Abstract
The aberrant expression of human microRNA-181a-1 (hsa-miR-181a) has been implicated in the pathogenesis of various cancers, serving as an oncogene or a tumor suppressor. However, the role of hsa-miR-181a in the pathogenesis of endometrial carcinoma (EC) and its clinical significance are unclear. This study aimed to search for the molecular targets of hsa-miR-181a using bioinformatic tools and then determine the expression levels of hsa-miR-181a in normal, hyperplasia, and EC samples from humans. To predict the targets of hsa-miR-181a, ten different algorithms were used, including miRanda-mirSVR, DIANA microT v5.0, miRDB, RNA22 v2, TargetMiner, TargetScan 6.2, PicTar, MicroCosm Targets v5, and miRWALK. Two algorithms, TarBase 6.0 and miRTarBase, were used to identify the validated targets of hsa-miR-181a-5p (a mature product of hsa-miR-181a), and the web-based Database for Annotation, Visualization and Integrated Discovery (DAVID) 6.7 was used to provide biological functional interpretation of the validated targets of hsa-miR-181a-5p. A total of 78 formalin-fixed, paraffin-embedded tissue specimens from 65 patients and 13 healthy subjects were collected and examined, including normal endometrium (n=13), endometrial hyperplasia (n=18), and EC (37 type I and 10 type II EC cases). Our bioinformatic studies have showed that hsa-miR-181a might regulate a large number of target genes that are important in the regulation of critical cell processes, such as cell fate, cell survival, metabolism, and cell death. To date, 313 targets of hsa-miR-181a have been validated, and 22 of these targets are cancer genes. The precision of predictions by all the algorithms for hsa-miR-181a-1’s targets was low. Many of these genes are involved in tumorigenesis of various cancers, including EC, based on the DAVID and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. In comparison with normal endometrial tissue, the expression level of hsa-miR-181a was significantly increased in type I and type II EC (P<0.05), and type II EC exhibited a significant higher expression level of hsa-miR-181a than that in type I EC (P<0.05). In addition, there was a significant increase in the expression level of hsa-miR-181a in type II EC compared with endometrial hyperplasia (P<0.05). Taken together, these results suggest that hsa-miR-181a may serve as an oncogene in endometrial tumorigenesis and that hsa-miR-181a might be used as a new biomarker in the prediction of prognosis of EC in clinical practice. More functional and mechanistic studies are needed to validate the role of hsa-miR-181a in the development, progression, and metastasis of EC.
Keywords: RT-PCR, endometrial cancer, development, progression
Introduction
microRNAs (miRNAs) are a large group of noncoding small RNAs with 20–25 nucleotides that have a capability of regulating gene expression at posttranscriptional levels by antisense complementarily to specific target messenger (m)RNAs.1,2 Based on miRBase version 21 released in June 2014 (http://www.mirbase.org/), there are 1,881 miRNA precursors and 2,588 mature miRNAs in humans. miRNAs are transcribed as ~70 nucleotide stem-loop precursors and subsequently processed by the cytoplasmic RNase III-type enzyme Dicer to generate ~22 nucleotide mature products that can target and modulate protein expression by inhibiting translation and/or inducing degradation of target mRNAs. The mature miRNA is incorporated into a RNA-induced silencing complex (RISC), which recognizes target mRNAs through imperfect base pairing with the miRNA. miRNAs act as adaptors that employ a silencing complex to target mRNAs by selective base-pairing, primarily in the 3′-untranslated region (3′-UTR). Target interaction does not require perfect complementarity between microRNA and mRNA sequences, although near-perfect base-pairing in a small region in the 5′-end (positions 2–8) of the microRNA (sometimes termed “seed”) appears to be one of the key determinants of target recognition. miRNAs regulate almost every signaling pathway and play crucial roles in diverse biological processes, such as development, differentiation, apoptosis, and proliferation.1–3 It has been shown that aberrant expression of miRNAs is involved in the development and progression of many types of cancer through regulation of functional proteins and the network of signaling pathways related to cell proliferation, cell migration and invasion, programmed cell death, and cell survival.3–7 It has been proposed that miRNAs can function as tumor suppressors or oncogenes, targeting other oncogenes and/or tumor-suppressors to modulate cancer development, progression, and metastasis.1,5–8
An extremely large number of potential target sites exist for any given miRNA, and the process of validating a potential miRNA target in the laboratory is time consuming and costly. A computational approach to prediction of miRNA targets facilitates the process of narrowing down potential target sites for experimental validation, which is a critical initial step in identifying miRNA–target interactions. Several useful algorithms/tools provide microRNA target predictions based on sequence complementarity to target sites, with emphasis on perfect or near-perfect base-pairing in the seed region and sequence conservation.9,10 These tools for miRNA target prediction, encompassing a range of different computational approaches, from the modeling of physical interactions to the incorporation of machine learning, are mostly based on seed match, conservation, free energy, and site accessibility.10
