Abstract
Staphylococcal cassette chromosome mec (SCCmec) is a mobile genetic element composed of the mec gene complex, which encodes methicillin resistance, and the ccr gene complex, which encodes the recombinases responsible for its mobility. The mec gene complex has been classified into four classes, and the ccr gene complex has been classified into three allotypes. Different combinations of mec gene complex classes and ccr gene complex types have so far defined four types of SCCmec elements. Now we introduce the fifth allotype of SCCmec, which was found on the chromosome of a community-acquired methicillin-resistant Staphylococcus aureus strain (strain WIS [WBG8318]) isolated in Australia. The element shared the same chromosomal integration site with the four extant types of SCCmec and the characteristic nucleotide sequences at the chromosome-SCCmec junction regions. The novel SCCmec carried mecA bracketed by IS431 (IS431-mecA-ΔmecR1-IS431), which is designated the class C2 mec gene complex; and instead of ccrA and ccrB genes, it carried a single copy of a gene homologue that encoded cassette chromosome recombinase. Since the open reading frame (ORF) was found to encode an enzyme which catalyzes the precise excision as well as site- and orientation-specific integration of the element, we designated the ORF cassette chromosome recombinase C (ccrC), and we designated the element type V SCCmec. Type V SCCmec is a small SCCmec element (28 kb) and does not carry any antibiotic resistance genes besides mecA. Unlike the extant SCCmec types, it carries a set of foreign genes encoding a restriction-modification system that might play a role in the stabilization of the element on the chromosome.
The spread of antibiotic resistance among Staphylococcus aureus strains is of great concern in the treatment of staphylococcal infections, since S. aureus has quickly acquired resistance to all antibiotics introduced for clinical use. The first clinical isolate of methicillin-resistant S. aureus (MRSA) was reported in 1961, only 1 year after the introduction of methicillin (18). MRSA produces penicillin-binding protein 2′ (PBP 2′), which has a reduced affinity for β-lactam antibiotics (11, 34, 42). The mecA gene, which encodes PBP 2′, and its regulatory genes, mecI and mecR1, were cloned and sequenced in the 1980s (27, 38). Those genes are located on the chromosome, and they have become widely distributed among many staphylococcal species (1, 12, 14, 23, 35, 37, 40, 41).
In the last few years, understanding of the genetic basis for methicillin resistance has advanced significantly. MRSA is produced when methicillin-susceptible S. aureus (MSSA) acquires a genetic element called staphylococcal cassette chromosome mec (SCCmec).
SCCmec is a genomic island (Gisland) that is inserted at the 3′ end of orfX and that is located near the replication origin of S. aureus (3, 24). Since our discovery of the first SCCmec element from pre-MRSA strain N315 in 1999, several types of SCCmec elements have been identified by determining their entire nucleotide sequences (16, 17, 26, 39). Pre-MRSA is a mecA gene-carrying MSSA strain in which mecA gene expression is strongly repressed by the presence of an intact mecI gene. The SCCmec element contains the mec gene complex (the mecA gene and its regulators) and the ccr gene complex, which encodes site-specific recombinases responsible for the mobility of SCCmec (22).
Four classes of the mec gene complex have been identified by PCR, using chromosomal DNA from methicillin-resistant coagulase-negative staphylococci as templates. The different mec gene complexes are structured as follows: class A, IS431-mecA-mecR1-mecI; class B, IS431-mecA-ΔmecR1-IS1272; class C, IS431-mecA-ΔmecR1-IS431; and class D, IS431-mecA-ΔmecR1 (21).
The ccr gene complex contains two site-specific recombinase genes, ccrA and ccrB, which are responsible for the mobility of SCCmec (16, 22). There are four allotypes in each of the ccrA and ccrB genes: ccrA1, ccrA2, ccrA3, and ccrA4 for ccrA and ccrB1, ccrB2, ccrB3, and ccrB4 for ccrB. SCCmec is classified into allotypes according to the combination of the mec gene complex class and the ccr gene complex type that it possesses (16, 26), as follows: type I SCCmec, class B mec gene complex and type 1 ccr gene complex; type II SCCmec, class A mec gene complex and type 2 ccr gene complex; type III SCCmec, class A mec gene complex and type 3 ccr gene complex; and type IV SCCmec, class B mec gene complex and type 2 ccr gene complex. The region other than the mec and ccr gene complexes is designated the J (junkyard) region. Each SCCmec type is further classified into subtypes on the basis of the J-region sequence (13).
SCC is a basic mobile genetic element that serves as the vehicle for gene exchange among staphylococcal species; it has been reported in some coagulase-negative staphylococci as well as in S. aureus (15, 20, 25). SCCmec is a member of the SCC family, the members of which specialize as carriers of methicillin resistance.
Both SCC12263 (found in S. hominis GIFU12263) and SCC476 (found in MSSA strain 476 [the seventh S. aureus strain whose whole genome sequence is being determined]) carried ccrA and ccrB genes, but they did not carry mecA or any other antibiotic resistance gene (20) (http://www.sanger.ac.uk/Projects/S_aureus/). SCCcap1 encodes a capsule gene cluster that confers a mucoid appearance because of overexpression of the capsule in S. aureus and is also a member of the SCC family (25).
MRSA has been a major causative agent of nosocomial infections (2). Recently, however, MRSA has become increasingly isolated from patients with community-acquired infections (4, 5, 9, 28, 30, 31). The SCCmec typing system that we described above has turned out to be an important marker for distinguishing these two categories of MRSA. Namely, by using the SCCmec typing system, we have provided strong evidence for the independent derivation of health care-associated MRSA (H-MRSA) and community-acquired MRSA (C-MRSA) clones (32).
The majority of H-MRSA strains carry one of the three types of SCCmec (type I, II, or III) (6, 16), whereas well-defined American C-MRSA and nonmultiresistant oxacillin-resistant S. aureus (NORSA) strains carry type IV SCCmec (32). Then, Vandenesch et al. (43) reported that majority of C-MRSA strains in France also carry type IV SCCmec. Type IV SCCmec is a small element that does not carry antibiotic resistance genes other than mecA and has multiple subtypes. The extreme heterogeneity of the chromosome genotypes in C-MRSA strains suggests that type IV SCCmec is highly transmissible. However, we have also noted several strains whose SCCmec elements are nontypeable (32). We propose that clarifying the unknown structures of these SCCmec elements is indispensable for increasing the typeability of strains for SCCmec-based epidemiology and, most importantly, for obtaining a better understanding of the role of SCC in the evolution of S. aureus, including the acquisition of multidrug resistance.
In this study, we determined the entire nucleotide sequence of the unknown element found in Australian C-MRSA strain WIS to characterize this SCCmec element and to relate it phylogenetically to other known SCCmec types. It turned out to be a distinct type of SCCmec with a distinct ccr gene homologue and with the mecA gene characteristically bracketed by two insertion sequences.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in the experiments described here are shown in Table 1. Brain heart infusion (BHI) broth and agar (Becton Dickinson, Sparks, Md.) were used as culture media for S. aureus. Tetracycline (Sigma Chemical Co., St. Louis, Mo.), tobramycin (Shionogi Co., Osaka, Japan), and ceftizoxime (Fujisawa Pharmaceutical Co. Ltd, Osaka, Japan) were used where appropriate at the concentrations indicated in the text.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains or plasmid | Description | Reference(s) or source |
|---|---|---|
| Strains | ||
| MRSA | ||
| WIS [WBG8318] | C-MRSA strain isolated in Australia | 31 (provided by W. B. Grubb) |
| 81/0342 | NORSA strain isolated in Australia | 32 (provided by J. D. Turnidge) |
| 91/2574 | NORSA strain isolated in Australia | 32 (provided by J. D. Turnidge) |
| N315 | Pre-MRSA strain isolated in Japan in 1982 | 12 |
| N315ex | SCCmec excised from strain N315 | 22 |
| Plasmids | ||
| pYT3 | Temperature-sensitive shuttle vector (tetracycline resistant) | 9a |
| pSR2 | pYT3 into which the ccrA2 and ccrB2 genes were cloned | 22 |
| pSR5w | pYT3 into which ccrCWis was cloned | This study |
| pSR5E | pYT3 into which ccrC0342 was cloned | This study |
| pYTattII | pYT3 into which attII (the presumptive attachment-sequence in type II SCCmec) was cloned | 22 |
| pYTattV | pYT3 into which attV (the presumptive attachment sequence in type V SCCmec) was cloned | This study |
| pSR2attII | pYTattII into which the ccrA2 and ccrB2 were genes cloned | 22 |
| pSR5EattV | pYTattV into which ccrC0342 was cloned | This study |
DNA manipulation and nucleotide sequencing.
The DNA fragments encompassing the entire SCCmec nucleotide sequence of strain WIS were amplified by long-range PCR with several sets of primers, as follows: the region from the orfX gene to the mecA gene was covered by primers cR1 and mA3, and the region from the mecA gene to the chromosomal region flanked to the left end of SCCmec was amplified by PCR with primers is1 and cLs1. The latter region was later amplified by PCR with primer sets mA2 and V3, is1 and V2, and V1 and cLs1, as indicated in Fig. 1. The PCR products were purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany), and their nucleotide sequences were determined. The amplification steps for PCR, long-range PCR, and nested PCR were performed as described previously (17).
