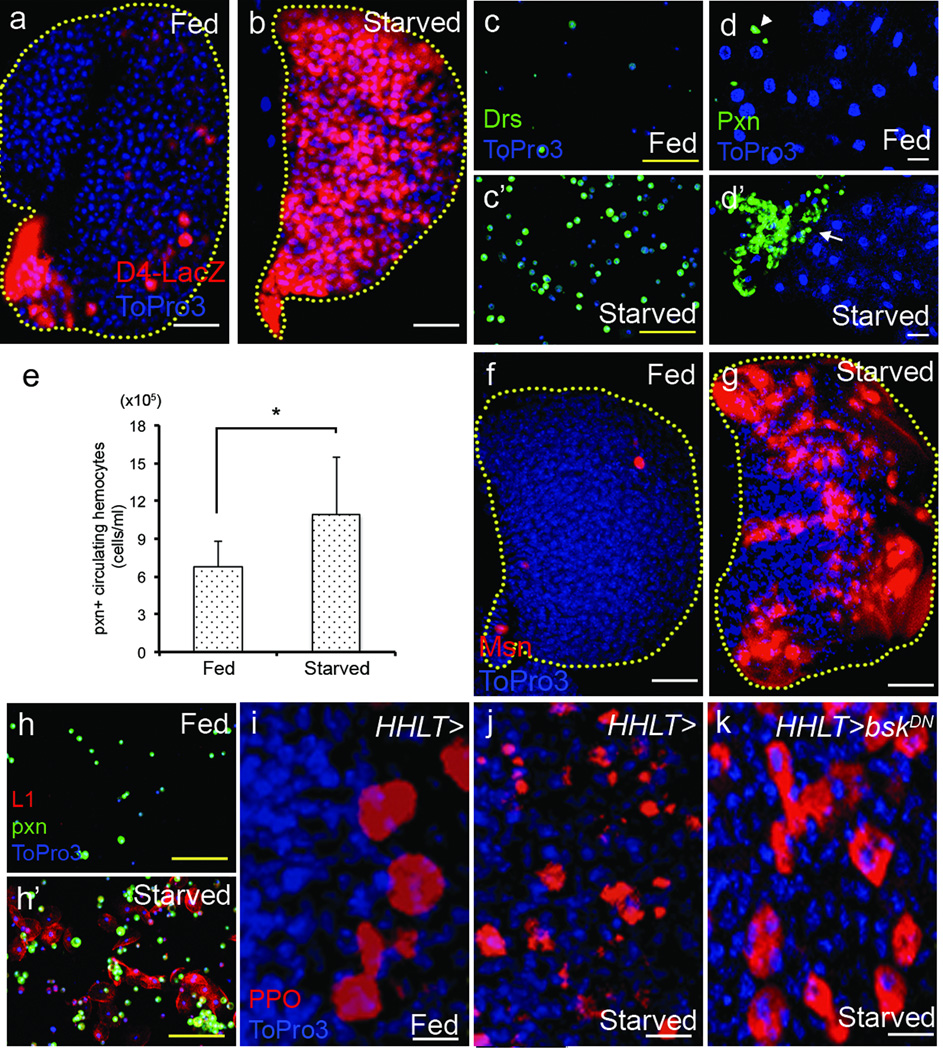
Similar to metabolically induced inflammation in mammals13, starvation in Drosophila larvae activates NFκB-like transcription factors, assessed by the expression of a targets, D4-LacZ14 (Fig 2a–b) and antimicrobial peptides15 in circulating hemocytes (Fig 2c–c’) and within the lymph gland (Supplementary Fig 1m–n). Starvation also causes an increase in circulating blood cells arising from the embryonic head mesoderm (Fig 2e), infiltration of Pxn+ plasmatocytes into the fat body (Fig 2d–d’), the Drosophila equivalent of the mammalian liver and adipose tissue, and differentiation of lamellocytes, another hallmark of inflammatory response, in both the lymph gland and in the circulating blood cell population (Fig 2f–h’). Finally, starvation induces the rupture of crystal cells (Fig 2i–j), a process known to release coagulation and melanization enzymes16. This rupture depends upon JNK signaling (Fig 2k), a stress signaling pathway required for crystal cell maintenance16. Thus, starvation alters the homeostatic balance between progenitors and differentiating blood cells through extensive progenitor differentiation, and also activates mature blood cells in a manner that is reminiscent of mammalian inflammatory response.
Figure 2. Starvation induces inflammatory response in blood cells.
Yellow dotted line indicates boundary of the lymph gland. Each lymph gland section is a middle section of a Z stack. In j–k, HHLT-gal4, UAS-GFP staining is omitted for clarity. White scale bars: 25 µm except in panels i–k where they are 10µm. Yellow scale bars: 50 µm. Blue indicates DNA marker, ToPro3. The number of lobes or larvae examined is indicated as n.
a–b, In well-fed animals, D4-lacZ (containing 4 Dorsal/Dif binding sites that are direct targets of NFκB-like transcription factors, red) is not expressed except in the PSC and scattered crystal cells (a). Starvation dramatically promotes D4-lacZ expression (red) in the entire lymph gland (n=18) (b).
c–c’, Circulating hemocytes from well-fed animals rarely express antimicrobial peptides, Drosomycin (abbreviated as Drs) (green) (c), whereas starvation abnormally induces Drs expression in circulating hemocytes (n=10) (c’).
d–d’, Starvation causes hemocyte infiltration into the fat body. In well-fed animals, the fat body (ToPro3, blue) is devoid of hemocytes (Pxn, green; arrowhead) (n=6) (d), while in starved animals, the fat body is infiltrated by large number of hemocytes (Pxn, green; arrow) (n=8) (d’).
e, Starvation increases the number of Pxn+ circulating hemocytes indicated as total number of cells per ml (n=8, *p=0.02). Error bars indicate s.d.
f–h’, Starvation causes abnormal differentiation of lamellocytes in the lymph gland (f–g) and in circulating cells (h–h’). Lamellocytes are marked either by Msn or L1 expression in red and plasmatocytes are marked by Peroxidasin in green.
f, Lymph glands from well-fed wild-type animals are devoid of lamellocytes (red, n=10).
g, Starvation promotes differentiation of lamellocytes in the lymph gland (red, n=12).
h, Circulating hemocytes do not include lamellocytes when isolated from well-fed larvae.
h’, Following starvation many lamellocytes (red, n=10) can be seen in circulation.
i–k, In well-fed animals, crystal cells remain round and intact in the lymph gland (PPO, red; n=20) (i). Upon starvation, crystal cells abnormally rupture (fragmented expression of PPO, red; n=20) (j). Expression of a dominant negative form of JNK (bskDN) in the lymph gland under the control of HHLT-gal4 rescues this phenotype (n=6) (k).