Endometrial cancer (EC) is the sixth most common cancer in women worldwide, with at least 320,000 new cases being diagnosed and 74,000 women who die from this disease every year.11 In the United States, there was an estimation of 52,630 new cases and 8,590 deaths due to EC in 2014.12 In the United Kingdom, there were 8,474 women diagnosed with EC and 1,914 deaths from EC in 2011.11 In the People’s Republic of China, the incidence of EC is much lower than Western countries. It contributes about 1% of the world’s new EC cases.11
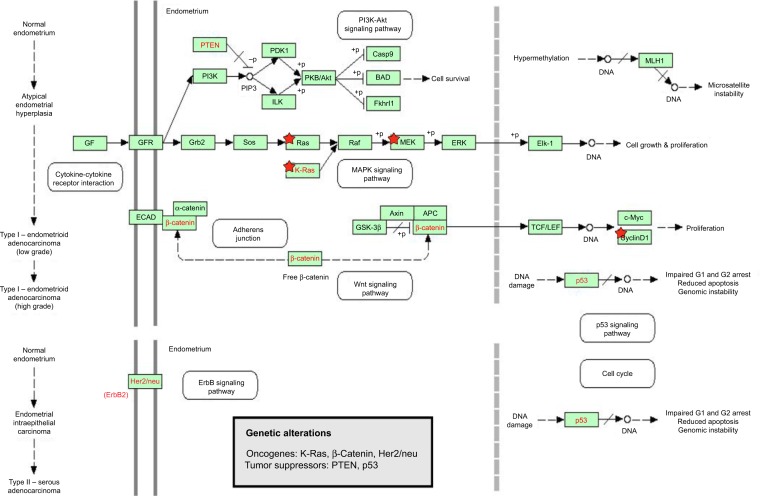
There are two types of EC, type I and type II, with different molecular expression profiles and histopathological and clinical behaviors.13,14 Type I EC, accounting for 75% of EC, is estrogen-dependent with endometrioid morphology, and the 5-year survival rate is 80%−85%.15 Type II EC exhibits poorly differentiated endometrioid and serous histological alterations with myometrial invasion and extra-uterine spread, and the 5-year survival rate is about 35%. Type I EC is related to hyperestrogenism, associated with endometrial hyperplasia, frequent expression of estrogen and progesterone receptors (ER and PR), and younger age, whereas type II EC is unrelated to estrogen and is associated with atrophic endometrium, frequent lack of ER and PR, and older age. The morphologic differences in type I and type II EC are mirrored in their molecular genetic profile, with type I showing defects in DNA-mismatch repair and mutations in phosphatase and tensin homolog (PTEN), phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α (PIK3CA), V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), and β-catenin/CTNNB1, and type II showing chromosomal instability and aneuploidy, p53 mutations, and human epidermal growth factor receptor 2 (HER2)/neu (erbB-2) amplification.16,17 Type I EC is associated with chronic exposure to unopposed estrogen and is often preceded by complex atypical hyperplasia. Current therapies for EC include surgery, chemotherapy, radiation therapy, immunotherapy, and biological therapy.18 Most cases of EC are diagnosed at an early stage, which has a 5-year survival rate of over 91%.11 However, the prognosis of EC does not meet the long-term survival expectation due to tumor metastasis, lack of effective treatment, and rarity of valid biomarkers to precisely predict therapeutic outcome.19 There are lines of evidence that a number of genetic and epigenetic factors have been implicated in the pathogenesis of EC, including abnormality in oncogenes, tumor suppressors, and miRNAs and related signaling pathways.13 Alterations in the expression profiles of oncogenes and tumor suppressor genes are the major contributing factors to the initiation, development, progression, and metastasis of EC. However, the association between such alterations and the clinical phenotypes of EC has not been conclusively established yet, and the underlying mechanism for EC etiology remains elusive.
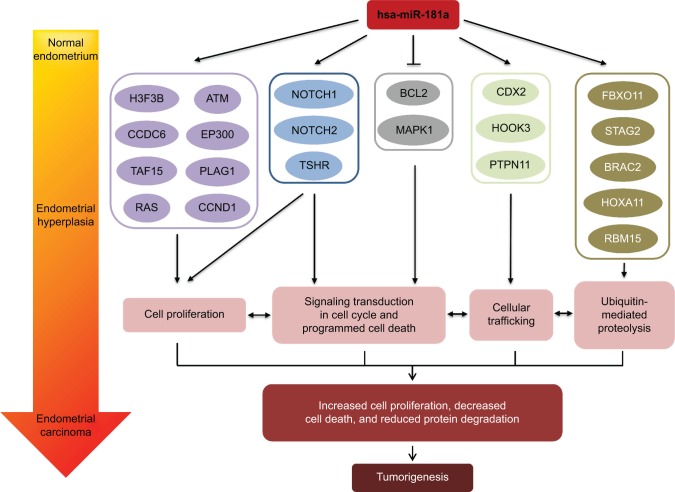
There is an increased interest in the discovery and identification of miRNAs as novel epigenetic biomarkers for early detection and prediction of therapeutic outcomes in cancer therapy.20–24 So far, there are a number of human miRNAs that have been identified to have a potential role in the development and progression of EC, including hsa-miR-181a, hsa-miR-185, hsa-miR-106a, hsa-miR-210, hsa-miR-423, hsa-miR-107, hsa-miR-let7c, and hsa-miR-221.3,4 hsa-miR-181a is one of the many miRNAs conserved among the vertebrates that is preferentially expressed in B lymphocytes of bone marrow, and its ectopic expression in hematopoietic stem/progenitor cells modulates blood cell development.25 hsa-miR-181a has been reported to be a key modulatory factor in the regulation of cell proliferation and differentiation at transcriptional and posttranscriptional levels in gastric cancer, acute myeloid leukemia, and other types of cancer,26,27 and hsa-miR-181a overexpression resulted in promotion of cell proliferation and migration but inhibition of apoptosis in colorectal cancer.28 RalA, one of the Ral family small G proteins, is directly regulated by hsa-miR-181a and plays an important role in the development chronic myelogenous leukemia.29 hsa-miR-181a and hsa-miR-181b act as tumor suppressors by inducing cell growth inhibition, apoptosis, and repression of invasion in glioma cells.30 So far, there have been only scattered reports about the role of a form of miR-181a in EC.4 Panda et al31 showed that the expression level of miR-181a-1 was higher in EC than in normal endometrial tissues. However, there is limited information on the association between the expression profile of miR-181a and the development and progression of EC. In this regard, we conducted a comprehensive bioinformatic study to predict the targets of miR-181a-1 and then validate these targets based on published experimental evidence. Finally, we examined the expression levels of miR-181a-1 in different types of EC and the association with the clinical progression of EC.