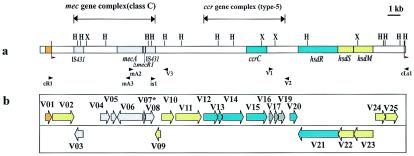
FIG. 1.
Genetic structure of the type V SCCmec element of strain WIS. The structure of the type V SCCmec element illustrated is based on the nucleotide sequence deposited in the DDBJ/EMBL/GenBank databases under accession no. AB121219. (a) Essential structure of type V SCCmec. The locations of the essential genes are illustrated. The colors of the essential genes are based on those for the ORFs described for panel b. The HindIII (H) and XbaI (X) restriction sites are indicated. Black arrowheads indicate primer-specific locations. The nucleotide sequences of primers cR1, mA2, mA3, is1, and cLs1 have been described previously (17, 26). The nucleotide sequences of primers V1, V2, and V3 are listed in Table 2. The locations of direct repeats are indicated by red arrowheads. (b) ORFs in and around type V SCCmec. The ORFs corresponding to sequences more than 100 bp are indicated by arrows, which also indicate the directions of the ORFs. Light gray arrows, ORFs conserved in the five types of SCCmec elements with identities of more than 99%; gray arrows, ORFs conserved in the five types of SCCmec elements with identities of 48.1 to 93.4%; blue arrows, ORFs commonly found in both the type V SCCmec element and the J region of type III SCCmec; yellow arrows, ORFs unique to type V SCCmec; orange arrow, orfX.
Construction of recombinant plasmids and excision assay.
The DNA fragments containing the ccrC gene of strain WIS (ccrCWIS) and the ccrC gene of strain 81/0342 (ccrC0342) were amplified by PCR with primers Vcc1 and Vcc2 and digested with BamHI. The fragments were cloned into the BamHI site of temperature-sensitive shuttle vector pYT3 to construct recombinant plasmids pSR5W and pSR5E, respectively. The fidelity of the cloning was ascertained by determining the nucleotide sequence of the cloned DNA fragment. Recombinant plasmids were introduced into S. aureus cells by electroporation, as described previously (22).
For the excision assay, approximately 103 cells of transformants and their parent strains were inoculated into 10 ml of BHI broth containing tetracycline (10 mg/liter), and the culture was incubated at 30°C with shaking. After 20 h, an aliquot of 0.8 ml was transferred to an Eppendorf tube and cells were collected by centrifugation. DNAs were extracted from the cells and used as templates for subsequent PCR experiments to identify the attBscc and attSCC sequences generated in the cells with the primer sets listed in Table 2.
TABLE 2.
Primers used in this study
| Genetic element(s) or purpose and primer | Nucleotide sequence | Expected product size (kb) | Reference or source |
|---|---|---|---|
| Type V SCCmec of WIS | |||
| V1 | 5′-TACCACTTTTACCACTTAGCTTT-3′ | This study | |
| V2 | 5′-ATGAGGCTTTAACATTTCCATCA-3′ | This study | |
| is1 | 5′-ACATTAGATATTTGGTTGCGT-3′ | 21 | |
| V3 | 5′-TATCATTACACTCTTGAGTCTCT-3′ | This study | |
| mA2 | 5′-AACGTTGTAACCACCCCAAGA-3′ | 21 | |
| mA3 | 5′-TGCTATCCACCCTCAAACAGG-3′ | 21 | |
| Detection of ccrC | |||
| γF | 5′-CGTCTATTACAAGATGTTAAGGATAAT-3′ | 0.52 | This study |
| γR | 5′-CCTTTATAGACTGGATTATTCAAAATAT-3′ | This study | |
| Construction of recombinant plasmids with ccrC | |||
| Vcc1 | 5′-AAAAGGATCCAAGTTGTTTGCTTAGCGTCATTA-3′ | 1.93 | This study |
| Vcc2 | 5′-AAAAGGATCCTAGTACTCATATGATTAAGTGGT-3′ | This study | |
| attSCC-V (attSCC of type V SCCmec) | |||
| mVR2 | 5′-AAAAAGTCGACTACCGTCGATATCAATTGCTTTTT-3′ | 0.90 | This study |
| mVL2 | 5′-AAAAAGTCGACTGGAGACGTAGTATAAATATAGCT-3′ | This study | |
| Detection of precise excision and closed circular DNA formation | |||
| attBscc (N315) | |||
| cL1 | 5′-ATTTAATGTCCACCATTTAACA-3′ | 0.28 | 17 |
| cR1 | 5′-AAGAATTGAACCAACGCATGA-3′ | 17 | |
| attBscc (WIS) | |||
| cLs1 | 5′-TGCCAATCACAGTTCAATCAATT-3′ | 0.31 | 26 |
| cR1 | 5′-AAGAATTGAACCAACGCATGA-3′ | 17 | |
| attSCC (N315) | |||
| mL1 | 5′-GAATCTTCAGCATGTGATTTA-3′ | 0.46 | 22 |
| mR8 | 5′-ATGAAAGACTGCGGAGGCTAACT-3′ | 22 | |
| attSCC (WIS) | |||
| mVL1 | 5′-TACTTTGGTTTCATATTAATAGCACT-3′ | 0.29 | This study |
| mVR1 | 5′-TCACTAGTGTAATTATCGAATGAT-3′ | This study | |
| Generation of chromosome-SCCmec (mini-SCC) junction | |||
| α | 5′-TTTCACACAGGAAACAGCTATGAC-3′ | 22 | |
| β | 5′-ATCACGATATTGCTTATAAGCA-3′ | 22 | |
| γ | 5′-ATGTTATTAAGCAGATTGCGTCAA-3′ | This study |
To investigate the strains from which DNA was excised for the generation of SCCmec, cells were further cultivated in BHI broth with tetracycline at 30°C, with one passage per day for 10 days. The cultures were diluted, and approximately 103 cells were inoculated for each passage. The cells were evaluated for the loss of SCCmec by replicating them onto agar plates with and without tobramycin at 10 mg/liter (for strain N315) and ceftizoxime at 5 mg/liter (for strain WIS).
Construction of a recombinant plasmid carrying the ccrC gene and attSCC formed in WIS(pSR5E) and integration assay.
Recombinant plasmid pSR5EattV, which carried ccrC0342 and attSCC formed in WIS(pSR5E), was constructed as a model of the closed circular form of type V SCCmec (mini-SCC). Briefly, the DNA fragment containing attSCC of type V SCCmec was amplified by PCR with primers mVR2 and mVL2 (Table 2). DNA extracted from WIS(pSR5E) was used as the template for the amplification of type V attSCC. A SalI-digested DNA fragment carrying attSCC type V was cloned into pYT3 to produce pYT3attV. Plasmid pSR5EattV was constructed by cloning a BamHI-digested DNA fragment carrying ccrC0342 into pYT3attV.
For the integration assay, recombinant plasmids and control plasmids were introduced into N315 by electroporation, and the cells were grown on BHI agar plates containing tetracycline at a concentration of 10 mg/liter at 30°C for 46 to 47 h. The colonies that grew on each tetracycline plate were resuspended in 0.5 ml of BHI broth, spread onto the BHI agar plates with or without tetracycline (10 mg/liter), and incubated at 30°C (permissive temperature for the replication of plasmid pYT3) and 43°C (a temperature nonpermissive for the replication of plasmid pYT3) for 18 h.
Computer analysis.
Open reading frames (ORFs) of more than 100 bp were identified with the GAMBLER software, and their functions were predicted by a search homology with the BLAST program. All the ORFs in and around type V SCCmec were compared to those of the four known types of SCCmec (type I in strain NCTC10442 [DDBJ/EMBL/GenBank accession no. AB033763], type II in strain N315 [DDBJ/EMBL/GenBank accession no. D86934], type III in strain 85/2082 [DDBJ/EMBL/GenBank accession no. AB037671], and type IVa in strain CA05 [DDBJ/EMBL/GenBank accession no. AB063172]). The homologies between the nucleotide sequence of the type V SCCmec element of strain WIS and those of the type I, II, III, and IVa SCCmec elements were studied as described previously (20).
Several types of analyses were carried out with the BLAST program at the website of the National Center for Biotechnology Information (http://www3.ncbi.nlm.nih.gov/BLAST/). The codon usage values for prokaryotes were taken from a database (http:www.kazusa.or.jp/codon). Codon usage was tabulated from international DNA sequence databases (sequence status for the year 2000) (29). The similarity of codon usage was evaluated by codon bias analysis (19).
Nucleotide sequence accession number.
The sequence of the type V SCCmec element of strain WIS has been deposited in the DDBJ/EMBL/GenBank databases under accession no. AB121219.
RESULTS
Type V SCCmec as a new member of the SCCmec family.
Our purpose was to investigate the genetic organization of the unknown element carrying the class C mec gene complex that was found in three strains, a C-MRSA strain (strain WIS) and two NORSA strains (strains 81/0342 and 91/2574). For that reason, we selected strain WIS and determined the nucleotide sequence of the region in and around the mec gene complex. The overall organization of the element and the strategy used to amplify the DNA fragment by long-range PCR with different sets of primers are shown in Fig. 1a.
The element carried the class C mec gene complex, which is composed of a copy of insertion sequence IS431, mecA, truncated mecR1 (ΔmecR1), and another copy of IS431 inserted in the opposite direction (21). From the size of ΔmecR1 and the direction of another copy of IS431, it was judged that the element carried the class C2 mec gene complex (21).
