Abstract
Background:
Macrolide resistance in Streptococcus pneumoniae (S. pneumoniae) is a worldwide problem.
Aims:
The aim of this work was to analyze the phenotypes, genotypes, and clonal relatedness among macrolide-resistant S. pneumoniae strains isolated from various clinical specimens in our hospital.
Study Design:
Cross-sectional study.
Methods:
80 non-duplicate S. pneumoniae strains were analyzed by polymerase chain reaction for both the erm (B) and mef (A) genes.
Results:
Macrolide resistance was observed in 22.5% (18 strains) of strains. Two (11.2%) isolates possessed mef (A), eight possessed erm (B) (44.4%) and eight strains (44.4%) were positive for both erm (B) and mef (A) genes. Although BOX-PCR of 18 macrolide-resistant strains revealed 11 band patterns, they clustered as seven clones with a genetic distance >10% to each other. Eight isolates possessed both erm (B) and mef (A) genes and belonged to a single clone (44.44% of all macrolide-resistant strains).
Conclusion:
Increased positivity rates for both resistance genes have also been reported from other hospitals in Turkey, but this is the first study from Turkey showing the clonal dissemination of both resistance genes.
Keywords: erm (B), BOX-PCR, Macrolide resistance, mef (A), Streptococcus pneumoniae
Streptococcus pneumoniae (S. pneumoniae) is a major pathogen of bacterial pneumonia, meningitis, sinusitis and otitis media. Antimicrobial resistance in S. pneumoniae, including macrolide resistance, has been a growing problem in recent years due to the increasing numbers of cases with treatment failures of infections caused by macrolide-resistant pneumococci (1).
Macrolide resistance in S. pneumoniae is mediated by: the erm (B) gene encodes methylation of the ribosomal macrolide target sites and mef (A) encodes the drug efflux. Deterioration of riboproteins L4 and L22 and mutations in the 23S rRNA genes are involved in the other less common mechanisms (2).
In this study, we analyzed the distribution of the phenotypes, genotypes, and clonal relatedness of macrolide-resistant S. pneumoniae strains isolated in our hospital.
MATERIALS AND METHODS
Eighty S. pneumoniae strains were collected between January 2008 and September 2009 in the Department of Microbiology of the Cerrahpaşa Faculty of Medicine. Ethics committee approval was received for this study from the Ethics Committee of Cerrahpaşa Faculty of Medicine. Strains were isolated mostly from sputum. The remaining isolates were from tracheal aspirates, bronchoalveolar lavage fluid, nasal swab and blood culture.
S. pneumoniae isolates were identified based on conventional microbiological methods. Antibiotic susceptibility testing was performed with the disc diffusion method (3).
Macrolide resistance phenotypes were determined by Montanari et al. (4) method in 2001.
MICs of macrolides (erythromycin, clarithromycin and azithromycin), lincosamides (clindamycin), streptogramins (quinupristin-dalfopristin) and penicillin G were measured by E-test (AB Biodisk; Solna, Sweden). The strain S. pneumoniae ATCC 49619 was used as a control strain.
Detection of erythromycin-resistance genes
A PCR method was used for the detection of erythromycin-resistance genes (5). Bacterial DNA was isolated using a DNA isolation kit (Roche Diagnostic; Mannheim, Germany). All PCR mixtures contain a volume of 25 µL with 5µL of target DNA, 0.5 µL of each primer (25 mM), 1.5 µL of 25 mM MgCl2, 0.5 µL of dNTP mixture (25 mM each), 0.125 µL of Taq DNA polymerase (5 U/µL) and 2.5 µL of 10X buffer (MBI; Fermentas, Lithuania). PCR for both genes was performed on PTC-200 (Peltier Thermal Cycler; MJ Research, USA). Cycling conditions for amplification were: 5 min at 94°C, followed by 40 cycles of 1 min at 94°C, 1 min at 48°C and 1.5 min at 72°C, with a final 10 min incubation at 72°C. PCR products were separated on ethidium bromide stained 1.5% agarose gels. The expected sizes of amplification products for erm (B) and mef (A) were 639 bp and 348 bp, respectively.
The positive control strains were ATCC 700673 (Hungary19A-6) for erm (B) and ATCC 51916 (Tennessee23F-4) for mef (A).