Material and methods
Gene nomenclature
The miR-181 family includes four members, namely miR-181a, miR-181b, miR-181c, and miR-181d. They are expressed in at least 70 species and various human cancers and are highly conserved in the seed-region sequence and RNA secondary structure. hsa-miR-181a-1 was retrieved from miRBase 21 (http://www.mirbase.org/). The hsa-miR-181a-1 gene (MIR181A1) has been mapped to 1q32.1. miRBase was established in 2002 as the public and central online repository for all published miRNA sequences and associated annotations, and the latest miRBase release of v21 (released in June 2014) contains 28,645 miRNA loci from 223 species (of which 1,881 precursors and 2,588 mature miRNAs are for humans), processed to produce 35,828 mature miRNAs. As compared with miRBase v20, a total of 4,196 new hairpin sequences and 5,441 novel mature products, mainly for bat, horse, goat, cobra, and salmon, have been added, with 72 dubious and misannotated entries removed from version 21. miRBase provides a user-friendly web interface for miRNA data, allowing the user to search using key words or sequences, trace links to the primary literature referencing the miRNA discoveries, analyze genomic coordinates and context, and mine relationships between miRNA sequences.32–34 Clusters of miRNA sequences in the genome are highlighted and can be defined and retrieved with any inter-miRNA distance. The mature forms of hsa-miR-181a include hsa-miR-181a-3p and hsa-miR-181a-5p (Table 1).
Table 1.
Basic information on hsa-miR-181a-1 gene (MIR181A1) retrieved from miRBase 21
| Gene | Accession number | Previous IDs | Sequence | Number of nucleotides | Predicted targets | Validated targets |
|---|---|---|---|---|---|---|
| hsa-miR-181a-1 | MI0000289 | hsa-miR-213 | UGAGUUUUGAGGUUGCUUCAGUGAACAUUCAACGCUGUCGGUGAGUUUGGAAUUAAAAUCAAAACCAUCGACCGUUGAUUGUACCCUAUGGCUAACCAUCAUC UACUCCA | 109 | – | – |
| hsa-miR-181a-5p | MIMAT0000256 | hsa-miR-181a | 24-AACAUUCAACGCUGUCGGUGAGU-46 | 23 | DIANA microT v3.0, miRanda-mirSVR (microRNA.org), miRDB, RNA22 v2, TargetMiner, TargetScan6.2, PicTar, MicroCosm, and miRWALK | miRTarBase and TarBase |
| hsa-miR-181a-3p | MIMAT0000270 | hsa-miR-213 and hsa-miR-181a* | 64-ACCAUCGACCGUUGAUUGUACC-85 | 22 | DIANA microT v3.0, miRanda- mirSVR (microRNA.org), miRDB, RNA22 v2, TargetMiner, MicroCosm, and miRWALK | – |
Prediction of the targets of hsa-miR-181a using various computational algorithms
Before starting the bench and clinical work, we conducted a bioinformatic study to predict the target genes regulated by hsa-miR-181a-3p and hsa-miR-181a-5p, using ten different algorithms, including miRanda-mirSVR (http://www.microrna.org/), DIANA microT v5.0 (http://diana.cslab.ece.ntua.gr/microT/), miRDB (http://mirdb.org/miRDB/), RNA22 v2 (https://cm.jefferson.edu/rna22v2.0/), Target-Miner (http://www.isical.ac.in/~bioinfo_miu/targetminer20.htm), TargetScan 6.2 (http://www.targetscan.org/), PicTar (http://pictar.mdc-berlin.de/), MicroCosm Targets v5 (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/), and miRWALK (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/index.html).
miRanda-mirSVR (http://www.microrna.org/) is an online tool that can be used to predict targets and score them.35 Unlike most miRNA target predictors, miRanda considers matching along the entire miRNA sequence, and it takes the seed region into account by weighting matches in the seed region more heavily; free energy is calculated by predicting the folding of the miRNA:mRNA hybrid, using the Vienna package. DIANA microT v5.0 currently hosts miRNA target predictions for Homo sapiens, Mus musculus, Drosophila melanogaster, and Caenorhabditis elegans, based on data from Ensembl release 69 and miRBase version 18.36,37 miRDB, a web-based database and tool, can predict miRNAs and their targets.38 All the targets are predicted by the bioinformatic tool MirTarget2, which has been developed by analyzing thousands of genes impacted by miRNAs, using a support vector machine (SVM) learning machine. RNA22 v2 can be used to predict the targets of miRNAs in human, mouse, roundworm, and fruit fly.39 It allows users to visualize the predictions within a complementary (c)DNA map and also find transcripts where multiple miRNAs of interest target.