Neither the ccrA gene nor the ccrB gene, both of which are responsible for the mobility of SCCmec, was found in the element. Instead, a novel ORF sequence, V15, encoding a protein similar to site-specific recombinases was found. The deduced amino acid sequence of V15 showed the highest degree of similarity to that of a type III SCCmec ORF, CZ072, with sequence identity of 93.2%.
We examined by PCR whether the novel ORF is commonly found in the other two NORSA strains. By amplifying DNA fragments with the primer set listed in Table 2 and determining the nucleotide sequences of the two DNA fragments, we found that the other two strains also carried identical ORFs. These ORFs were actually highly similar to that of WIS (ORF V15), with a nucleotide sequence identity of 91.4%.
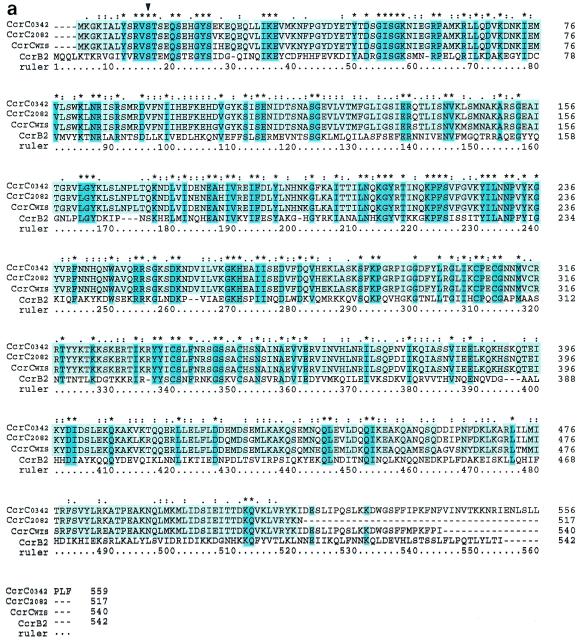
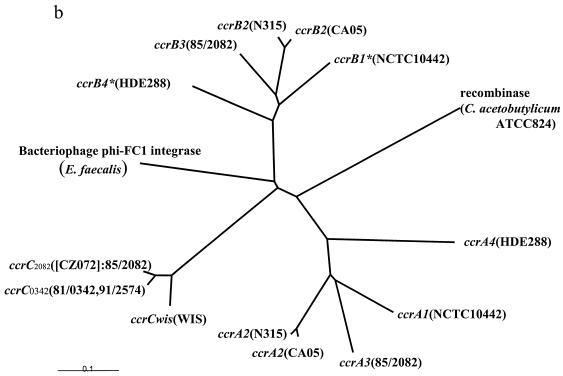
The deduced amino acid sequences of the four ORFs, ORF V15 of strain WIS, the corresponding ORFs of strains 81/0342 and 91/2574, and ORF CZ072 of strain 85/2082, showed high degrees of similarity, with identities of 93.2 to 96.9%, although their C-terminal portions were dissimilar. Figure 2a shows the alignments of the deduced amino acid sequences of four ORFs: ORF V15, the corresponding ORF of 81/0342, CZ072 of 85/2082, and ccrB of N315. All four ORFs have a catalytic motif at the N-terminal domain, which is characteristic of recombinases of the invertase-resolvase family. They were basic proteins with pI values of 9.68 to 9.85. These features are similar to those of the CcrA and CcrB proteins that have been reported previously. We tried to relate the newly found ccrC genes phylogenetically to previously reported ccr genes: the three types of each of the ccrA and ccrB genes and another set of ccrA and ccrB genes (16, 33). In order to investigate the phylogenetic relations of those ccr genes, we reconstituted the putative ccrB1 (ccrB1*) and ccrB4 (ccrB4*) genes of 1626 and 1,629 bp, respectively, by adding back an adenine to the deleted point of the truncated ccrB1 and ccrB4 genes, since ccrB1 of NCTC10442 and ccrB4 of HDE288 are truncated. Figure 2b illustrates the phylogenetic relations of those ORFs, extant Ccr proteins, and other site-specific recombinases such as the integrase of Enterococcus faecalis bacteriophage φ FC1 and the site-specific recombinase of Clostridium acetobutylicum ATCC 824 (Fig. 2b). A phylogenetic tree showed that the four ORFs constitute a novel group of ccr genes distinct from the ccrA and ccrB genes. Accordingly, we designated the ORFs cassette chromosome recombinase C (ccrC) as a new putative site-specific recombinase and the elements as type V SCCmec. Type V SCCmec is defined by the carriage of the class C2 mec gene complex and the ccrC gene, which is located in the type 5 ccr gene complex, as described below.
FIG. 2.
Characteristics of deduced amino acid sequences of CcrC. (a) The deduced amino acid sequences of the three ccr genes were aligned with that of ccrB2 of strain N315. The alignment was performed by using ClustalX software with the protein weight matrix PAM series. The three Ccr proteins were CcrCWIS (CcrC of WIS), CcrC0342 (CcrC of strain 81/0342; the sequence of the strain 91/2674 CcrC was exactly identical to that of CcrC0342), and CcrC2082 (CcrC of strain 85/2082). The three Ccr proteins were highly homologous. Of the 517 amino acids corresponding to CcrC2082, 484 amino acids were conserved in the three Ccr proteins (pale blue) and 30 amino acids were conserved in two of three Ccr proteins. One hundred thirty-eight amino acids were conserved in CcrC0342, CcrWIS, and CcrB2 (blue). The consensus sequence shown above the amino acid sequences indicates conserved amino acids, as follows: asterisks, identical or conserved residues in all sequences in the alignment; colons, conserved substitutions; dots, semiconserved substitutions; hyphens, gaps. A black arrowhead indicates the presumptive serine involved in the phosphoseryl linkage conserved in the NH2-terminal catalytic domain of site-specific recombinases of the invertase-resolvase family. (b) Phylogenetic relationships among ccrA genes, ccrB genes, ccrC genes, and two site-specific recombinases. The two site-specific recombinases used were the integrase (int) of bacteriophage φ FC1 found in E. faecalis (1,216 bp; DDBJ/EMBL/GenBank accession no. AF124258) and the site-specific recombinase found in C. acetobutylicum ATCC 824 (1,635 bp; DDBJ/EMBL/GenBank accession no.AE007636). The ccrA and ccrB genes used were as follows: ccrA1 and ccrB1*, respectively, found in strain NCTC10442 (DDBJ/EMBL/GenBank databases accession no. AB033763); ccrA2 and ccrB2, respectively, found in strains N315 (DDBJ/EMBL/GenBank databases accession no. D86934) and CA05 (DDBJ/EMBL/GenBank databases accession no. AB063172); ccrA3 and ccrB3, respectively, found in strain 85/2082 (DDBJ/EMBL/GenBank databases accession no. AB037671); and ccrA4 and ccrB4*, respectively, found in strain HDE288 (DDBJ/EMBL/GenBank databases accession no. AF411935). Since ccrB1 of NCTC10442 and ccrB4 of HDE288 were truncated, we reconstituted large ORFs of 1,626 bp (ccrB1*) and 1,629 bp (ccrB4*) by adding an adenine and used them for the comparison. The nucleotide sequences of five ccrA genes, five ccrB genes, four ccrC genes, and two site-specific recombinases were aligned by using the ClustalX program. The phylogenetic tree was generated by the neighbor-joining method by creating 2,000 bootstrap replicates. The tree was visualized with Tree View software, which was obtained from the website http://taxonomy.zoology.gla.ac.uk/rod/treeview.htm. The branch length indicates the distance, which is expressed as the number of substitutions per 100 bases.
The boundaries of type V SCCmec.
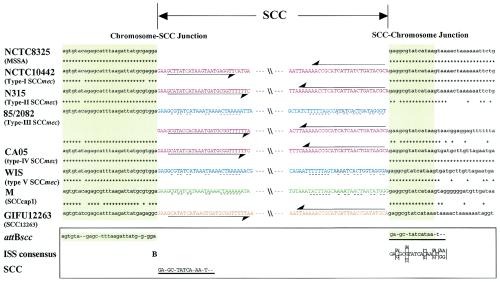
Both the left and right boundaries of type V SCCmec were determined by comparing their nucleotide sequences with those of previously reported SCCmec and SCC elements. Determination of the nucleotide sequence of attBscc on the chromosome and attSCC on closed circular SCC, generated by precise excision of the element, supported the predicted positions of the boundaries. Type V SCCmec was integrated at exactly the same nucleotide position at the 3′ end of orfX (shown as a yellow square in Fig. 3), where the four types of SCCmec were integrated (Fig. 3). Two SCC elements which did not carry the mecA gene, SCCcap1 and SCC12263, were also integrated at the same position. SCCcap1, which is found in S. aureus M, is an element that carries a capsule gene cluster (25). SCC12263 was found in S. hominis GIFU12263, and it is an element that carries active ccrA1 and ccrB1 genes (20). Previously, we could not judge the exact extremities of the SCCmec elements only by comparison of the nucleotide sequences of SCCmec elements found in S. aureus strains. This was possible because of the availability of new data from the nucleotide sequences of the attBscc elements from strains M and GIFU12263, generated by precise excision of the SCC element, and they were estimated to be at the positions shown in Fig. 3 (20, 25). The integration site sequence for SCC (ISS), which is uniquely present at the 3′ end of orfX in MSSA NCTC8325, is conserved in all strains examined so far. The ISS contains the consensus sequence 5′-BGA(A/G)GC(A/G/T)TATCA(C/T)AA(A/G)T(A/G)(A/G)-3′ (where the cutting site is between the B [B signifies A, C, or G] and the G at the 5′ end). The nucleotide sequences of the ISSs and the nucleotide sequences of the left extremities of the elements were nearly identical and constitute directly repeated sequences. These directly repeated sequences at the chromosome-SCC junction were found in all the elements, as shown in Fig. 3. In contrast, the degenerate inverted repeats found at both extremities of the four types of SCCmec and SCC12263 were not found at the extremities of type V SCCmec or SCCcap1 (Fig. 3).