In Drosophila, systemic level of glucose is regulated by insulin-like peptides (Dilps) that are produced and secreted by neuroendocrine cells in the brain, much like insulin production by pancreatic β-cells of mammals17. As in mammals, insulin signaling in Drosophila plays a conserved role in regulating metabolism and growth, and the levels of nutrients, such as amino acids, regulates secretion of Dilps12,18,19. We find the effects of starvation on Drosophila blood particularly interesting given the connection between myeloid cell function and insulin signaling in human metabolic diseases13. We delineate here the mechanisms by which a systemic signal, namely insulin signaling, controls maintenance and differentiation of progenitors in the hematopoietic organ.
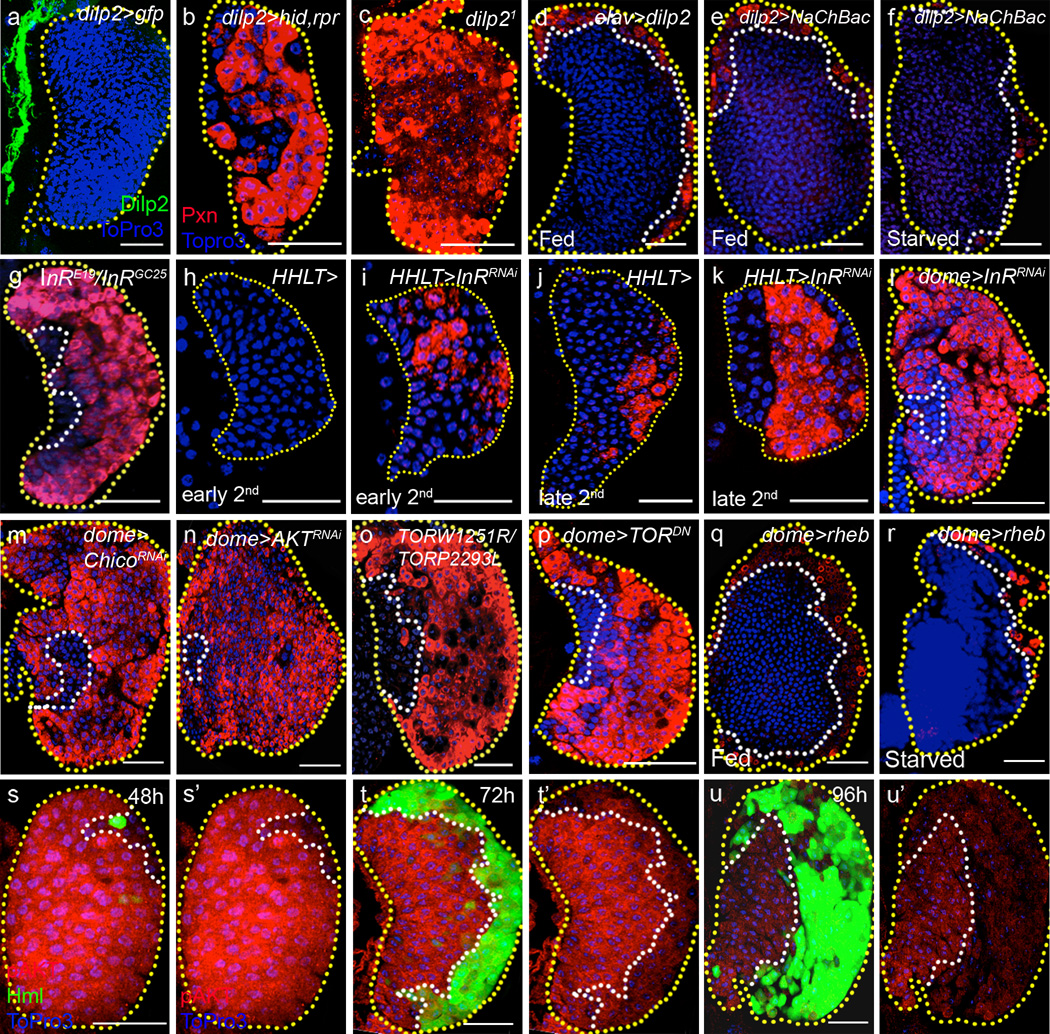
We specifically ablated the insulin-producing cells (IPCs) by inducing cell death with the expression of the pro-apoptotic genes, hid and rpr, and found robust differentiation of blood cells in the lymph gland (Fig 3a–b, also see Supplementary Fig 2 for quantification of data presented in Fig 3) similar to that seen upon starvation (Fig 1b). Although several larval Dilps, including dilp2,3 and 518, are produced by IPCs, further analysis showed that deficiency mutants containing dilp2 lesion or a specific deletion of the dilp2 gene using the null dilp21 mutant allele causes blood cell differentiation (Fig 3c, Supplementary Fig 3a–b). Depletion of any of the other dilps, including dilp6 or 7, does not cause this phenotype (Supplementary Fig 3c–d, data for dilp6 and 7 not shown). We do not detect Dilp2 expression in the lymph gland cells and propose that the ligand source is the IPC neurons in the brain. Consistent with previous findings12, we find that starvation blocks Dilp2 release from the IPCs (Supplementary Fig 3e–e’). Furthermore, forced depolarization of the IPCs by expressing the bacterially derived voltage-gated sodium channel (NaChBac)20, which will cause increase in Dilp secretion, suppresses blood cell differentiation under both well-fed and starved conditions (Fig 3e–f). Finally, over-expression of Dilp2 using the neuronal driver, elav-gal4 causes suppression of differentiation of mature blood cells (Fig 3d). Taken together, we conclude that Dilp2 expression from the IPC neurons is essential for progenitor maintenance and loss of Dilp2 release during starvation results in excessive differentiation of blood cells.