TargetMiner is a robust tool for microRNA target prediction with systematic identification of negative examples.40 In this algorithm, ~300 tissue-specific negative examples have been identified, using a novel approach that involves expression profiling of miRNAs and mRNAs, miRNA–mRNA structural interactions, and seed-site conservation. TargetScan 6.2 can predict biological targets of miRNAs by searching for the presence of conserved 8-mer and 7-mer sites matching the seed region of each miRNA,41 with non-conserved sites being predicted as well. TargetScan is the first computational method used for human miRNA target prediction, that uses mouse, rat, and fish genomes for conservation analysis. In mammals, the prediction is ranked based on the predicted efficacy of targeting as calculated using the context+ scores of the sites.42 The context score for a specific site is the sum of the contribution of four features: site-type contribution, 3′-pairing contribution, local nucleobases adenine and uracil contribution, and position contribution. In the current work, the sum of the context scores for each miRNA was calculated, and the most favorable (lowest) was shown. PicTar is an algorithm for the identification of miRNA targets.43 In addition, MicroCosm Targets v5 was used to predict the targets that might be regulated by hsa-miR-181a. In this tool, there are 851 miRNAs (711 native) with 34,788 targets for humans.
miRWALK is a comprehensive database that provides information on miRNAs from the human, mouse, and rat, on their predicted as well as validated binding sites on their target genes (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/index.html).44 miRWALK predicts miRNA binding sites on the complete sequence of all known genes, including all transcripts and mitochondrial genes of the human, mouse, and rat, based on a comparison of identified miRNA binding sites with ten established miRNA-target prediction programs: miRWALK, DIANA-microT v3.0, miRanda, miRDB, PicTar 4 and PicTar 5, PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html), RNA22, RNAhybrid (http://bibiserv.techfak.uni-bielefeld. de/rnahybrid/submission.html), and TargetScan. In addition, it provides predicted miRNA binding sites on genes associated with 449 human biological pathways and 2,356 Online Mendelian Inheritance in Man (OMIM) disorders.44 This algorithm also presents information on experimentally validated miRNA interaction information associated with genes, pathways, diseases, organs, OMIM disorders, cell lines, and literature on miRNAs.
Validated targets of hsa-miR-181a based on TarBase 6.0 and miRTarBase 4.0
Two algorithms were used to identify the validated targets of miR-181a-5p: TarBase 6.0 and miRTarBase 4.0. TarBase 6.0 is a database that houses a manually curated collection of experimentally supported miRNA targets in 21 species, including human, rat, mouse, virus, Caenorhabditis elegans, Danio rerio (zebrafish), Drosophila, and plant (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index).45 TarBase 6.0 is the largest available manually curated target database, indexing more than 65,000 miRNA-gene interactions, 16.5- to 175-fold more than any other available implementation. The database includes targets derived from specific as well as high-throughput experiments, such as microarrays and proteomics. Specific attention was paid in the inclusion of targets derived from sequencing experiments, such as high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) and photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP). TarBase 6.0 hosts data derived from three CLIP-Seq and 12 Degradome-Seq studies, significantly more than any other available database. DIANA TarBase 6.0 offers a significant amount of crucial information to the user, including detailed description of the involved genes and miRNAs, a list of publications supporting each interaction, and the experimental methods used for validations, along with their outcomes. The database also provides links to related Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, as well as to other external databases, such as Ensembl, Uniprot, and RefSeq. It is also equipped with powerful searching and filtering capabilities.
miRTarBase 4.0 has collected 51,460 miRNA–target interactions from 18 species having experimental evidence (http://mirtarbase.mbc.nctu.edu.tw/).46 Generally, the collected miRNA–target interactions are validated experimentally by reporter assay, Western blotting, microarray, and next-generation sequencing experiments. There are 4,572 miRNA–target interactions validated by reporter assays or Western blotting experiments.
Cancer genes are defined as “mutated genes that are causally implicated in oncogenesis” based on the criteria by Futreal et al47 (Table S1 gives the full list of cancer genes). The proteins that are encoded by cancer genes normally regulate cell proliferation, cell differentiation, and cell death. Mutations underlying oncogenesis also occur in genes that mediate DNA-repair processes. Currently, more than 1% of all human genes have been implicated, via mutation, in cancer. Of these, approximately 90% have been implicated in somatic mutations in cancer, 20% bear germline mutations that predispose to cancer, and 10% show both somatic and germline mutations.48
Pathway analysis by the Database for Annotation, Visualization and Integrated Discovery (DAVID)
The web-based DAVID 6.7 (http://david.abcc.ncifcrf.gov/)49,50 was used to provide biological functional interpretation of the validated targets of hsa-miR-181a-5p, based on TarBase 6.0 and miRTarBase 4.0. DAVID 6.7 systematically maps a large number of interesting genes in a list to associated Gene Ontology (GO) terms, and then statistically highlights the most overrepresented (enriched) GO terms out of a list of hundreds or thousands of terms.47,48 The DAVID Knowledgebase has comprehensively integrated more than 20 types of major gene/protein identifiers and more than 40 well-known functional annotation categories from dozens of public databases, to address the enriched and redundant relationships among many genes to many terms. The protein IDs of the validated targets of hsa-miR-181a-5p from the National Center for Biotechnology Information (NCBI), Protein Information Resource (PIR), and UniProtKB were converted into gene lists, using the Gene ID Conversion Tool in DAVID. By doing so, interesting genes derived from one identifier system can be quickly translated to other gene identifier types preferred by a given annotation resource. The DAVID database adds biological function annotation including GO terms, protein–protein interactions, protein functional domains, disease associations, gene clustering, biopathways, sequence general features, homologies, gene functional summaries, and gene tissue expressions in a network context.47,48 The genes of interest were visualized using BioCarta and KEGG pathway maps. The highest classification stringency was selected for functional annotation clustering. Enrichment scores and Fisher’s exact test P-values (and corresponding false discovery rate [FDR]) were then calculated to identify which functionally related gene groups are significantly enriched in the target list.