FIG. 3.
Chromosome-SCC junction sequences. The nucleotide sequences at the left and right boundaries of the SCCmec element of strain WIS are aligned with those of six previously reported SCC elements: type I SCCmec of S. aureus NCTC10442 (DDBJ/EMBL/GenBank databases accession no. AB033763), type II SCCmec of S. aureus N315 (DDBJ/EMBL/GenBank databases accession no. D86934), type III SCCmec of S. aureus 85/2082 (DDBJ/EMBL/GenBank databases accession no. AB037671), type IV SCCmec of S. aureus CA05 (DDBJ/EMBL/GenBank databases accession no. AB063172), SCCcap1 of S. aureus M (DDBJ/EMBL/GenBank databases accession no. U10927), and SCC12263 of S. hominis GIFU12263 (DDBJ/EMBL/GenBank databases accession no. AB063171). Two sets of nucleotide sequences are listed for the type III SCCmec of S. aureus 85/2082. One set is composed of the nucleotide sequence of the left extremity (direct repeat 1) and the nucleotide sequence in the midst of the element flanked by direct repeat 2. The other set is composed of the nucleotide sequence containing direct repeat 2 and the right extremity of the element flanked by direct repeat 3 (15). Thin arrows indicate inverted repeats at both extremities of SCCmec elements carrying the ccrA and ccrB genes. Dotted lines indicate the characteristic nucleotide sequence conserved at both extremities of type V SCCmec or SCC elements carrying the ccrC gene. The 3′ ends of orfX are indicated with light green shading and lowercase bases. The consensus sequences of attBscc and ISS are boxed. Direct repeat sequences in ISS and the left end of the SCC element are indicated by thick lines. Nucleotide sequences of both extremities of SCC or SCCmec elements are colored as follows: red, type I, II, and IV SCCmec elements and the region between direct repeat 2 and direct repeat 3 in type III SCCmec element; orange, SCC12263; blue, type V SCCmec and the region between direct repeat 1 and direct repeat 2 in type III SCCmec element; green, SCCcap1.
Structure of type V SCCmec.
Judging from the nucleotide sequences at both extremities of the type V SCCmec shown in Fig. 3, type V SCCmec was estimated to be 27,624 bp. This is slightly larger than the type IV SCCmec elements (21 to 25 kb) but smaller than the type I SCCmec element of NCTC10442 (34 kb), the type II SCCmec element of N315 (53 kb), and the type III SCCmec element of 85/2082 (67 kb).
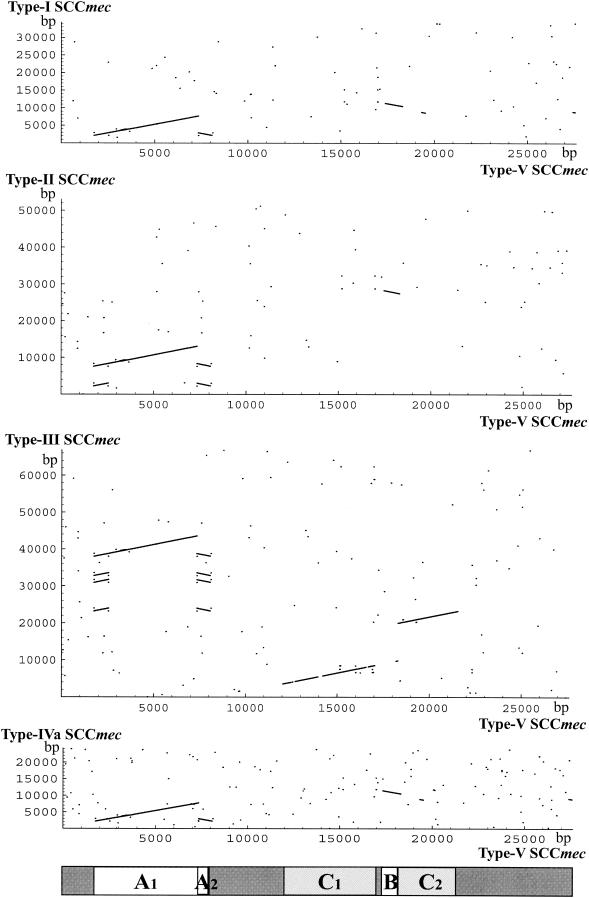
A total of 23 ORFs larger than 100 bp were found in the type V SCCmec element (Fig. 1b and Table 3). No antibiotic resistance gene other than mecA was found in the element. Figure 4 compares the nucleotide sequences of the five types of SCCmec elements obtained with the BLAST program. Type V SCCmec is composed of the regions conserved in other types of SCCmec and the regions unique to type V SCCmec. The regions in type V SCCmec similar to extant SCCmec elements are denoted A, B, and C in Fig. 4. Region A contained seven ORFs conserved in all five types of SCCmec elements with very high degrees of identity. The identities between region A1 of type V SCCmec and the corresponding regions in other SCCmec elements were more than 99.7%. Region A1 contained six ORFs in the mec gene complex, a transposase for IS431 (V03), a glycerophosphoryldiester phophodiesterase homologue (V04), a hypothetical protein (V05), mecA (V06), and ΔmecR1 (V07) (shown in light gray in Fig. 1b). ΔmecR1 (V07) was smaller than ΔmecR in the class B mec gene complex. Region A2 corresponded to IS431, which was found to be closely associated with the deletion point of ΔmecR1. This insertion of a copy of IS431 whose direction is opposite that of IS431 mec is characteristic of the class C2 mec gene complex. The transposases V03 and V08 encoded by two IS431 copies were not identical. The transposase V03 was nearly identical to that of IS431mec elements of four other types of SCCmec elements, whereas the transposase V08 showed slightly lower identity to that of IS431mec elements.
TABLE 3.
ORFs in and around type V SCCmec of WIS with deduced products showing similarities to extant proteins
| ORFa | Value for CDSb
|
|||||
|---|---|---|---|---|---|---|
| Starting nucleotide | Ending nucleotide | Size (bp) | Length (amino acids) | Gene | Product | |
| V01 | 418 | 897 | 480 | 159 | orfX | Conserved hypothetical protein orfX |
| V02 | 952 | 2652 | 1,701 | 566 | Hypothetical protein | |
| V03 | 3367 | 2693 | 675 | 224 | tnp | Transposase for IS431 |
| V04 | 4709 | 5452 | 744 | 247 | Glycerophosphoryl diester phosphodiesterase homolog | |
| V05 | 5549 | 5977 | 429 | 142 | Conserved hypothetical protein | |
| V06 | 8029 | 6023 | 2,007 | 668 | mecA | PBP 2′ |
| V07* | 8129 | 8239 | 111 | 36 | Δmec R1 | Truncated signal transducer protein MecR1 |
| V08 | 8277 | 8951 | 675 | 224 | tnp | Transposase for IS431 |
| V09 | 9439 | 9011 | 429 | 142 | Hypothetical protein | |
| V10 | 9520 | 10449 | 930 | 309 | Hypothetical protein | |
| V11 | 10611 | 12599 | 1,989 | 662 | Hypothetical protein | |
| V12 | 12794 | 13903 | 1,110 | 369 | Hypothetical protein | |
| V13 | 13896 | 14264 | 369 | 122 | Hypothetical protein | |
| V14 | 14264 | 15907 | 1,644 | 547 | Hypothetical protein | |
| V15 | 16132 | 17754 | 1,623 | 540 | ccrC | Cassette chromosome recombinase C |
| V16 | 17915 | 18253 | 339 | 112 | Hypothetical protein | |
| V17 | 18332 | 18658 | 327 | 108 | Hypothetical protein | |
| V19 | 18674 | 19177 | 504 | 167 | Hypothetical protein | |
| V20 | 19563 | 20129 | 567 | 188 | Hypothetical protein | |
| V21 | 23325 | 20206 | 3,120 | 1039 | hsdR | Type 1 restriction-modification system endonuclease |
| V22 | 24547 | 23309 | 1,239 | 412 | hsdS | Specificity subunit of type I restriction-modification system |
| V23 | 26051 | 24537 | 1,515 | 504 | hsdM | Modification subunit of type I restriction-modification system |
| V24 | 26242 | 27054 | 813 | 270 | Hypothetical protein | |
| V25 | 27054 | 27986 | 933 | 310 | Hypothetical protein | |
ORFs in parentheses were located outside of the type V SCCmec. Incomplete ORFs that are potentially defective genes or pseudogenes containing frameshift mutations are designated with asterisks.