BOX-PCR
DNA amplifications for BOX-PCR were performed in 25 µl final volumes with 5 µL of purified bacterial DNA, 1.5 µL of primer AR1 (CTACGGCAAGGCGACGCTGACG) (25 mM), 1.5 µL of MgCl2 (25 mM), 0.75 µLof dNTPs (25 mM), 0.125 µL of Taq DNA polymerase (5 U/µL) and 2.5 mL of 10X reaction buffer (6).
After the initial denaturation at 95°C for 5 min, this step was followed by 30 cycles of 1 min at 90°C, 1 min at 52°C, and 2 min at 72°C, with a final extension at 72°C for 5 min (7). Amplification products were run on 1.5% agarose gels and detected by staining with ethidium bromide. The band patterns were converted to series of 1s and 0s where 1s indicates the presence and 0s the absence of any band.
The analysis of BOX-PCR results and the phylogenetic analysis were performed using FreeTree and TreeView software packages (8, 9).
RESULTS
Of the total 80 isolates, 22.5% (18/80) were resistant to erythromycin, azithromycin and clarithromycin. Twenty percent of strains were clindamycin-resistant and four (5%) strains had quinupristin-dalfopristin intermediate resistance. According to Clinical Laboratory Standards Institute (CLSI) criteria, penicillin resistance in S. pneumoniae isolates was found to be 7.5% (6/80) and the intermediate resistance rate was 20% (16/80) for oral penicillin criteria. Penicillin intermediate resistance was found to be 2.5% (2/80) for non-meningitis parenteral penicillin criteria. No meningeal isolate was included in the study. Tetracycline, levofloxacin and trimethoprim-sulphomethoxazole resistance rates were as follows: 18.75%, 1.25% and 62.5%, respectively.
According to the erythromycin-clindamycin double-disk test, 16 (88.8%) of the 18 test strains had the cMLSB phenotype and two (11.2%) strains possessed the M phenotype.
PCR of the 18 macrolide-resistant S. pneumoniae strains showed that 44.4% (n=8) strains harbored only the erm (B) gene and 11.2% (n=2) only the mef (A) gene. Eight (44.4%) strains were positive for both the erm (B) and mef (A) genes (Figure 1 and 2). All erm (B) and erm (B)+mef (A) positive strains were resistant to erythromycin, clarithromycin, azithromycin and clindamycin, while mef (A) positive strains had lower erythromycin, clarithromycin and azithromycin MICs and remained susceptible to clindamycin (Table 1).
FIG. 1.
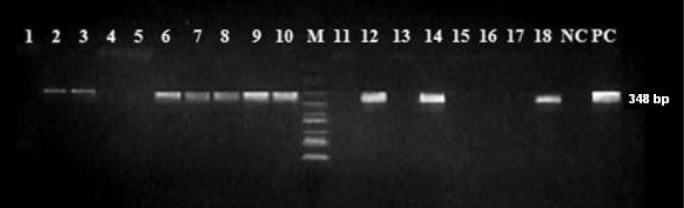
PCR results for mef (A)/(E) genes. Lanes 1, 4, 5, 11, 13, 15, 16, 17 negative strains; Lanes 2, 3, 6, 7, 8, 9, 10, 12, 14, 18 positive strains; NC, Negative control; PC, Positive control; M, DNA size marker
FIG. 2.
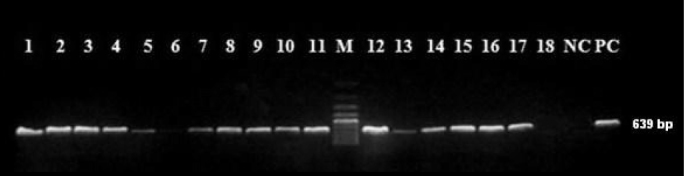
PCR results for erm (B) genes. Lanes 1, 2, 3, 4, 5, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 positive strains; Lanes 6 and 18 negative strains; NC, Negative control; PC, Positive control; M, DNA size marker
TABLE 1.