Reagents and antibodies
The RNA inhibitor and Moloney murine leukemia virus reverse transcriptase were obtained from Promega Inc. (Madison, WI, USA). The antigen retrieval solution (ethyl-enediaminetetraacetic acid [EDTA] method) and Dolichos biflorus agglutinin kit were purchased from Maixin Biological Co. Ltd., Fuzhou, Fujian, People’s Republic of China. Human monoclonal primary antibodies against (ER, PR), and horseradish peroxidase-conjugated secondary antibodies were bought from Linked-Biotech Pathology Co. Ltd. (Guangzhou, Guangdong, People’s Republic of China). MiR-easy FFPE Kit and Syber® Green PCR mix were purchased from QIAGEN Inc. (Venlo, the Netherlands).
Sample collection
Fresh tissue samples were obtained from 65 patients who received uterusectomy and 13 healthy subjects at Xiaolan People’s Hospital or Zhongshan People’s Hospital, Zhongshan, Guangdong, or at Nanfang Hospital of Southern Medical University, Guangzhou, Guangdong, People’s Republic of China, with a mean age of 48 years (range 24–69). We collected the endometrial samples from 47 patients with EC, 18 patients with endometrial hyperplasia, and 13 healthy subjects. There was no preoperative radiotherapy, chemotherapy, or endocrine therapy performed in any of the recruited participants. All samples were fixed with formalin and embedded with paraffin. International Federation of Gynecology and Obstetrics (FIGO) staging was performed according to the FIGO classification.15,51 Histological classification of tissue samples was performed according to the World Health Organization (WHO) criteria (www.iarc.fr/en/.../BB2.pdf), and samples were classified as G1 (well differentiated), G2 (moderately differentiated), or G3 (poorly differentiated). The study design was approved by the Ethics Committees of Xiaolan People’s Hospital, Zhongshan People’s Hospital, and Nanfang Hospital. Written informed consent was obtained from each participant.
Immunohistochemistry
Dewaxed and dehydrated sections were first washed with phosphate-buffered saline (PBS) and then incubated with 3% peroxyl in methanol for 15 minutes to terminate the activity of endogenous peroxidases. The sections were washed with PBS, and antigen retrieval was performed. The sections were immersed into boiled citrate-buffered solution for 10 minutes and blocked with 5% bovine serum albumin in PBS for 20 minutes at room temperature. Thereafter, the sections were probed with primary antibody against ER or PR overnight in a humidified chamber at 4°C. On the following day, sections were incubated with biotinylated anti-Rabbit antibody (Boster Biotechnology Ltd., Wuhan, Hubei, People’s Republic of China) for 30 minutes at room temperature and then coupled with diaminobenzidine to visualize the expression of the targeted proteins. After all sections were counterstained with hematoxylin, they were dehydrated in ascending ethanol and then mounted using neutral resins. Samples with both ER-positive and PR-positive were classified as type I EC, while samples with both ER-negative and PR-negative were classified as type II EC.4,25
Primer design
The sequences of target gene were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/genbank/) and miRBase (http://www.mirbase.org/). The primers were designed using Primer Designer 2.0, and the sequences are shown in Table 2. All primers were synthesized by Beijing Liu He Synthetic Genomics Ltd., Beijing, People’s Republic of China. U6 was used as the internal control.
Table 2.
Sequences of the primers for the determination of hsa-miR-181a and U6
| Gene | Forward primer | Reverse primer |
|---|---|---|
| hsa-miR-181a | GTCGTATCCAGTGCGTGTCGTGGAGTCG | GCAATTGCACTGGATACGACACTCAC |
| U6 | GTCGTATCCAGTGCGTGTCGTGGAGTCGG | CAATTGCACTGGATACGACAAAATATG |
Total RNA extraction
A series of sections of thickness 10 μm was obtained, and the paraffin was dissolved by xylene treatment. Ten slides were prepared for each sample. Sections with cell content more than 50% of the area were selected for total RNA extraction. Briefly, sections were washed twice with ethanol in a 1.5 mL centrifuge tube to remove residual xylene. Total RNA was extracted, and the purity and integrity of the total RNA were examined using a miRNeasy FFPE kit according to the manufacturer’s instruction. The purity of the total RNA was tested using an ultraviolet (UV) spectrophotometer. The ratio of A260/A280 between 1.8 and 2.1 was considered as high purity. Electrophoresis was performed to detect the RNA integrity in 1.0% agarose denaturing gel.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
RNA was reversely transcribed into cDNA, using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc., Waltham, MA, USA) according to the manufacturer’s instruction. The resultant cDNA was subject to qRT-PCR analysis using a Bio-Rad Real-time PCR System (Bio-Rad Laboratories Inc., Hercules, CA, USA) and Syber green PCR mix. The conditions for RT-PCR were 95°C for 6 minutes and then 50 cycles of 95°C for 10 seconds, 55°C for 10 seconds, and 72°C for 30 seconds. The dissolution curve was analyzed to determine the specificity of the real-time PCR amplification. The relative expression level of hsa-miR-181a was calculated by the comparative cycle threshold method, with U6 as the internal reference and expressed as the percentage change relative to untreated controls. Quantification of the relative expression levels of hsa-miR-181a was achieved by the following formula: 2−ΔΔCt, where ΔΔCt equals (Ct of hsa-miR-181a- Ct of U6)experiment minus (Ct of hsa-miR-181a- Ct of U6)control. 2−ΔΔCt was presented as the relative change of hsa-miR-181a expression.