CDS, coding sequence. The nucleotide positions are given from the nucleotide sequence deposited in the DDBJ/EMBL/GenBank databases under accession no. AB121219, and the sizes were measured from the 5′ (starting nucleotide) to the 3′ (ending nucleotide) direction.
Identity of the amino acid sequence to each ORF.
FIG. 4.
Homologous regions in nucleotide sequences of the type V SCCmec and extant SCCmec elements. The nucleotide positions of the type V SCCmec element are indicated on the horizontal axes, and those of extant SCCmec elements (types I, II, III, and IVa) are indicated on the vertical axes. The following regions of the nucleotide sequences of the SCCmec elements were used: type V SCCmec, nucleotides 880 to 28503 of the nucleotide sequence with DDBJ/EMBL/GenBank databases accession no. AB121219; type I SCCmec, the region from nucleotides 4504 to 38867 of the nucleotide sequence with DDBJ/EMBL/GenBank databases accession no. AB033763; type II SCCmec, nucleotides 4687 to 57653 of the nucleotide sequence with DDBJ/EMBL/GenBank databases accession no. D86934; type III SCCmec, nucleotides 899 to 67794 of the nucleotide sequence with DDBJ/EMBL/GenBank databases accession no. AB037671; and type IVa SCCmec, nucleotides 975 to 25222 of the nucleotide sequence with DDBJ/EMBL/GenBank databases accession no. AB063172. The regions showing the high degrees of similarity between two SCCmec elements are indicated with lines. Rising lines signify that homologous regions between two elements are located in the opposite direction. Short homologous regions appear as dots. The regions through A (A1 and A2), B, and C (C1 and C2) were designated by the extent of nucleotide sequence identity described in the text. The locations of the regions in the nucleotide sequence of type V SCCmec are as follows: A1, nucleotide positions 1778 to 7341; A2, nucleotide positions 7342 to 8131; B, nucleotide positions, 16964 to 18296; C1, nucleotide positions 12041 to 17040; and C2, nucleotide positions 18309 to 21540.
Region B represents the region conserved in type I, II, and IV SCCmec elements, with nucleotide sequence identities of 79.7 to 82.6%. Two ORFs, V17 and V19, were located in this region of type V SCCmec (Table 3). Two ORFs showed high degrees of similarity to the ORFs in the ccr gene complex of type I, II, and IV SCCmec elements.
The sequence of region C of type V SCCmec was homologous only to the corresponding region of the type III SCCmec element of strain 85/2082. Two regions of the type V SCCmec sequence, C1 and C2, showed high degrees of similarity to the corresponding region of the type III SCCmec element of strain 85/2082, with nucleotide sequence identities of 86.6 and 94.4%, respectively. Region C1 contained four ORFs (V12 to V15) whose sequences showed high degrees of similarity to the sequences of CZ072 (ccrC), CZ073, CZ074, and CZ075 located downstream of orfX in 85/2082. Although the region corresponding to region B was not identified in the type III SCCmec element, two sets of three ORFs whose sequences were similar to those of V16, V17, and V19, respectively, and which had amino acid identities ranging from 48.1 to 67.1% were found in the element. The first set of three ORFs (Z011, Z013, and Z014) was located downstream of the ccrA3 and ccrB3 genes, and these three ORFs were constituents of the type 3 ccr gene complex. The other set of three ORFs (CZ070, CZ069, and CZ068) was located downstream of ccrC (CZ072). It was noted that seven ORFs (three ORFs upstream of ccrC, ccrC, and three ORFs downstream of ccrC) were conserved in type V SCCmec and the J region of the type III SCCmec of 85/2082 (the amino acid identities of the corresponding ORFs were greater than or equal to 48.1%).
Thus, they were unified as a ccr gene complex together with the ccr gene, similar to the constructions of the type I to IV SCCmec elements.
Region C2 of type V SCCmec contained two ORFs (V20 and V21) which showed high degrees of similarity to CZ053 and CZ059 (truncated hsdR), respectively, in type III SCCmec. Although two regions, C1 and C2, were located near each other in type V SCCmec, the regions homologous to C1 and C2 in type III SCCmec were separated (Fig. 4).
The deduced amino acid sequence of ORF V21 (hsdR) showed a high degree of similarity (identity, 98.0%) to that of ORF Z059 (truncated hsdR) in the type III SCCmec of 85/2082, whereas it showed a very low degree of similarity (identity, less than 21%) to HsdR proteins encoded by hsdR genes in the S. aureus genome (hsdRaur).
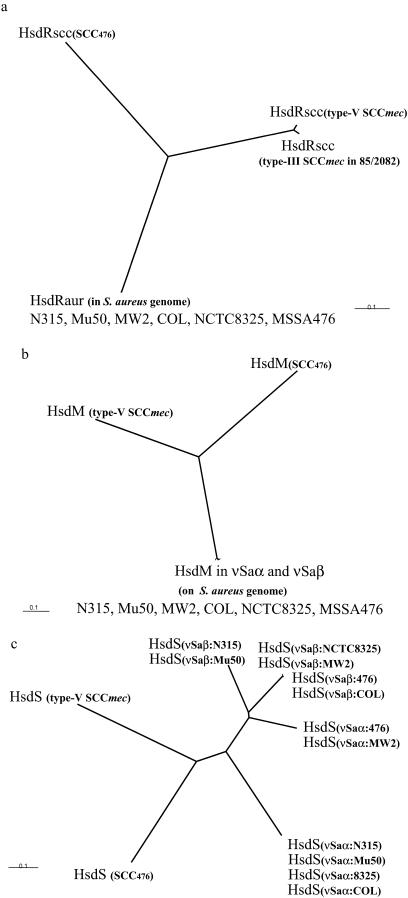
The regions other than those described above are unique in type V SCCmec. Two ORFs, V22 (hsdS) and V23 (hsdM), encoding the restriction-modification system were located at the right end of the element. Two type I restriction-modification DNA specificity domains, one of which is known as the target recognition domain and the other of which is the region conserved in the hsd subunit, were identified in ORF V22. The deduced amino acid sequence of ORF V23 showed the highest degree of similarity to the putative HsdM protein of Lactococcus sakei, with an identity of 65.3%. On the other hand, both the HsdS (V22) and the HsdM (V23) proteins showed low levels of similarity to the HsdS and HsdM proteins encoded by the corresponding genes in staphylococcal G islands, with identities of less than 26 and 29%, respectively (3, 24). Figure 5 shows the phylogenetic relationships of the HsdR proteins (Fig. 5a), the HsdS proteins (Fig. 5b), and the HsdM proteins (Fig. 5c). Phylogenetic trees clearly showed that three ORFs, V21 (HsdR), V22 (HsdS), and V23 (HsdM), belonged to a group distinct from those found in the S. aureus genome or the G island.
FIG. 5.
Phylogenetic relations of the constituents of the hsd system. Type V SCCmec carries three genes, hsdR, hsdM, and hsdS, which encode a type I restriction-modification system. To look for the derivations of those genes, the phylogenetic relationships among the genes found in the type V SCCmec element and previously reported hsd genes were investigated by creating phylogenetic trees for the hsdR (a), hsdM (b), and hsdS (c) genes. Phylogenetic trees were generated as described in the legend to Fig. 2b. The nucleotide sequence of the truncated hsdR gene in type III SCCmec was obtained from the DDBJ/EMBL/GenBank databases (accession no. AB037671). All other nucleotide sequences of the hsd genes analyzed here were obtained from websites that provide the whole genome sequences of the following S. aureus strains: N315 and MW2 (http://www.bio.nite.go.jp/), Mu50 (http://w3.grt.kyushu-u.ac.jp/VRSA/), MSSA 476 and strain 252 (http://www.sanger.ac.uk/Projects/S_aureus/), COL (http://www.tigr.org/tdb/mdb/mdbinprogress.html), and NCTC8325 (http://www.genome.ou.edu/staph.html).
The G+C content of type V SCCmec was 30.5%. This value is lower than that for S. aureus (32.8 to 32.9%).
Further analysis of the G+C content of the third nucleotide in the codon (GC3) revealed that the GC3 values for the ORFs in the region unique to type V SCCmec were very low (range, 14.2 to 19.7%; average, 18.6%). When we applied the definition of the criteria that we used for S. aureus genome analysis, the ORFs were regarded as possible alien genes; i.e., their GC3 values differed by more than 1.5 standard deviations from the average GC3 values for all ORFs longer than 150 bp in the S. aureus genome (3, 24). They were transposases for IS431 (V02 and V08), mecA (V07), ΔmecR1 (V08), V12, V17, V18, V19, V20, and hsdS (V22).
CcrC-mediated precise excision and closed circular SCCmec formation.