Genotypic and phenotypic data of 18 macrolide resistant S. pneumoniae isolates
| Number of isolates | Phenotype | Genotype | MIC (μg/mL) | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| E | Cd | C | A | P | Q/D | |||
| 1 | cMLSB | erm (B) | >256 | >256 | >256 | >256 | 0.016 | 0.5 |
| 2 | cMLSB | erm (B)+mef (A)/(E) | >256 | >256 | >256 | >256 | 0.5 | 2 |
| 3 | cMLSB | erm (B)+mef (A)/(E) | >256 | >256 | >256 | >256 | 1 | 0.5 |
| 4 | cMLSB | erm (B) | >256 | >256 | >256 | >256 | 1 | 0.75 |
| 5 | cMLSB | erm (B) | >256 | >256 | >256 | >256 | 1 | 1 |
| 6 | iMLSB+M | mef (A)/(E) | 4 | 0,125 | 6 | 24 | 0.75 | 0.38 |
| 7 | cMLSB | erm (B)+mef (A)/(E) | >256 | >256 | >256 | >256 | 1 | 1.5 |
| 8 | cMLSB | erm (B)+mef (A)/(E) | >256 | >256 | >256 | >256 | 1 | 2 |
| 9 | cMLSB | erm(B)+mef (A)/(E) | >256 | >256 | >256 | >256 | 1 | 0.75 |
| 10 | cMLSB | erm (B)+mef (A)/(E) | >256 | >256 | >256 | >256 | 2 | 1.5 |
| 11 | cMLSB | erm (B) | >256 | >256 | >256 | >256 | 0.23 | 2 |
| 12 | cMLSB | erm (B)+mef (A)/(E) | >256 | >256 | >256 | >256 | 2 | 2 |
| 13 | cMLSB | erm (B) | >256 | >256 | >256 | >256 | 1 | 1 |
| 14 | cMLSB | erm (B)+mef (A)/(E) | >256 | >256 | >256 | >256 | 1.5 | 1.5 |
| 15 | cMLSB | erm (B) | 16 | >256 | 6 | >256 | 0.016 | 0.5 |
| 16 | cMLSB | erm (B) | >256 | >256 | >256 | >256 | 1 | 0.75 |
| 17 | cMLSB | erm (B) | >256 | >256 | >256 | >256 | 2 | 0.75 |
| 18 | M | mef (A)/(E) | 24 | 0,094 | 24 | >256 | 1.5 | 0.38 |
E: erythromycin; Cd: clindamycin; C: clarithromycin; A: azithromycin; P: penicillin; Q/D: quinupristin/dalfopristin
BOX-PCR
Through DNA fingerprint analysis by BOX-PCR, seven clones were identified among 18 macrolide-resistant strains. All of the erm (B) and mef (A) positive isolates were shown to belong to a single clone. The eight strains carrying erm (B) were grouped into four clones. Two mef (A) carrying strains were unrelated (Figure 3 and 4).
FIG. 3.
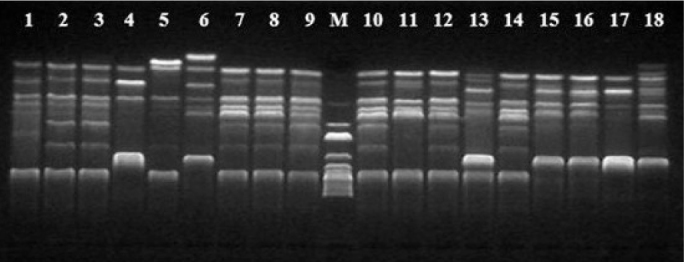
BOX-PCR results of erythromycin-resistant S. pneumoniae isolates. The numbers above the lanes correspond to the strain numbers. M, DNA size marker
FIG. 4.
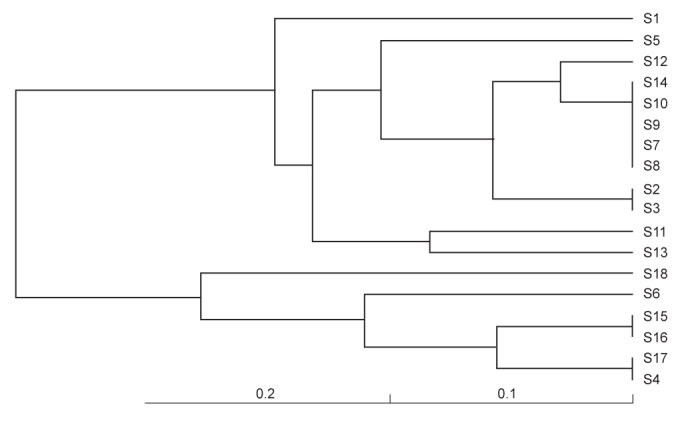
Dendrogram depicting the phylogenetic relatedness of macrolide-resistant S. pneumoniae containing ermB and/or mefA genes (by FreeTree program, UPGMA cluster analysis with Nei and Li/Dice algorithm)
Among erythromycin-susceptible S. pneumoniae strains, the penicillin resistance rate was very low (5 isolates were intermediate and only one strain was fully resistant). In contrast, only two of the 18 erythromycin-resistant isolates were penicillin susceptible. All erm (B) and mef (A) positive strains were penicillin-resistant according to oral criteria; two of these strains showed intermediate resistance according to non-meningitis parenteral penicillin criteria. Three of the four quinupristin-dalfopristin intermediate resistance isolates were positive for erm (B)+mef (A) genes.