Statistical analysis
Data are expressed as the mean ± standard deviation (SD). Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison to determine statistical significance among multiple groups. Student’s t-test was used to compare the difference between two groups when appropriate. P<0.05 was considered as statistically significant.
Results
Predicted targets of hsa-miR-181a-5p and hsa-miR-181a-3p using various algorithms
Predicted targets of hsa-miR-181a-5p
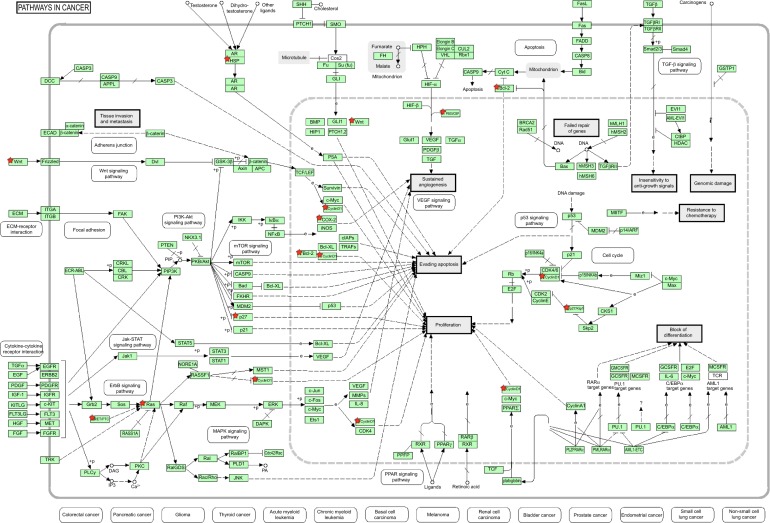
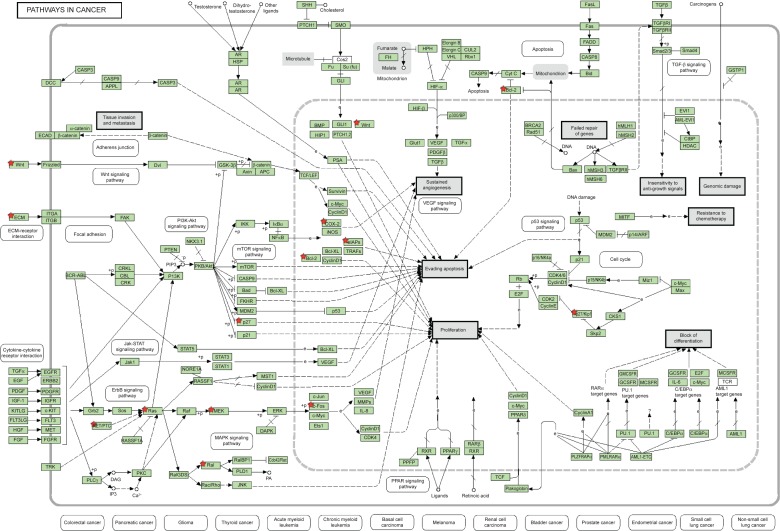
Using DIANA microT v3.0, hsa-miR-181a-5p was predicted to regulate 522 targets when the threshold was set to 0.7, including ATP1B1, BHLHE40, CLASP1, CRE-BRF, FBXO33, GLS, KLHL5, LRBA, MAP1B, MTF2, NMT2, PAPD5, REPS2, RLF, SESN3, SLC2A3, SLITRK1, TMEM131, TRIM2, ZFP90, ZNF268, ZNF441, ZNF527, ZNF570, ZNF704, ZNF717, ZNF781, ZNF788, ZNF799, ZNF846, etc (Table 3 and Table S2). Many of these targets regulate a number of important cellular processes, such as cell proliferation, cell death, cell division, mitosis, metabolism of glucose, lipids, nuclear acids, and amino acids, and transport. Among the predicted targets, 30 of the predicted targets (5.74%) were cancer genes, including ABI1, ARID2, ATM, BCL11B, BCL6, CARD11, CCDC6, CHN1, CREB1, GNAQ, HMGA2, LCP1, MAF, MAP2K1, MLLT10, MSI2, NOTCH2, NR4A3, PAX5, PBX1, PHOX2B, PMS1, PRDM1, PTEN, SS18L1, STAG2, TRIM33, WHSC1, WIF1, and XPO1 (Table 3 and Table S2).
Table 3.