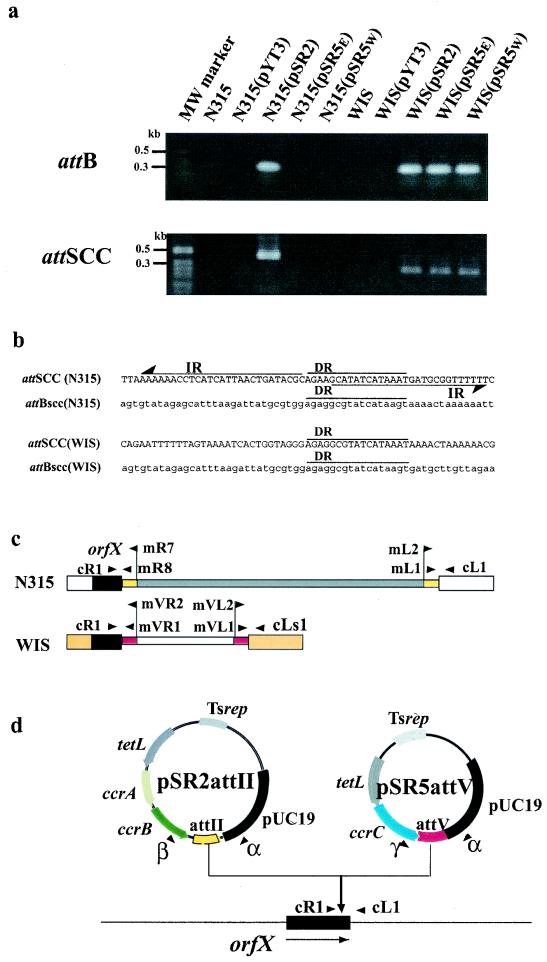
To investigate whether CcrC can catalyze the precise excision of type V SCCmec, we constructed two recombinant plasmids, pSR5W and pSR5E, which carried ccrCWIS and ccrC0342, respectively, and introduced them into WIS and N315, respectively, by electroporation.
As controls, plasmids pSR2 (a plasmid formerly called pSR that carries the ccrA2 and ccrB2 genes) and pYT3 were also each introduced into WIS and N315. After cultivation of the eight purified transformants, WIS(pSR5W), WIS(pSR5E), WIS(pSR2), WIS(pYT3), N315(pSR5W), N315(pSR5E), N315(pSR2), and N315(pYT3), as well as recipient strains, for 20 h at 30°C, the DNAs were extracted from the cultures. We tested by PCR whether attBscc was generated and whether the closed circular form of SCCmec was formed in each strain. The strategy and the primer sets used for the detection of attBscc and the closed circular form of SCCmec, which carries attSCC, are shown in Fig. 6c and Table 2, respectively.
FIG. 6.
Precise excision and site- and orientation-specific integration of SCCmec. (a) Detection of ccr-mediated SCCmec excision and appearance of attSCC. Template DNAs for PCR were extracted from a culture incubated in BHI broth with tetracycline (10 mg/liter) for 20 h. Four sets of primers were used to detect the precise excision and the closed circular form of SCCmec in strain N315 or WIS cells. The locations of the primers used are shown in panel b. For N315, primers cL1 and cR1 were used to detect attBscc (276 bp), and primers mL1 and mR8 were used to detect attSCC (456 bp). For WIS, primers cLs1 and cR1 were used to detect attBscc (314 bp), and primers mVL1 and mVR1 were used to detect attSCC (292bp). Lane MW, molecular size marker (1-kb ladder; Invitrogen, Carlsbad, Calif.), and only the relevant sizes are indicated. (b) Generation of attBscc and attSCC. The nucleotide sequences of attBscc in the DNA fragments amplified from N315ex and N315(pSR2) and from WIS(pSR2), WIS(pSR5W), and WIS(pSR5E) are indicated attBscc(N315) and attBscc(WIS), respectively. The nucleotide sequences of attSCC in DNA fragments amplified from N315(pSR2) and from WIS(pSR2), WIS(pSR5W), and WIS(pSR5E) are indicated attSCC(N315) and attSCC(WIS), respectively, above the nucleotide sequence of attBscc. (c) Locations of the primers used to detect attBscc generated from N315 (primers cR1 and cL1) and WIS (primers cR1 and cLs1) and attSCC generated from N315 (primers mR8 and mL1) and WIS (primers mVR1 and mVL1). The locations of the primers used to clone attSCC-II (primers mR7 and mL2) and attSCC-V (primers mVR2 and mVL2) are also illustrated. The drawings are not to scale. (d) Identification of site- and orientation-specific integration of recombinant plasmids (mini-SCCs) into the chromosome. The recombinant plasmids carrying ccr gene and attSCC of each type, pSR2attII and pSR5EattV, were introduced into N315ex by electroporation as models of SCCmec elements. DNAs were extracted from cultures of the transformants grown at 43°C on BHI agar plates containing tetracycline (10 mg/liter). The locations and directions of the four primers (primers α, β, cL1, and cR1) used to detect the integration of plasmid pSR2attII as well as four primers (primers α, γ, cL1, and cR1) used to detect the integration of plasmid pSR5attV are illustrated. We could amplify DNA fragments of the expected sizes by the site- and orientation-specific integration of the plasmids. The expected sizes of the DNA fragments are 1,255 bp (cL1-β) and 735 bp (cR1-α) for pSR2attII and 826 bp (cL1-γ) and 719 bp (cR1-α).
When we used the chromosomal DNAs of WIS(pSR5W) and WIS(pSR5E) as templates, a 0.31-kb DNA fragment containing attBscc and a 0.29-kb DNA fragment containing attSCC were successfully amplified, whereas no DNA fragment was amplified with DNAs extracted from WIS and WIS(pYT3) (Fig. 6a). By nucleotide sequencing of the amplified DNA fragments, we have verified that a novel nucleotide sequence, attSCC of type V SCCmec, is generated by the head-to-tail ligation of both termini and that attBscc is generated by the precise excision of type V SCCmec (Fig. 6b). These results show that CcrC serves as a site-specific recombinase responsible for the precise excision and formation of the closed circular form of type V SCCmec. Since no visible DNA fragment was amplified with DNAs extracted from N315(pSR5W) and N315(pSR5E), we could not be certain whether CcrC works on other types of SCCmec elements.
In contrast, DNA fragments of 0.28 and 0. 31 kb containing attBscc were amplified with DNAs extracted from N315(pSR2) and WIS(pSR2), respectively. Furthermore, DNA fragments of 0.46 and 0.29 kb containing attSCC were amplified with DNAs extracted from the two strains, respectively. Subsequent nucleotide sequencing showed that the fragments contained attBscc and attSCC. The data indicate that the sets of ccrA and ccrB genes cause precise excision and formation of the closed circular forms of both type II SCCmec and type V SCCmec.
CcrC catalyzes site- and orientation-specific integration of type V SCCmec.
To examine whether the putative closed circular form of type V SCCmec serves as a substrate for integration similar to that of type II SCCmec, we constructed experimental plasmid pSR5EattV, in which ccrC0342 and the presumptive attachment sequence of type V SCCmec (attV) are subcloned, and used the plasmid as a model of the closed circular form of type V SCCmec (mini-SCC). Recombinant plasmids pSR2attII (a plasmid formerly called pSRatt that carries ccr2 genes and the presumptive attachment sequence of type II SCCmec [attSCC-II]), pYTattII (a plasmid carrying attSCC-II), pYTattV (a plasmid that carries the presumptive attachment sequence of type V SCCmec [attSCC-V]), pSR2, pSR5E, and pYT3 were used as controls. The recombinant plasmids were introduced into N315ex by electroporation, followed by selection on plates with tetracycline at 30°C. A colony of each transformant was respread onto BHI agar plates containing tetracycline (10 mg/liter) and incubated for 18 h at 30 and 43°C (the latter of which is nonpermissive for plasmid replication). The numbers of colonies that grew on each plate were counted. Two strains, N315ex(pSR5EattV) and N315ex(pSR2attII), generated significant number of colonies at 43°C compared with the numbers that grew at 30°C (2.2 and 4.4%, respectively), whereas the other strains did not generate colonies when they were grown at 43°C. The result shows that integration of the plasmids into the chromosome occurs when the attSCC and the ccr genes are present on the plasmids and that CcrC may mediate the integration of pSR5EattV (a model of the closed circular form of type V SCCmec) at an efficiency nearly equal to that for the integration of pSR2attII (a model of the closed circular form of type II SCCmec) mediated by a set of CcrA and CcrB proteins.
The integration sites of the plasmids and their directions in the chromosome were examined by PCR experiments with primers as follows: cL1 and cR1, whose sequences flank that of the attB region on the chromosome of N315ex; α (RV in pUC119) and β (in the ccrB gene), whose sequences flank that of the attSCC-II region of plasmid pSR2attII; and γ (in the ccrC gene), whose sequence flanks the attSCC-V region of plasmid pSR5EattV (Fig. 6d and Table 2). The results show that two plasmids, pSR2attII and pSR5EattV, generated plasmid-chromosome integration junctions which were detected by two combinations of primers: primers cL1-β and cR1-α and primers cL1-γ and cR1-α, respectively. By determining the nucleotide sequences of the amplified DNA fragments, we found that the fragment contains sequences which are identical to those of the chromosome-SCCmec junction regions of N315 or artificially constructed chromosome (N315)-type V SCCmec junction regions.
These results indicate that the recombinant plasmid is integrated in the chromosome only when it carries the ccrA and ccrB genes and attSCC-II or the ccrC gene and attSCC-attV and that ccrC mediates the site- and orientation-specific integration of type V SCCmec in a way similar to that for a set of ccrA and ccrB genes.
DISCUSSION
Characteristics of a new site-specific recombinase, CcrC.