DISCUSSION
Macrolide resistance rates among pneumococci have increased over the last two decades. Reported resistance rates were high among strains isolated in the Far East (80%) and South Africa (54%). Low rates were reported from South America (15%), Australia (18%) and Northern Europe (18%). In Europe, the highest resistance rate was reported in France and Greece (10). In Turkey, macrolide resistance among S. pneumoniae has increased from 7% to 29% (11–15). Although Telli et al. (16) reported higher percentages (40%) in 2007–2009, the macrolide resistance rate in this study was 22.5%, which is similar to the result of EARRSS, but higher than the rates reported previously in our hospital (13, 14).
The percentage of isolates positive for erm(B)+mef(A) genotype is increasing and this is a serious public health problem (17). In Germany, Bley et al. (18) reported the presence of combined erm (B)+mef (A) genotype in 4.1% of pneumococcal isolates. Similar results have been increasingly reported in North America, South Asia and South Africa during the last decade (17, 19). In Russia, dual resistance genes among isolates collected between 2003 and 2005 were found in 30.3% (20). In China, McGee et al. (21) found both the erm and mef genes in 6% of isolates. In South Africa, the percentage of strains possessing the resistance genes erm and mef was 30.5% in 2001 and 30 (83%) of them belonged to a single, multi-resistant clone (19). In the USA, Di Persio et al. (22) reported that 71% of macrolide-resistant pneumococci possessed dual resistance genes. Among these strains, two predominant and possibly related clones were detected. A study in Arizona revealed that 23.6% of 592 clinical isolates were macrolide-resistant from 1999 to 2008. More than 50% of the macrolide-resistant population were erm (B)+mef (A) positive (23). In Canada, erm (B) and mef (A) positive isolates represent 4% in 1998 and 12% in 2004 and one major cluster containing 36 (72%) of the 50 isolates was described (24).
In Turkey, Şener et al. (25) studied 669 S. pneumoniae strains isolated in Ankara, Turkey, between 1994 and 2002. They found that 57 (62.6%) of the 91 erythromycin-resistant pneumococci had the cMLSB phenotype, 19 (20.9%) had the iMLSB phenotype and 15 (16.5%) had the M phenotype. Overall, 83.5% of isolates had the erm (B) and 16.5% the mef (A) genotype. Gulay et al. (15) analyzed a total of 151 randomly selected S. pneumoniae isolates from seven centers in Turkey between 1998 and 2002. The erythromycin resistance rate was 26.4%. Only one strain was found to possess both erm (B) and mef (A) genes (2.5%) and a clonal relationship could not be demonstrated by BOX-PCR. Sağıroğlu et al. (26) analyzed 50 erythromycin-resistant pneumococci in Istanbul; 86% had the cMLSB and 14% had the M phenotype. Overall, 42% of strains were erm (B) and mef (A) positive. Clonal dissemination wasn’t studied. Telli et al. (15) analyzed 89 S. pneumoniae strains isolated between 2007 and 2009 in Aydın, Turkey; of these, 74%, 14% and 9% were the cMLSB phenotype, the M phenotype and the iMLSB phenotype, respectively. In addition, 20% were found to be erm (B)+mef (A) positive. No clonal dissemination was found according to PFGE analysis.
In conclusion, this study show that the macrolide resistance of S. pneumoniae isolates is relatively high and the cMLSB phenotype is the most prevalent resistance mechanism in our hospital. The percentage of isolates harboring both erm (B) and mef (A) genes is the highest reported from the studies conducted in our country to date. Dual erm (B) and mef (A) positive isolates tended to have higher resistance rates against penicillin and quinupristin-dalfopristin. In this study, we also showed the clonal relationship among macrolide-resistant S. pneumoniae strains possessing both erm (B) and mef (A) in Turkey for the first time.