Predicted targets of hsa-miR-181a-5p by various predicting tools
| Tool | Website (URL) | Number of predicted targets/transcripts | Examples of predicted targets | Cancer genes |
|---|---|---|---|---|
| DIANA microT v5.0 | http://diana.cslab.ece.ntua.gr/microT/ | 522 (threshold set at 0.7) | ATP1B1, BHLHE40, CLASP1, CREBRF, FBXO33, GLS, KLHL5, LRBA, MAP1B, MTF2, NMT2, PAPD5, REPS2, RLF, SESN3, SLC2A3, SLITRK1, TMEM131, TRIM2, ZFP90, ZNF268, ZNF441, ZNF527, ZNF570, ZNF704, ZNF717, ZNF781, ZNF788, ZNF799, ZNF846, etc | ABI1, ARID2, ATM, BCL11B, BCL6, CARD11, CCDC6, CHN1, CREB1, GNAQ, HMGA2, LCP1, MAF, MAP2K1, MLLT10, MSI2, NOTCH2, NR4A3, PAX5, PBX1, PHOX2B, PMS1, PRDM1, PTEN, SS18L1, STAG2, TRIM33, WHSC1, WIF1, and XPO1 (n=30) |
| miRanda-mirSVR | http://www.microrna.org/ | 7,847 | ZNF527, ZNF439, ZNF781, ZNF559, ZNF204P, BAZ2B, ZNF844, C15orf29, FBXO34, C5orf41, KIAA0528, ZNF594, EIF4A2, ZNF833, GATM, ZNF440, MARK1, OSBPL3, AP1G1, GABRA1, DDX3X, HCN1, CPOX, TMEM87B, RPE65, BIRC6, NOVA1, LOC442421, ZNF780A, etc | ABI1, ABL1, ABL2, AKAP9, AKT2, APC, ARHGEF12, ARID2, ARNT, ATF1, ATM, ATP2B3, ATRX, BAP1, BCL2, BCL6, BCL7A, BCL9, BCOR, BCR, BRCA1, BTG1, C16orf75, CALR, CAMTA1, CANT1, CARD11, CASP8, CBFA2T3, CBL, CBLB, CCDC6, CCNE1, CD274, CDC73, CDH1, CDK6, CDX2, CEBPA, CHCHD7, CHN1, CLTC, CREB1, CREB3L2, CREBBP, CRTC3, CTNNB1, CYLD, DAXX, DDX10, DDX5, DEK, DICER1, DNM2, ECT2L, EGFR, EIF4A2, EML4, EPS15, ERG, ETV1, ETV6, EXT2, EZH2, FAM46C, FANCA, FANCD2, FANCF, FANCG, FAS, FBXO11, FGFR1, FGFR1OP, FGFR2, FGFR3, FLT3, FNBP1, FOXP1, FSTL3, FUS, GAS7, GATA2, GNAS, GOPC, H3F3B, HERPUD1, HEY1, HLF, HMGA2, HNRNPA2B1, HOOK3, HOXA11, HOXC11, HOXC13, IDH1, IGL@, IL2, IL21R, IL6ST, IL7R, IRF4, JAK2, JAZF1, JUN, KCNJ5, KDM5A, KDM5C, KDM6A, KDR, KIAA1549, KLK2, KRAS, LASP1, LCP1, LIFR, LMO1, LPP, MAF, MAFB, MALAT1, MALT1, MAP2K1, MAP2K4, MDM2, MDM4, MDS2, MET, MITF, MKL1, MLF1, MLH1, MLL, MLL3, MLLT10, MLLT3, MLLT4, MLLT6, MN1, MPL, MSH2, MSI2, MYB, MYCN, MYH11, NCOA1, NCOA2, NF1, NF2, NFE2L2, NFIB, NIN, NOTCH2, NPM1, NR4A3, NRAS, NSD1, NT5C2, NUP98, PAFAH1B2, PAX3, PAX5, PAX7, PBRM1, PBX1, PDE4DIP, PDGFRA, PDGFRB, PHF6, PHOX2B, PICALM, PIK3R1, PLAG1, PML, PMS1, PPARG, PRDM1, PTEN, PTPRC, RAC1, RAD21, RALGDS, RANBP17, RAP1GDS1, RB1, RNF43, RPL5, RUNX1, SDC4, SETBP1, SF3B1, SH2B3, SLC34A2, SMARCA4, SMARCE1, SRGAP3, SS18L1, STAT3, SUFU, SUZ12, SYK, TAF15, TAL2, TBL1XR1, TCF7L2, TCL6, TET2, TFRC, TMPRSS2, TOP1, TPM3, TRA@, TRIM27, TRIM33, TRRAP, TSC1, TSHR, U2AF1, UBR5, VTI1A, WHSC1, WIF1, YWHAE, and ZNF521 |
| miRDB | http://mirdb.org/miRDB/ | 1,065 | PDE5A, ZNF439, PRTG, BRWD1, ZNF549, NFAT5, SH3TC2, OSBPL3, GFPT1, ZNF781, TNPO1, PAPD5, FIGN, S1PR1, TMEM87B, DDX3X, ZNF559, ZNF844, CLMN, GPR26, CTDSPL, ANKRD13C, FUT9, RAB3IP, DLGAP2, BIRC6, ZNF268, C5orf41, PAM, KIAA0528, ARHGEF3, etc | ABI1, ATF1, ATM, BCL11A, CBLB, CREB1, EIF4A2, ETV6, FAS, FOXP1, GAS7, HLF, HOOK3, HOXA11, IL2, JAZF1, KDM5A, LIFR, LPP, MAP2K1, MAP2K4, MDM4, MET, MLL, MLL3, MLLT10, NCOA2, NFIB, NOTCH2, NR4A3, NRAS, PBX1, PDGFRA, PLAG1, RAD21, SH2B3, TCF7L2, TET2, TFRC, TMPRSS2, VHL, VTI1A, and WIF1 |