We demonstrated in this study that type V SCCmec carries a recombinase gene, ccrC, which can mediate type V SCCmec recombination events (integration and excision), whereas a set of ccrA and ccrB genes was previously found to be required for this function in other types of SCCmec elements. It was curious that both ccrC gene variants, ccrCWIS (1,623 nucleotides) and ccrC0342 (1,680 nucleotides), have been demonstrated to be active in the precise excision function. Approximately one-fourth of the deduced amino acid sequence from the NH2 terminus, which contains the serine residue that catalyzes DNA strand exchange (36), was well conserved among the CcrB variant and two CcrC variants (Fig. 2a). Therefore, a lack of some amino acids in the COOH-terminal domain may not much affect the activities of Ccr proteins. However, when we compared the frequency of precise excision by quantitative PCR, we observed that attBscc was present in slightly larger amounts in WIS(pSR5E) than in WIS(pSR5W) (X. X. Ma, unpublished data). That was why we used ccrC0342 to construct the recombinant plasmid used for the integration experiment.
Mini-SCC plasmid pSR5EattV was integrated in the N315ex chromosome at exactly the same nucleotide position at which pSR2attII integrated. The data indicate that CcrC recognizes the ISS in a way similar to that in which a set of CcrA and CcrB proteins in combination does.
Interestingly, when ccr genes were introduced into host cells carrying preexisting SCCmec to test ccr gene-mediated excision, the frequency of excision differed depending on the combination of the types of SCCmec elements and ccr genes. The types of SCCmec elements greatly influenced the efficiency of excision mediated by ccrC. The recombinase encoded by ccrC appeared to be inactive in excising type II SCCmec from the N315 chromosome, while it was active in the excision of type V SCCmec from the WIS chromosome. It is tempting to speculate that the difference resides in the nucleotide sequences at the extremities of SCCmec. Characteristic inverted repeat sequences were located at the extremities of the extant types of SCCmec carrying ccrA and ccrB genes, whereas they were not found at the extremities of type V SCCmec. They have also not been found in the extremities of SCCcap1, which carries a broken homologue of the ccrC gene (25). A similar preference between the types of ccr and SCCs was also observed in the integration experiment. The efficiency of integration decreased when the recombinant plasmids carried the attSCC sequences of different types of SCCmec elements. The efficiency of integration observed with N315ex(pSR5EattII) and N315ex(pSR2attV) was smaller than that observed with N315ex(pSR5EattV) and N315ex(pSR2attII). Those data suggest that both ccr proteins have specificity for the recognition of the nucleotides located at the extremities of the SCC elements.
Origin of hsd system and its role in maintenance of the element.
The restriction-modification system seems to play an important role in the stability of certain regions of the S. aureus chromosome (3, 24). A set of hsdS and hsdM genes is located in each of the two Gislands, vSaα and vSaβ, in the genomes of all seven S. aureus isolates that have been completely sequenced (isolates COL, MRSA 252, MSSA 476, MW2, Mu50, N315, and NCTC8325). The sequences of the HsdM proteins encoded by the two Gislands are nearly identical to each other, whereas those of the HsdS proteins in the target recognition domain regions differ significantly from each other. The hsdRaur gene (the hsdR gene commonly found on the genomes of the seven S. aureus strains) encodes the restriction function and is found on the S. aureus genome at the same locus, which is far from the two Gislands. The sequences of the HsdR proteins of the seven strains were nearly identical. It is speculated that the Gislands, which carry enterotoxins and exotoxin-like genes, are protected from deletional loss by the copresence of the modification function (3).
Unlike the other G islands, the type V SCCmec is unique in that it carries a complete set of the genes involved in the type I restriction-modification system, composed of hsdR, hsdS, and hsdM. A complete set of the genes involved in the restriction-modification system is also found on an SCC in MSSA strain476 (SCC476) (http://www.sanger.ac.uk/Projects/S_aureus/).The restriction-modification system encoded by those elements was judged to be distinct from that encoded by the hsdRaur, hsdM, and hsdS genes on the S. aureus chromosome, according to data from phylogenetic trees as well as codon usage patterns. This may signify that those mobile genetic elements were formed in a species different from S. aureus and that S. aureus acquired those elements by horizontal transfer. We tried to excise SCCmec elements from strain WIS. Although we could not obtain them from either WIS(pSR5E) or WIS(pSR5W), large amounts of DNA fragments containing attBscc and attSCC were amplified by PCR from cultures of both strains. It is reported that carriage of type II restriction-modification system genes contributes to plasmid stability (7, 8). Handa et al. (10) showed that Escherichia coli cells harboring a plasmid that carries the type II restriction-modification system die due to restriction cleavage of the chromosome after they lose the plasmid. This postsegregational killing phenomenon may explain why we could not excise SCCmec elements, despite the PCR amplification results indicating that an apparent excision event occurred in cultured cells of strain WIS. The restriction enzymes (encoded by either hsdRaur or hsdRscc) that remain in the cells after the loss of type V SCCmec might have cleaved the chromosome and killed the cells from which the element was excised. Thus, the type I restriction-modification system may serve as a stabilizer of the type V SCCmec element integrated in the S. aureus chromosome.
Type V SCCmec as an element found in C-MRSA.
MRSA clones are defined by the types of SCCmec and the genotypes of the MSSA chromosomes into which the SCCmec element is integrated. Multiple MRSA clones carrying type IV SCCmec were identified in C-MRSA strains from both the United States and Australia by multilocus sequence typing for chromosome genotyping and the PCR technique for SCCmec typing (32). On the basis of that observation, it was proposed that C-MRSA strains have been generated de novo from S. aureus populations more diverse than H-MRSA strains (32). The identification of a new type of SCCmec in C-MRSA strains in the present study further supports that proposal. Type V SCCmec was structurally similar to type IV SCCmec, in that it contains mecA as the only gene encoding antibiotic resistance. Its size (28 kb) was also comparable to that of type IV SCCmec and was much smaller than those of the type I to III SCCmec elements (34 to 67 kb) found in H-MRSA strains. Although the type V SCCmec element was slightly larger than the smallest subtype of type IV SCCmec (type IVb; 21 kb), the only difference was the presence of a restriction-modification system in type V SCCmec.
The most unique feature of type V SCCmec is the carriage of a type 5 ccr gene complex composed of the ccrC gene and its surrounding ORFs (Fig. 1). After sequence determination, we realized that the type 5 ccr gene complex is also found in SCC elements other than the type V SCCmec. SCCcap1, which encodes capsule formation but which has no mec gene, carried a gene complex similar to the type 5 ccr gene complex, although most of the ORFs in SCCcap1 were truncated. In addition, the Ψccr gene complex that we previously reported in the type III SCCmec element was closely related to the type 5 ccr gene complex: ORF CZ072 was a ccrC gene homologue, and the sequences of the surrounding ORFs were highly homologous to those of the corresponding ORFs in the type 5 ccr gene complex (Table 3). A copy of type 3 ccr gene complex and three ISS copies, in addition to a copy of type 5 ccr gene complex, are present in the type III SCCmec element (13, 15). This configuration now indicates that the long DNA region downstream of orfX, previously designated the J3 region of type III SCCmec, is in fact an SCC element independent of the rest of the region. We now consider the latter region to constitute a true type III SCCmec. If this is the case, the SCC—in which a truncated copy of hsdR, the mercury resistance operon, and Tn554, in addition to the type 5 ccr gene complex, are carried—must have been integrated prior to or in succession with the type III SCCmec element.
The class C2 mec gene complex, which, along with the type 5 ccr gene complex, defines the type V SCCmec element, is distributed among coagulase-negative staphylococcal species, especially S. haemolyticus. Among 38 S. haemolyticus strains tested, 30 strains carried the class C2 mec gene complex and the other 8 strains carried mec gene complexes of class A (5 strain), class B (2 strain), or class C1 (1 strain) (21). The deletion point of ΔmecR1 in the class C2 mec gene complex of the type V SCCmec element was identical to that in the class C2 mec gene complex found in S. haemolyticus strain JB16 and S. epidermidis strain JK8 (21). The data suggest the horizontal transfer of a certain molecular version of the class C2 mec gene complex among staphylococcal species. On the other hand, only 20 of 30 S. haemolyticus strains carrying the class C2 mec gene complex had the ccrC gene. Three other strains carried ccrA2 and ccrB2 genes, and the remaining seven did not carry any of the type 1 to 4 ccr genes (X. X. Ma, H. Yuzawa, and M. Kapi, unpublished observations). Thus, class C2 may not always be linked with the type 5 ccr gene complex, and additional types of SCCmec elements may be existent in coagulase-negative staphylococci. We have also reported that many subtypes defined by J1-region polymorphisms exist in the type IV SCCmec elements prevalent in C-MRSA strains. Type IV SCCmec elements with diverse J1 regions are distributed in more than 30% of community-acquired S. epidermidis isolates in Japan (K. Hisata, K. Hiramatsu et al., unpublished data). We think that it is quite likely that the mec gene complex and the ccr gene complexes (of diverse SCC elements) go through complex recombination and rearrangement processes in the genomes of coagulase-negative staphylococci; thus, novel types of SCCmec elements are incessantly generated, and only a fraction of them are transferred to S. aureus strains in the community. Thus, we may be witnessing only the tip of the iceberg of the SCCmec element diversity displayed by C-MRSA isolates. The type IV and type V SCCmec elements that have come onto the scene, however, may be the most refined molecular products and may have high competitive capabilities in terms of their transferability among the cells of staphylococcal species and strains.