Figure 3. Systemic dilp2 is directly sensed by hematopoietic progenitors.
Borders of the lymph glands are marked with yellow dotted lines; the progenitors, white dotted lines. In h–k, HHLT-gal4, UAS-GFP staining, and in l–n and p–r, dome-gal4, UAS-2xEYFP staining has been omitted for clarity. Each panel shows a single section from the middle of a Z stack of images from a lymph gland except h–k, which show super-positions of 3 middle sections. Blue indicates DNA marker, ToPro3. Scale bars: 25 µm. AED : After Egg Deposition. Quantification of each genotype is presented in Supplementary Fig 2, and the p values obtained from that quantitation for ten random samples and compared with corresponding wild types are quoted here for each genotype. The number of lobes examined is indicated as n.
a, Neuronal projections from the insulin producing cells (marked by dilp2-gal4; UAS-GFP, green) are adjacent to the lymph gland, but do not penetrate the lymph gland (n=10).
b–c, Maturing blood cell marker, Pxn (red) staining is increased in the lymph gland when IPCs are ablated in the brain (dilp2-gal4; UAS-hid,rpr) (n=14; p=1.8×10−8) (b) or in dilp2 deficiency, dilp21/ dilp21 animals (n=8; p=3.3×10−7) (c) (compare with Fig 1a).
d, Constitutive expression of Dilp2 using elav-gal4; UAS-dilp2 greatly reduces differentiation (Pxn, red) (n=16; p=2.3×10−10).
e–f, Forced secretion of larval insulins using dilp2-gal4; UAS-NaChBac, which depolarizes Dilp2-expressing IPCs, either under well fed (n=30; p= 4.8×10−14) (e) or starved condition (n=10; p=6.9×10−12) (f) also greatly reduces differentiation (Pxn, red).
g, The heteroallelic mutant combination InRE19/InRGC25 promotes differentiation of maturing blood cells (Pxn, red, n=14; p=2.7×10−7).
h–i, In early second instar (36h AED at 29°C), the wild-type lymph gland does not express Pxn (n=6) (h), while specific knockdown of InR in the lymph gland (using HHLT-gal4; UAS-InRRNAi) causes precocious expression of this maturing cell marker (Pxn, red, n=8, p=0.0001) (i).
j–k, In mid to late second instar (50h AED at 29°C), maturing cells are first seen in the normal wild-type lymph gland (Pxn, red, n=8) (j). Loss of InR (HHLT-gal4; UAS-InRRNAi) causes increased Pxn+ cells to appear at this stage (n=8, p=0.0001) (k).
l–n, Insulin receptor maintains progenitors. Specific knockdown of InR in the progenitor population (dome-gal4; UAS-InRRNAi) causes a loss of progenitors and a robust differentiation phenotype in the late third instar (n=40; p=1.6×10−8) (l) (Pxn, red). RNAi targeting chico (dome-gal4; UAS-chicoRNAi) (n=5; p=1.2×10−7) (m) or dAKT (dome-gal4; UAS-dAKTRNAi ) (n=14; p=2.1×10−5) (n) leads to the same phenotype (Pxn, red).
o–p, dTOR regulates maintenance of progenitors. The heteroallelic mutant combination dTORW1251R/dTORP2293L (n=14; p=3.0×10−9) (o) or expression of dTORDN (dome-gal4; UAS-dTORDN) (n=10; p=3.1×10−8) (p) causes increased differentiation (Pxn, red).
q–r, Activation of dTOR by overexpression of Rheb (dome-gal4; UAS-rheb) greatly inhibits differentiation of maturing cells (Pxn, red) under both fed (n=8; p=4.4×10−6) (q) and starved conditions (n=8; p=1.0×10−3) (r).
s–u’, Differential expression profile of phospho-AKT (abbreviated as pAKT) during development of the lymph gland. At 48 hour AED (s–s’), pAKT (membrane, red) is uniformly and highly expressed throughout the lymph gland except in a few mature cells at the periphery that are marked by hmlΔ-gal4,UAS-2xEGFP (green). At 72 hour AED (t–t’), high levels of pAKT are seen only within the progenitor population (red), and the hmlΔ-gal4,UAS-2xEGFP (green) expressing maturing blood cells have relatively low pAKT. At 96 hour AED (u–u’), expression level of pAKT is significantly reduced and the pattern is even more restricted (red) to the progenitors, as mature blood cells completely exclude pAKT expression.
The loss of function, heteroallelic combination, InRE19/InRGC25, for the Drosophila insulin receptor (InR), is viable and larvae from this genotype also exhibit extensive differentiation of the progenitor population (Fig 3g). As Dilp2 is a secreted protein, its target receptor could, in principle, be functional in any tissue that then, in turn, signals to the hematopoietic organ using a secondary pathway. However, we found that disrupting InR function directly in the lymph gland with the use of the lymph gland specific driver, HHLT-Gal4 (C. Evans & U. Banerjee, unpublished) causes precocious differentiation of the progenitors from an earlier stage in development than is seen in wild type (Fig 3h–k and Supplementary Fig 3f–g). As HHLT-Gal4 is also expressed in the heart (dorsal vessel), we examined possible involvement of cardiogenic cells by disrupting InR using the heart-specific driver Mef2-Gal4; this does not induce abnormal differentiation in the lymph gland (Supplementary Figure 3h), indicating that HHLT-Gal4 driven phenotype is due to its expression in the hematopoietic system. Within the lymph gland, down-regulation of InR in the progenitor cell population (using dome-gal4) causes their robust differentiation (Fig 3l), whereas loss of InR in the already differentiating cells of the CZ (using hml-gal4) or the niche cells of the PSC (using antp-gal4) does not affect the progenitor population (Supplementary Fig 4a–b). Consistent with these findings, we detect high levels of InR transcript in the progenitors (Supplementary Fig 4c–d), and mutant clones of InR (InRE19/InRE19) within the MZ region induce precocious differentiation (Supplementary Figure 4e–e’’ and f–f’). Additionally, down-regulation of chico, a downstream effector of insulin receptor signaling21, in the progenitor population recapitulates the InRRNAi phenotype (Fig 3m), while activation of PI3K kinase inhibits differentiation of blood cells (Supplementary Figure 4g–g’). These results establish that the hematopoietic progenitor directly responds to brain IPC-derived Dilp2 by activating InR signaling, which serves to maintain the progenitor cell population within the lymph gland.