Acknowledgments
We thank Zeynep Gulay for providing ATCC 700673 (Hungary19A-6) and ATCC 51916 (Tennessee23F-4) standard strains.
Footnotes
Ethics Committee Approval: Ethics committee approval was received for this study from the ethics committee of İstanbul University Cerrahpaşa Faculty of Medicine.
Informed Consent: N/A.
Peer-review: Externally peer-reviewed.
Author contributions: Concept - N.G.; Design - N.G., K.M., L.S.; Supervision - N.G.; Resource - L.S., M.K.; Materials - L.S., M.K.; Data Collection&/or Processing - L.S.; Analysis&/or Interpretation - M.K., S.S.; Literature Search - L.S.; Writing - L.S., N.G.; Critical Reviews - N.G., K.M.
Conflict of Interest: No conflict of interest was declared by the authors.
Financial Disclosure: The authors declared that the present work was supported by the Research Fund of İstanbul University, project no. 3357/2009.
REFERENCES
- 1.Daneman N, McGeer A, Green K, Low DE, Toronto Invasive Bacterial Diseases Network Macrolide resistance in bacteremic pneumococcal disease: implications for patient management. Clin Infect Dis. 2006;43:432–8. doi: 10.1086/505871. [DOI] [PubMed] [Google Scholar]
- 2.Calatayud L, Ardanuy C, Tubau F, Rolo D, Grau I, Pallarés R, et al. Serotype and genotype replacement among macrolide-resistant invasive pneumococci in adults: mechanisms of resistance and association with different transposons. J Clin Microbiol. 2010;48:1310–6. doi: 10.1128/JCM.01868-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute . Performance standards for antimicrobial susceptibility testing M100-S23. Wayne, Pa: USA: 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montanari MP, Mingoia M, Giovanetti E, Varaldo PE. Differentiation of resistance phenotypes among erythromycin-resistant pneumococci. J Clin Microbiol. 2001;39:1311–5. doi: 10.1128/JCM.39.4.1311-1315.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 1996;40:2562–6. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Belkum A, Sluijter M, de Groot R, Verbrugh H, Hermans PW. Novel BOX repeat PCR assay for high-resolution typing of Streptococcus pneumoniae strains. J Clin Microbiol. 1996;34:1176–9. doi: 10.1128/jcm.34.5.1176-1179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGee L, McDougal L, Zhou J, Spratt BG, Tenover FC, George R, et al. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J Clin Microbiol. 2010;39:2565–71. doi: 10.1128/JCM.39.7.2565-2571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavlicek A, Hrda S, Flegr J. FreeTree-Freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Application in the RAPD analysis of the genus Frenkelia. Folia Biol (Praha) 1999;45:97–9. [PubMed] [Google Scholar]
- 9.Page RD. TreeView: An application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–8. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 10.Linares J, Ardanuy C, Pallares R, Fenoll A. Changes in antimicrobial resistance, serotypes and genotypes in Streptococcus pneumoniae over a 30-year period. Clin Microbiol Infect. 2010;16:402–10. doi: 10.1111/j.1469-0691.2010.03182.x. [DOI] [PubMed] [Google Scholar]
- 11.Borg MA, Tiemersma E, Scicluna E, van de Sande-Bruinsma N, de Kraker M, Monen J, et al. Prevalence of penicillin and erythromycin resistance among invasive Streptococcus pneumoniae isolates reported by laboratories in the southern and eastern Mediterranean region. Clin Microbiol Infect. 2009;15:232–7. doi: 10.1111/j.1469-0691.2008.02651.x. [DOI] [PubMed] [Google Scholar]
- 12.European Antimicrobial Resistance Surveillance System EARSS Annual Report 2008. http://www.rivm.nl/earss/Images/EARSS%202008_final_tcm61-65020.pdf.