Table 3a.
| Data indicating homology to ORF in SCCmec of:
| |||||||
|---|---|---|---|---|---|---|---|
| NCTC10442 (type I)
|
N315 (type II)
|
85/2082 (type III)
|
CA05 (type IV)
|
||||
| % Identityc | Corresponding ORF (size [bp]) | % Identity | Corresponding ORF(s) (size [bp]) | % Identity | Corresponding ORF(s) (size [bp]) | % Identity | Corresponding ORF (size [bp]) |
| 100 | orfX (480) | 100 | orfX (480) | 100 | orfX (480) | 100 | orfX (480) |
| 99.10 | E040 (675) | 99.10 | N062, N070 (675) | 99.1, 98.7, 98.2, 98.2 | Z035, Z041, Z046, Z058 (675) | 99.10 | Q012 (675) |
| 100 | CE026 (744) | 99.00 | CN039 (633) | 100 | CZ030 (744) | 100 | CQ007 (744) |
| 99.30 | CE025 (429) | 100 | CN038 (429) | 100 | CZ029 (429) | 100 | CQ005 (429) |
| 99.70 | mecA10442 (2007) | 99.90 | mecAN315 (2007) | 99.60 | mecA2082 (2007) | 99.90 | mecACA05 (2007) |
| 100 | ΔmecR110442 (987) | 100 | mecR1N315 (1758) | 88.60 | ΔmecR12082 (114) | 100 | ΔmecR1CA05 (987) |
| 97.80 | E040 (675) | 97.80 | N062, N070 (675) | 97.8, 97.3, 97.8, 97.8 | Z035, Z041, Z046, Z058 (675) | 97.80 | Q012 (675) |
| 76.70 | CZ075 (1101) | ||||||
| 84.30 | CZ074 (372) | ||||||
| 82.10 | CZ073 (1644) | ||||||
| 93.60 | CZ072 (1554) | ||||||
| 54.50 | E031 (351) | 53.60 | N041 (351) | 50.9, 48.6 | Z011 (351), CZ070 (342) | 55.90 | Q007 (351) |
| 91.70 | E032 (327) | 89.30 | N042 (312) | 48.1, 48.1 | Z013 (395), CZ069 (312) | 87.40 | Q008 (312) |
| 91.60 | E033 (510) | 92.80 | N043 (318) | 61.7, 67.1 | Z014 (522), CZ068 (507) | 93.40 | Q009 (510) |
| 97.20 | CZ053 (323) | ||||||
| 98.00 | Z059* (2244) | ||||||
REFERENCES
- 1.Archer, G. L., D. M. Niemeyer, J. A. Thanassi, and M. J. Pucci. 1994. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob. Agents Chemother. 38:447-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayliffe, G. A. 1997. The progressive international spread of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 24:74-79. [DOI] [PubMed] [Google Scholar]
- 3.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997-1999. JAMA 282:1123-1125. [PubMed] [Google Scholar]
- 5.Chambers, H. F. 2001. The changing epidemiology of Staphylococcus aureus. Emerg. Infect. Dis. 7:178-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enright, M. C., D. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerdes, K., P. B. Rasmussen, and S. Molin. 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 83:3116-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillen, J. R., D. K. Willis, and A. J. Clark. 1981. Genetic analysis of the RecE pathway of genetic recombination in Escherichia coli K-12. J. Bacteriol. 145:521-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groom, A. V., D. H. Wolsey, T. S. Naimi, K. Smith, S. Johnson, D. Boxrud, K. A. Moore, and J. E. Cheek. 2001. Community-acquired methicillin-resistant Staphylococcus aureus in a rural American Indian community. JAMA 286:1201-1205. [DOI] [PubMed] [Google Scholar]
- 9a.Hanaki, H., K. Kuwahara-Arai, S. Boyle-Vavra, R. S. Daum, H. Labischinski, and K. Hiramatsu. 1998. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 42:199-209. [DOI] [PubMed]
- 10.Handa, N., A. Ichige, K. Kusano, and I. Kobayashi. 2000. Cellular responses to postsegregational killing by restriction-modification genes. J. Bacteriol. 182:2218-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartman, B. J., and A. Tomasz. 1984. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158:513-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiramatsu, K., Asada, E. Suzuki, K. Okonogi, and T. Yokota. 1992. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA). FEBS Lett. 298:133-136. [DOI] [PubMed] [Google Scholar]
- 13.Hiramatsu, K., Y. Katayama, H. Yuzawa, and T. Ito. 2002. Molecular genetics of methicillin-resistant Staphylococcus aureus. Int. J. Med. Microbiol. 292:67-74. [DOI] [PubMed] [Google Scholar]
- 14.Hurlimann-Dalel, R. L., C. Ryffel, F. H. Kayser, and B. Berger-Bachi. 1992. Survey of the methicillin resistance-associated genes mecA, mecR1-mecI, and femA-femB in clinical isolates of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 36:2617-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, T., K. Okuma, X. X. Ma, H. Yuzawa, and K. Hiramatsu. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug. Resist. Update 6:41-52. [DOI] [PubMed] [Google Scholar]
- 16.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jevons, M. P. 1961. “Celbenin”-resistant staphylococci. Br. Med. J. 124:124-125. [Google Scholar]
- 19.Karlin, S., J. Mrazek, and A. M. Cambell. 1998. Codon usages in different gene classes of the Escherichia coli genome. Mol. Microbiol. 29:1341-1355. [DOI] [PubMed] [Google Scholar]
- 20.Katayama, Y., F. Takeuchi, T. Ito, X. X. Ma, Y. Ui-Mizutani, I. Kobayashi, and K. Hiramatsu. 2003. Identification in methicillin-susceptible Staphylococcus hominis of an active primordial mobile genetic element for the staphylococcal cassette chromosome mec of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 185:2711-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katayama, Y., T. Ito, and K. Hiramatsu. 2001. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 45:1955-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcal cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhl, S. A., P. A. Pattee, and J. N. Baldwin. 1978. Chromosomal map location of the methicillin resistance determinant in Staphylococcus aureus. J. Bacteriol. 135:460-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 25.Luong, T. T., S. Ouyang, K. Bush, and C. Y. Lee. 2002. Type 1 capsule genes of Staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J. Bacteriol. 184:3623-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuhashi, M., M. D. Song, F. Ishimoto, M. Wachi, M. Doi, M. Inoue, K. Ubukata, N. Yamashita, and M. Konno. 1986. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to β-lactam antibiotics in Staphylococcus aureus. J. Bacteriol. 167:975-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naimi, T. S., K. H. LeDell, D. J. Boxrud, A. V. Groom, C. D. Steward, S. K. Johnson, J. M. Besser, C. O'Boyle, R. N. Danila, J. E. Cheek, M. T. Osterholm, K. A. Moore, and K. E. Smith. 2001. Epidemiology and clonality of community-acquired methicillin-resistant Staphylococcus aureus in Minnesota, 1996-1998. Clin. Infect. Dis. 33:990-996. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nimmo, G. R., J. Schooneveldt, G. O'Kane, B. McCall, and A. Vickery. 2000. Community acquisition of gentamicin-sensitive methicillin-resistant Staphylococcus aureus in southeast Queensland, Australia. J. Clin. Microbiol. 38:3926-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Brien, F. G., J. W. Pearman, M. Gracey, T. V. Riley, and W. B. Grubb. 1999. Community strain of methicillin-resistant Staphylococcus aureus involved in a hospital outbreak. J. Clin. Microbiol. 37:2858-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliveira, D. C., A. Tomasz, and H. Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 34.Reynolds, P. E., and D. F. J. Brown. 1985. Penicillin-binding proteins of beta-lactam resistant strains of Staphylococcus aureus. FEBS Lett. 192:28-32. [DOI] [PubMed] [Google Scholar]
- 35.Ryffel, C., W. Tesch, I. Birch-Machin, P. E. Reynolds, L. Barberis-Maino, F. H. Kayser, and B. Berger-Bachi. 1990. Sequence comparison of mecA genes isolated from methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Gene 94:137-138. [DOI] [PubMed] [Google Scholar]
- 36.Sherratt, D. 1989. Tn3 and related transposable elements: site-specific recombination and transposition, p. 163-184. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 37.Sjostrom, J. E., S. Lofdahl, and L. Phillipson. 1975. Transformation reveals a chromosomal locus of the gene(s) for methicillin resistance in Staphylococcus aureus. J. Bacteriol. 123:905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song, M. D., M. Wachi, M. Doi, F. Ishino, and M. Matsuhashi. 1987. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 221:167-171. [DOI] [PubMed] [Google Scholar]
- 39.Sousa, M. A., and H. Lencastre. 2003. Evolution of sporadic isolates of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals and their similarities to isolates of community-acquired MRSA. J. Clin. Microbiol. 41:3806-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki, E., K. Hiramatsu, and T. Yokota. 1992. Survey of methicillin-resistant clinical strains of coagulase-negative staphylococci for mecA gene distribution. Antimicrob. Agents Chemother. 36:429-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki, E., K. Kuwahara-Arai, J. F. Richardson, and K. Hiramatsu. 1993. Distribution of mec regulator genes in methicillin-resistant Staphylococcus clinical strains. Antimicrob. Agents Chemother. 37:1219-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Utsui, Y., and T. Yokota. 1985. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 28:397-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]