We next examined the function of AKT, which acts downstream of InR as a protein kinase, by down-regulating its expression in the progenitors, and this too promotes progenitor differentiation (Fig 3n), identical to loss of InR (Fig 3g) and dilp2 (Fig 3c). Likewise, loss of the TORC1 components, dTOR (using the dominant negative mutant protein, dTORDN; Fig 3o–p) or Raptor (using RNAi; Supplementary Fig 4h), which together function downstream of AKT, causes loss of progenitors due to their differentiation. Feeding rapamycin, which blocks dTOR function22, also phenocopies this effect (Supplementary Fig 4i). Consistent with a role for this pathway in progenitor maintenance, over-expression of rheb, which activates dTOR23, strikingly inhibits differentiation of blood cells under both normally fed and starved conditions (Fig 3q–r). Interestingly, mammalian HSCs also respond to mTOR signaling24. Overall, it is evident that the canonical Dilp-InR and Rheb-dTOR signaling pathways play a critical role in the maintenance of hematopoietic progenitors, and this maintenance role is overridden during metabolic stress caused by starvation.
Dilp2 levels rise during early instars and then gradually decrease during the third instar of larval development25, suggesting a possible mechanism for maintaining InR signaling through the third instar in well-fed larvae. To determine whether InR signaling is modulated during normal development, we assessed levels of phospho-AKT at different developmental stages in the lymph gland. Using this approach, we found two distinct phenomena. First, phospho-AKT expression in progenitor cells is high during the second instar and gradually decreases in these cells during the third instar. Second, phospho-AKT is low, relative to progenitor levels, in differentiating cells at all stages when they are present (Fig 3s–u’). These observations suggest that during the course of normal development, InR signaling is modulated in progenitors, thereby differentially promoting maintenance at different stages. It is also apparent that once cells are committed to differentiate, little, if any, InR signaling occurs, consistent with lower levels of InR expression (Supplementary Fig 4c) and the lack of a phenotype associated with InR loss-of-function in these cells (Supplementary Fig 4a).
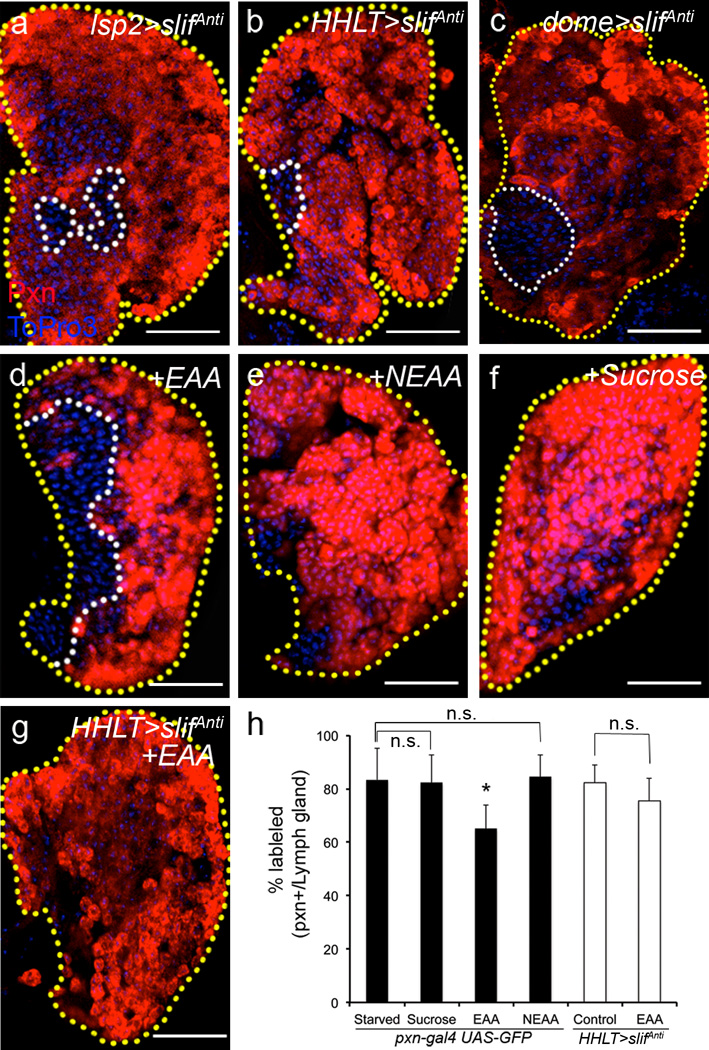
In mammals, glucose levels control insulin secretion26. This is less clear in Drosophila, but it is well established that amino acid levels are sensed by the fat body through the mediation of the amino acid transporter protein Slimfast (Slif)27 and that the fat body indirectly controls insulin secretion from the brain IPCs12. As expected, we found that slifAnti expressed in the fat body mimics the starvation phenotype in the lymph gland (Fig 4a), likely due to decreased Dilp2 secretion from the brain. More interestingly, however, knocking down slif expression directly in the lymph gland (Fig 4b), but not in the dorsal vessel (Supplementary Fig 3i and j–j’), and specifically in the progenitor population within the lymph gland (Fig 4c), accelerates differentiation of mature cells similar to that seen with starvation (Fig 1b). As with insulin signaling, this result shows that the hematopoietic progenitors themselves directly sense amino acid levels to maintain their stem-like fate. Taken together, these findings suggest a dual control of hematopoietic homeostasis by systemic levels of insulin and amino acids. Amino acids are sensed by the fat body, which then controls insulin secretion from the brain12. Insulin is then directly sensed by the blood progenitors. Additionally, amino acids are also directly sensed by the blood progenitors to maintain their undifferentiated state (Fig 5g).
Figure 4. Systemic essential amino acids are directly sensed by hematopoietic progenitors.