- 13.Gönüllü N, Berkiten R. Antimicrobial resistance of clinical isolates of Streptococcus pneumoniae in Istanbul. Int J Antimicrob Agents. 2000;16:77–8. doi: 10.1016/s0924-8579(00)00162-x. [DOI] [PubMed] [Google Scholar]
- 14.Gonullu N, Catal F, Kucukbasmaci O, Ozdemir S, Torun MM, Berkiten R. Comparison of in vitro activities of tigecycline with other antimicrobial agents against Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis in two university hospitals in Istanbul, Turkey. Chemotherapy. 2009;55:161–7. doi: 10.1159/000214144. [DOI] [PubMed] [Google Scholar]
- 15.Gulay Z, Ozbek OA, Bicmen M, Gur D. Macrolide resistance determinants in erythromycin-resistant Streptococcus pneumoniae in Turkey. Jpn J Infect Dis. 2008;61:490–3. [PubMed] [Google Scholar]
- 16.Telli M, Eyigör M, Gültekin B, Aydın N. Evaluation of resistance mechanisms and serotype and genotype distributions of macrolide-resistance strains in clinical isolates of Streptococcus pneumoniae in Aydın, Turkey. J Infect Chemother. 2011;17:658–64. doi: 10.1007/s10156-011-0238-x. [DOI] [PubMed] [Google Scholar]
- 17.Farrell DJ, Jenkins SG, Brown SD, Patel M, Lavin BS, Klugman KP. Emergence and spread of Streptococcus pneumoniae with erm (B) and mef (A) resistance. Emerg Infect Dis. 2005;11:851–8. doi: 10.3201/eid1106.050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bley C, van der Linden M, Reinert RR. Mef (A) is the predominant macrolide resistance determinant in Streptococcus pneumoniae and Streptococcus pyogenes in Germany. Int J Antimicrob Agents. 2011;37:425–31. doi: 10.1016/j.ijantimicag.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 19.McGee L, Klugman KP, Wasas A, Capper T, Brink A. Serotype 19F multiresistant pneumococcal clone harboring two erythromycin resistance determinants [erm (B) and mef (A)] in South Africa. Antimicrob Agents Chemother. 2001;45:1595–8. doi: 10.1128/AAC.45.5.1595-1598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reinert RR, Filimonova OY, Al-Lahham A, Grudinina SA, Ilina EN, Weigel LM, et al. Mechanisms of macrolide resistance among Streptococcus pneumoniae isolates from Russia. Antimicrob. Agents Chemother. 2008;52:2260–2. doi: 10.1128/AAC.01270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGee L, Wang H, Wasas A, Huebner R, Chen M, Klugman KP. Prevalence of serotypes and molecular epidemiology of Streptococcus pneumoniae strains isolated from children in Beijing, China: identification of two novel multiply-resistant clones. Microb Drug Resist. 2001;7:55–63. doi: 10.1089/107662901750152800. [DOI] [PubMed] [Google Scholar]
- 22.DiPersio LP, DiPersio JR, Beach JA, DeFine LA. Rise of Streptococcus pneumoniae isolates containing both erm (B) and mef (E) genes from an adult tertiary care community hospital system. Diagn Microbiol Infect Dis. 2006;55:327–31. doi: 10.1016/j.diagmicrobio.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 23.Bowers JR, Driebe EM, Nibecker JL, Wojack BR, Sarovich DS, Wong AH, et al. Dominance of multidrug resistant CC271 clones in macrolide-resistant Streptococcus pneumoniae in Arizona. BMC Microbiol. 2012;12:12–22. doi: 10.1186/1471-2180-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wierzbowski AK, Nichol K, Laing N, Hisanaga T, Nikulin A, Karlowsky JA, et al. Macrolide resistance mechanisms among Streptococcus pneumoniae isolated over 6 years of Canadian Respiratory Organism Susceptibility Study (CROSS) (1998–2004) J Antimicrob Chemother. 2007;60:733–40. doi: 10.1093/jac/dkm273. [DOI] [PubMed] [Google Scholar]
- 25.Sener B, Koseoglu O, Gur D, Bryskier A. Mechanisms of macrolide resistance in clinical pneumococcal isolates in a university hospital, Ankara, Turkey. J Chemother. 2005;17:31–5. doi: 10.1179/joc.2005.17.1.31. [DOI] [PubMed] [Google Scholar]
- 26.Sağıroğlu P, Aksu B, Hasdemir MU. Investigation of macrolide resistance mechanisms in Streptococcus pneumoniae: results of Marmara University hospital between 2005–2008. Marmara Med J. 2011;24:15–20. [Google Scholar]