All lymph glands are outlined with yellow dotted lines, and border of progenitors, white dotted line. In b and g, HHLT-gal4, UAS-GFP staining, and in c, dome-gal4,UAS-2xEYFP expression staining is omitted for clarity. In a–c and g, maturing blood cells are marked with Pxn (red); in d–f, pxn-gal4, UAS-GFP is shown in red for consistency. Each panel shows a single middle section from a lymph gland derived Z stack. Blue indicates DNA marker, ToPro3 in all panels. The number of lobes examined is indicated as n. Scale bars: 25 µm.
a, slimfast knock-down (using an anti-sense construct, slifAnti) in the fat body with lsp2-gal4 induces non-autonomous differentiation (Pxn, red) of maturing blood cells in the lymph gland (n=11).
b–c, Specific knockdown of slif either in the entire lymph gland (with the use of HHLT-gal4; UAS-slifAnti) (n=40; p=1.8×10−11) (b), or in the MZ progenitors using dome-gal4 (n=40; p=2.2×10−5) (c) increases maturation of blood cells (Pxn, red). The controls for each case (a–c) are as in Fig 1a. Quantification of the phenotype is shown in Supplementary Fig 2b–c.
d–f, Supplementing essential amino acids back to the starvation media is sufficient to restore progenitors (n=12) (d) (non-red area, outlined by white), whereas neither supplementation of non-essential amino acids (abbreviated as NEAA) (n=13) (e) nor sucrose (n=12) (f) restores the progenitors.
g, Supplementation of essential amino acids to the lymph gland without slif (HHLT-gal4; UAS-slifAnti) does not restore progenitors and block differentiation (n=9), compare with d.
h, Quantification of lymph gland phenotypes from d–g. Genotypes are as indicated. When Pxn+ ratio upon starvation is compared with essential amino acid (EAA) supplementation, the difference is significant (n=9; *p=1.2×10−8), while with sucrose supplementation or non-essential amino acids (NEAA) supplementation the difference is not significant (n.s.: not significant; n=12 and n=13; p=0.3 and 0.4, respectively). When HHLT-gal4; UAS-slifAnti is compared with HHLT-gal4; UAS-slifAnti+EAA, the difference is not significant (n.s.: not significant, p=0.06; The small decrease could be due to indirect effect of EAA supplementation). Error bars indicate s.d.
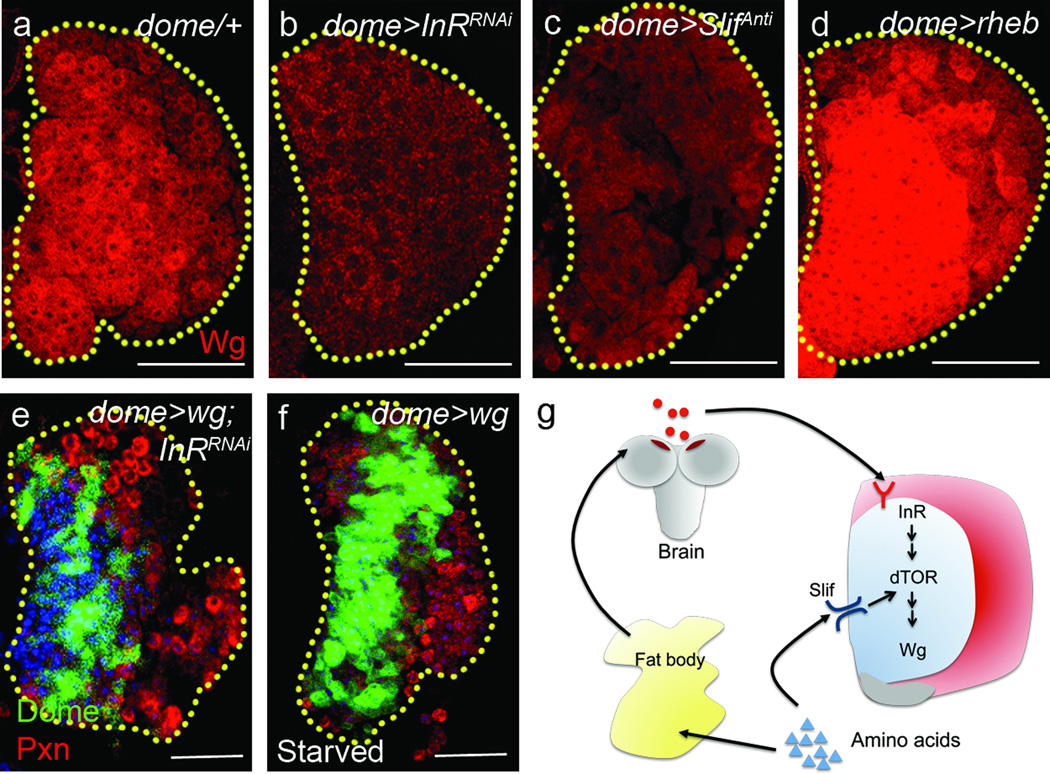
Figure 5. wingless as a target of Dilp2/InR/dTOR signaling.
In all panels, yellow dotted lines indicate borders of the lymph gland. In a–d, dome-gal4, UAS-2xEYFP staining, and in e–f, HHLT-gal4, UAS-GFP staining is omitted for clarity. Each panel shows a single section from the middle of a Z stack of images from a lymph gland except e–f, which are super-positions of 3 middle sections. Blue indicates DNA marker, ToPro3. The number of lobes examined is indicated as n. Scale bars: 25 µm.
a–d, Expression of Wingless (Wg) is regulated by InR, dTOR and slif. In mid-second instar, the progenitors exhibit high levels of Wg (red, n=12) (a), while knockdown of InR (dome-gal4; UAS-InRRNAi, n=12; p=2.5×10−13) (b) or slif (dome-gal4; UAS-slifAnti, n=10; p=1.5×10−7) (c) significantly reduces Wg expression. Overexpression of Rheb (dome-gal4; UAS-rheb, n=16; p=3.7×10−3) (d) strongly enhances the expression of Wg. Quantification of Wg expression is shown in Supplementary Fig 2e. Error bars indicate s.d.
e–f, Constitutive expression of Wg (using dome-gal4 gal80ts, UAS-EYFP;UAS-wg) is sufficient to restore the progenitors (dome-gal4 gal80ts,UAS-EYFP, green) and restrict differentiation of maturing blood cells (Pxn, red) in the lymph gland expressing InRRNAi (n=32; p=2.7×10−7) (e) and during starvation (n=8; p=1.0×10−7) (f). Quantification of the phenotype is shown in Supplementary Fig 2b.
g, Model for dual control of systemic maintenance by insulin and essential amino acids. Systemic Dilp2 (insulin) from brain IPCs and dietary essential amino acids are directly sensed by InR and Slif, respectively, in the progenitors. Amino acids are sensed by the fat body, which promotes secretion of insulin from the brain IPCs, and insulin is then directly sensed by the blood progenitors. In addition, amino acids are also directly sensed by progenitors, which consequently cause the progenitor maintenance in the lymph gland.
Supplementation of essential amino acids (EAA) partially restores the progenitor population during an otherwise starved condition (Fig 4d, h), whereas neither sucrose nor non-essential amino acids (NEAA) rescues the progenitors from differentiation (Fig 4e–f, h). Loss of slif in the lymph gland prevents progenitor maintenance despite EAA supplementation (Fig 4g–h), further establishing that the progenitors directly sense EAA and use the signal to promote their maintenance.
In wild-type lymph glands, the progenitors (expressing Domeless; Dome+) rarely overlap with the maturing cells (expressing Pxn; Pxn+), however, down-regulation of InR or expression of dominant-negative TOR (TORDN) in the progenitors causes a significant increase in the number of double-positive cells (Dome+ and Pxn+) that are in transition towards differentiation7 (Supplementary Fig 5). An increase in this particular cell type is reminiscent of the phenotype seen upon down-regulation of the wingless (wg) signaling pathway, which has been previously linked to the process of progenitor maintenance28. Wg is dynamically expressed in the lymph gland with higher levels at earlier stages that then decreases during the third instar28. Wg is expressed at high levels by progenitors at these stages and is withdrawn from differentiating cells, which is reminiscent of phospho-AKT staining patterns (Fig 3s–u’) and the expression of InR in the third instar (Supplementary Fig 4c). We found that downregulation of InR (Fig 5b) or Slif (Fig 5c) in the progenitors causes a significant decrease in Wg expression, while Rheb overexpression significantly increases Wg levels (Fig 5d) as compared to that seen in wild type (Fig 5a), suggesting that Dilp2-dTOR and Slif-dTOR activities positively regulate the expression of Wg within the progenitors. Importantly, over-expression of Wg restores the progenitor population in both starvation conditions and in the presence of reduced InR levels (Fig 5e–f), demonstrating that Wg is likely to be the most direct downstream targets of Dilp2-InR signaling, which maintains progenitors within the progenitors. However, our studies do not rule out either direct or indirect involvement of additional pathways downstream of InR-dTOR in this process.
A model describing the systemic and nutritional control of myeloid-like progenitors via insulin and amino acids is shown in Fig 5g. Our results demonstrate that metabolic changes are perceived by blood progenitors and this causes alteration of their cell-fate determination program. We find that a major consequence of reduced InR and amino acid levels is the reduction of Wg expression in the lymph gland, which functions to promote progenitor maintenance. In addition to accelerated differentiation of myeloid progenitors, starvation also causes a response similar to the inflammatory response (Fig 2) typically associated with metabolic disorders29. These responses suggest that metabolically induced inflammatory responses in mammals have an ancestral origin that arose to balance an organisms’ ability to withstand an unfavorable environment and the normal development of myeloid cells. Nutrient/insulin signaling has been linked to the homeostatic control of various Drosophila stem cell populations30–35. Given the highly conserved nature of the blood system in flies and mammals, and the known functional role of metabolism and insulin signaling in myeloid cells36, it will be important to determine if the direct metabolic and nutritional regulation mechanisms uncovered in these studies might also be relevant for the mammalian common myeloid progenitors (CMPs). Such studies will likely yield insights into chronic inflammation and the myeloid cell accumulation seen in patients with type II diabetes, and other metabolic disorders.
Materials and Methods
Drosophila stocks and genetics
The following Drosophila stocks were used in the experiments: domeless-gal4 (S.Noselli), peroxidasin-gal4 (P.Martin), hmlΔ-gal4 UAS-2xeGFP (S.Sinenko), dilp2-gal4 (E.Rulifson), lsp2-gal4 (C.Antoniewski), antp-gal4/TM3,Sb (S.Cohen) and elav-gal4, mef2-Gal4 (Bloomington Stock Center) are gal4 drivers used in this study. We obtained UAS-rpr, y1w*; P{UAS-TORTED}, w*; P{UAS-Rheb.Pa}, w*; P{UAS-wg}, y1w*; P{UAS-NaChBac}, InRE19/TM2, InR(3R)GC25, InR93Dj-4/TM3,Sb1, y1w1118;P{UAS-InRA1325D}, y1w1118; P{UAS-InRK1409A}, w1118 P{UAS-BskDN}, P{tub-Gal80ts},w*/FM7c, P{hs-FLP}, w*; P{neoFRT}82B P{ubi-GFP}83, P{neoFRT}82B from Bloomington stock center. w1118; ilp21, w1118; ilp31 and w1118; ilp51 were isolated by L. Partridge’s lab and obtained from Bloomington stock center. RNAi lines are from Vienna Drosophila RNAi Center. y,w; P{UAS-dilp2} was a kind gift from E.Hafen (ETS-IMSB, Switzerland), y,w;UAS-slifAnti, from P.Leopold (Universite de Nice, France), Df[dilp1-5]/TM6B,Tb and Df[dilp1-3]/TM6B,Tb from L.Pick (University of Maryland, USA), y,w;TORW1251R/SM6B-TM6B and y,w;TORP2293L/SM6B-TM6B from T.P.Neufeld (University of Minnesota, USA), P{msn-GFP.F9} and eater-GFP from R.A.Schulz (University of Notre Dame, USA), and D4-lacZ from A.Courey (UCLA), drs-GFP from J-M. Reichhart (Universite de Strasbourg, France). Stocks and crosses were maintained at 25°C, except for RNAi and GAL4/UAS expression used experiments, for which crosses were maintained at 29°C. For GAL80ts experiments, crosses were maintained at permissive 18°C for 3 days, and shifted to 29°C. For synchronization of larvae, eggs were collected for an hour and newly hatched larvae were transferred onto fresh food plates after 24h and aged for specified time periods at 25°C. For InRE19 clones in the lymph gland, we generated FRT82B InRE19/TM6B Tb, and confirmed the recombinant with ey-FLP. Larvae containing hs-FLP;FRT82B InRE19/FRT82B ubi-GFP were heat-shocked at 38°C for 40 min in each instar.
Immunohistochemistry and imaging
Lymph glands were dissected and stained as previously described37. For fat body samples, anterior fat bodies were gently dissected, and fixed with 3.7% formaldehyde for 20 minutes. After visualizing DNA with ToPro3, samples were mounted in Vectashield (Vector Labs). For anti-Wg antibody, a modified protocol was used28. The following antibodies were used in this study: anti-Pxn (1:1500, J.Fessler and L.Fessler), anti-phosphoAKT (1:200, Cell signaling #4054), anti-Wg (1:10, DSHB), anti-L1 (1:100, I.Ando), mouse anti-βgal (1:200, Promega), anti-Dilp2 (1:1000, P. Leopold) and anti-PPO (1:250, H.Muller). Nuclear DNA was visualized with ToPro3 (Invitrogen). Cy3 conjugated secondary antibodies were used (Jackson Laboratory) for immunohistochemistry. Samples were mounted with Vectashield (Vector Laboratories). Images were obtained using a Zeiss LSM700 confocal system using either ×20 or ×40 immersion oil objective. Images used for comparison were always captured using identical optical settings.
Quantification of the lymph gland phenotype
Samples were scanned using LSM700 confocal microscope and captured using Zen 2009 software. Images were processed using Image J (NIH). To estimate the expression of differentiation markers within the MZ, the middle 3 optical sections from the Z stack were merged into a single section. For wild type, this enables the clearest view of the Medullary zone surrounded by the differentiating cells of the Cortical zone. Depending on the genotypes studied, the merged image may contain images at two or three different wavelength channels indicating different markers used for the lymph gland analysis. Each individual channel/marker was analyzed separately. For the zone specific markers (Pxn, Hml and Dome), the colored areas were recalibrated into an identical threshold by using the Binary tool (Process-Binary-Make binary) as an automatic routine. The area with identical threshold was automatically captured with the Wand tool, and the size was measured using the Measure tool (Analyze-Measure). To measure total size of the lobe, ToPro3 expressing area was recalibrated by Threshold tool until overlaid with identical threshold color. This total area was selected using the Wand tool, and measured. Percentage of differentiation was calculated by dividing size of the marker-expressing area by the total size of the lobe (ToPro3 area). Ten randomly selected lymph glands were analyzed per genotype, and statistical significance was calculated with standard t test.
To quantify the expression of Wingless in different genotypes, we followed the protocol from (http://sciencetechblog.files.wordpress.com/2011/05/measuring-cell-fluorescence-using-imagej.pdf).
Hemocyte collection and counting
For hemocyte collection, staged larvae were washed with water and bled in 20µl of Schneiders insect media. The cells were gently redistributed onto 5mm 14-well slides (Fisher Scientific), allowed to settle down for 15min at room temperature, and fixed with 3.7% Formaldehyde for 20min. ToPro3 was used for visualizing DNA38.
For hemocyte counting, cells prepared in 20µl Schneiders medium were transferred to a Neubauer improved hemocytometer, and counted and analyzed as previously described38.
Starvation assay, nutrient supplementation assay and rapamycin treatment
Larvae were synchronized and allowed to grow at 25°C until early third instars (90–96h after egg deposition (AED)). Staged larvae were collected and washed with water, and finally rinsed with Ultrapure distilled water (Gibco). These larvae were then divided into two groups; one group of larvae was placed on regular food media (control) and the second was placed on 1% soft agar media (starvation). Control larvae were handled identically to the ones exposed to starvation conditions. Those raised on agar plates were also maintained with or without yeast to control for any possible effects due to handling and growth on agar plates. Per 60 mm dish, 15–20 larvae were used. Both the standard food media and 1% agar media were treated with 1× antibiotic-antimycotic solution (Invitrogen). The larvae were reared for 24 hours at 25°C after which they were collected, dissected and processed for antibody staining.
For aseptic starvation, collections of embryos were sterilized by rinsing in 0.25% Clorox, 0.04% n-alkyl dimethyl benzyl ammonium chloride, washed twice in 70% ethanol following two brief washes in Ultra pure distilled water (Gibco) and autoclaved distilled water, and then transferred onto axenic food plates. The animals were staged to 90–96h AED, and larvae were selected with sterilized forceps and washed with Ultrapure distilled water (Gibco). Normally fed larvae were reared on axenic food media with antibiotic-antimycotic solution, and starved larvae, on sterilized 1% agar plate with antibiotic-antimycotic solution as the standard starvation protocol12. Both food/agar and homogenized larvae from each plate were plated onto antibiotic-free bacterial LB agar plate, and assessed for lack of growth to verify axenic conditions. No bacterial colony grew with food/agar or homogenized larvae grown in axenic culture conditions. For nutrient supplementation assays, carbohydrate complexes, lipid complexes, and amino acids were added to sterilized 1% agar solution and processed for standard starvation method as described12. For yeast supplementation, 34g/l of inactivated yeast powder was added to 1% agar solution, and sterilized, processed for standard starvation method as described above. For rapamycin treatment, 90–96h AED larvae were transferred to standard fly medium containing 2µM rapamycin39 and kept for 24 hours.
In situ hybridization
For anti-InR probe, a 205 bp cDNA fragment from LD06045 (GDRC, cDNA library) was amplified with InR-for (CGA TGG CGG TGT TAT GGA GAG) and InR-rev primers (CGC TCC TTT TCC CGA TGC TGC AGA ). Dissection and in situ hybridization procedures were performed as described4.
Supplementary Material
Acknowledgement
We thank C.J.Evans, K.T.Jones, other members of the Banerjee laboratory, and J.A.Martinez-Agosto for helpful comments and discussions. We thank Ruby Erdmann for help with amino acid supplementation assay and Steven Pham, for confirming experiments with anti-hml in different genetic backgrounds. We acknowledge P. Leopold, E. Hafen, E.J.Rulifson, L.Pick, T.P.Neufeld, R.A.Schulz, A. Courey, J-M. Reichhart, the VDRC stock center and the Bloomington stock center for fly stocks and, Developmental Studies Hybridoma Bank (University of Iowa), Drosophila Genomics Resource Center and John Fessler for anti-Peroxidasin. We also thank M.Crozatiers’ group for providing in situ hybridization protocol. This work was supported by NIH grant (5R01 HL067395) to U.B. and the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research Training Grant at UCLA to J.S.
Footnotes
Author contributions
J.S designed and performed experiments, and U.B. supervised the project. T.M. performed experiments. J.S., T.M. and U.B. discussed and analyzed results and wrote the manuscript.
References
- 1.Meister M. Blood cells of Drosophila: cell lineages and role in host defence. Current opinion in immunology. 2004;16:10–15. doi: 10.1016/j.coi.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Jung SH, Evans CJ, Uemura C, Banerjee U. The Drosophila lymph gland as a developmental model of hematopoiesis. Development. 2005;132:2521–2533. doi: 10.1242/dev.01837. [DOI] [PubMed] [Google Scholar]
- 3.Mandal L, Martinez-Agosto JA, Evans CJ, Hartenstein V, Banerjee U. A Hedgehog- and Antennapedia-dependent niche maintains Drosophila haematopoietic precursors. Nature. 2007;446:320–324. doi: 10.1038/nature05585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krzemien J, et al. Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre. Nature. 2007;446:325–328. doi: 10.1038/nature05650. [DOI] [PubMed] [Google Scholar]
- 5.Mukherjee T, Kim WS, Mandal L, Banerjee U. Interaction between Notch and Hif-alpha in development and survival of Drosophila blood cells. Science. 2011;332:1210–1213. doi: 10.1126/science.1199643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 7.Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461:537–541. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson RE, et al. Peroxidasin: a novel enzyme-matrix protein of Drosophila development. EMBO J. 1994;13:3438–3447. doi: 10.1002/j.1460-2075.1994.tb06649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goto A, et al. A Drosophila haemocyte-specific protein, hemolectin, similar to human von Willebrand factor. Biochem J. 2001;359:99–108. doi: 10.1042/0264-6021:3590099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kocks C, et al. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. 2005;123:335–346. doi: 10.1016/j.cell.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 11.Becker T, et al. FOXO-dependent regulation of innate immune homeostasis. Nature. 2010;463:369–373. doi: 10.1038/nature08698. [DOI] [PubMed] [Google Scholar]
- 12.Geminard C, Rulifson EJ, Leopold P. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 2009;10:199–207. doi: 10.1016/j.cmet.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 14.Flores-Saaib RD, Jia S, Courey AJ. Activation and repression by the C-terminal domain of Dorsal. Development. 2001;128:1869–1879. doi: 10.1242/dev.128.10.1869. [DOI] [PubMed] [Google Scholar]
- 15.Tzou P, et al. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity. 2000;13:737–748. doi: 10.1016/s1074-7613(00)00072-8. [DOI] [PubMed] [Google Scholar]
- 16.Bidla G, Dushay MS, Theopold U. Crystal cell rupture after injury in Drosophila requires the JNK pathway, small GTPases and the TNF homolog Eiger. Journal of cell science. 2007;120:1209–1215. doi: 10.1242/jcs.03420. [DOI] [PubMed] [Google Scholar]
- 17.Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- 18.Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, et al. Deletion of Drosophila insulin-like peptides causes growth defects and metabolic abnormalities. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19617–19622. doi: 10.1073/pnas.0905083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren D, et al. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- 21.Bohni R, et al. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 1999;97:865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000;14:2712–2724. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saucedo LJ, et al. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 24.Lee JY, et al. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell stem cell. 2010;7:593–605. doi: 10.1016/j.stem.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slaidina M, Delanoue R, Gronke S, Partridge L, Leopold P. A Drosophila insulin-like peptide promotes growth during nonfeeding states. Dev Cell. 2009;17:874–884. doi: 10.1016/j.devcel.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutter GA. Nutrient-secretion coupling in the pancreatic islet beta-cell: recent advances. Molecular aspects of medicine. 2001;22:247–284. doi: 10.1016/s0098-2997(01)00013-9. [DOI] [PubMed] [Google Scholar]
- 27.Colombani J, et al. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- 28.Sinenko SA, Mandal L, Martinez-Agosto JA, Banerjee U. Dual role of wingless signaling in stem-like hematopoietic precursor maintenance in Drosophila. Dev Cell. 2009;16:756–763. doi: 10.1016/j.devcel.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annu Rev Immunol. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 30.Sousa-Nunes R, Yee LL, Gould AP. Fat cells reactivate quiescent neuroblasts via TOR and glial insulin relays in Drosophila. Nature. 2011;471:508–512. doi: 10.1038/nature09867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chell JM, Brand AH. Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell. 2010;143:1161–1173. doi: 10.1016/j.cell.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi NH, Lucchetta E, Ohlstein B. Nonautonomous regulation of Drosophila midgut stem cell proliferation by the insulin-signaling pathway. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18702–18707. doi: 10.1073/pnas.1109348108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu HJ, Drummond-Barbosa D. Insulin levels control female germline stem cell maintenance via the niche in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1117–1121. doi: 10.1073/pnas.0809144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLeod CJ, Wang L, Wong C, Jones DL. Stem cell dynamics in response to nutrient availability. Current biology : CB. 2010;20:2100–2105. doi: 10.1016/j.cub.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Brien LE, Soliman SS, Li X, Bilder D. Altered modes of stem cell division drive adaptive intestinal growth. Cell. 2011;147:603–614. doi: 10.1016/j.cell.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 37.Lebestky T, Chang T, Hartenstein V, Banerjee U. Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science. 2000;288:146–149. doi: 10.1126/science.288.5463.146. [DOI] [PubMed] [Google Scholar]
- 38.Zettervall CJ, et al. A directed screen for genes involved in Drosophila blood cell activation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14192–14197. doi: 10.1073/pnas.0403789101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hennig KM, Colombani J, Neufeld TP. TOR coordinates bulk and targeted endocytosis in the Drosophila melanogaster fat body to regulate cell growth. The Journal of cell biology. 2006;173:963–974. doi: 10.1083/jcb.200511140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.