| RNA22 v2 | https://cm.jefferson.edu/rna22v2.0/ | 7,042 | NIPAL3, PAX7, METTL13, FMO1, CLCN6, CLCA1, CLCA1, ATP1A2, KPNA6, SLAMF7, ZZZ3, USH2A, TRIT1, TPR, VPS13D, PER3, COL9A2, HHAT, TNFRSF9, KIF1B, ATP2B4, ZC3H11A, ZC3H11A, TMEM48, TARBP1, DDX20, GNAI3, SPEN, TIE1, IARS2, KCNAB2, TGFBR3, etc | AKAP9, ARHGEF12, ARID2, ARNT, ASXL1, ATM, ATP1A1, ATRX, AXIN1, BAP1, BCL11A, BCL11B, BCL3, BCL6, BCOR, BCR, BLM, BMPR1A, BRAF, BRCA1, BRD4, BTG1, BUB1B, C15orf55, C2orf44, CAMTA1, CARD11, CARS, CBLB, CCND1, CD74, CDH1, CDK4, CHCHD7, CHEK2, CHN1, CIITA, CLTC, COL1A1, CREB1, CREB3L2, CRTC3, CTNNB1, CYLD, DDX5, DICER1, DNM2, DNMT3A, ECT2L, EGFR, ELF4, ELL, ERBB2, ERCC2, ETV5, EXT1, EZH2, FANCA, FANCD2, FBXO11, FBXW7, FGFR1, FGFR1OP, FGFR2, FHIT, FNBP1, FOXP1, GAS7, GATA2, GATA3, GNAQ, GNAS, GOLGA5, GPHN, HERPUD1, HIP1, HMGA1, HMGA2, HRAS, IDH1, IDH2, IL6ST, IL7R, ITK, JAK3, JAZF1, JUN, KCNJ5, KDM5A, KIAA1549, KIT, KRAS, LASP1, LCP1, LIFR, LPP, MAF, MALT1, MAML2, MAP2K1, MAX, MDM2, MDM4, MED12, MITF, MKL1, MLF1, MLH1, MLL, MLL3, MLLT10, MLLT3, MLLT4, MLLT6, MN1, MSH6, MSI2, MSN, MYD88, MYH11, MYST4, NACA, NCOA1, NDRG1, NF1, NF2, NIN, NONO, NOTCH1, NOTCH2, NT5C2, NTRK3, NUP214, NUP98, OLIG2, P2RY8, PAFAH1B2, PALB2, PAX5, PAX7, PBRM1, PBX1, PDE4DIP, PDGFRA, PDGFRB, PER1, PHF6, PHOX2B, PIK3CA, PIK3R1, PLAG1, PML, POU2AF1, PPARG, PRDM16, PTEN, PTPN11, PTPRC, RAD21, RAF1, RALGDS, RANBP17, RET, ROS1, RPN1, SBDS, SDC4, SDHD, SETBP1, SETD2, SFPQ, SLC45A3, SMO, SOX2, SRGAP3, SS18, SS18L1, SSX1, SSX2, SSX4, STAG2, STAT3, SUZ12, TAF15, TAL1, TBL1XR1, TCL1A, TERT, TET2, TFRC, THRAP3, TMPRSS2, TNFAIP3, TNFRSF14, TOP1, TRAF7, TRIM33, TRIP11, TRRAP, TSC1, TSC2, TSHR, UBR5, VHL, WHSC1, WHSC1L1, WRN, WWTR1, ZNF331, ZNF384, and ZNF521 |
| TargetMiner | http://www.isical.ac.in/~bioinfo_miu/targetminer20.htm | 108 | GPD2, THRB, DIO2, GABRA4, KITLG, PGR, SMAD5, LDLRAD4, MFAP3L, MTX3, CREB5, KCNMA1, RAB3IP, AP1G1, FOXK1, GK5, CREBZF, CHIC1, PAPD5, CYLD, KDM5A, ACVR2B, LOC124389, CALCR, AFF2, ITGA2, etc | CYLD, KDM5A, KRAS, LIFR, and TET2 |
| TargetScan 6.2 | http://www.targetscan.org/ | 1,194 transcripts (626 genes) | ZNF780A, PPIP5K2, NUDT12, HOXC8, MARK1, TOM1L1, CLVS1, ZNF563, S1PR1, ZNF568, FLT1, BTBD3, TCERG1, CTDSPL, SLC25A37, DDX3Y, RPS6KB1, METAP1, FGD4, PBMUCL1, CDON, DDX3X, ZFP62, CLMN, TMEM165, PAPD5, ZFP82, CLIP1, SLC7A2, etc | ABL2, ARID2, ATP2B3, ATXN1, BCL2, BCL9, BCR, CALR, CBFA2T3, CBLB, CCDC6, CDC73, CEBPA, CREBL2, CYLD, EIF4A2, ERG, FGFR3, H3F3B, HLF, HOXA11, KDM5A, KIAA1549, LMO1, LPP, MAP2K1, NCOA2, NFIB, NOTCH2, PAFAH1B2, PBX1, PRDM1, SH2B3, SS18L1, TCF7L2, and WHSC1 |
| PicTar | http://pictar.mdc-berlin.de/ | 510 transcripts (399 genes) | KIAA0195, OSBPL3, CTDSPL, HIC2, GRIK2, ATXN1, ADAM11, ZBTB4, KIAA0802, FBXO33, PIP3AP, EYA3, CBX7, TARSH, CPEB4, LRRC5, MMP14, RLF, AKAP7, ZIC2, CLASP1, ATP2B2, SEMA4G, YTHDF3, FLJ23548, ALS2CR3, HOXC8, RSN, SOX6, TCERG1, COPEB, etc | ATXN1, CARD11, CBFA2T3, CBLB, CHN1, COPEB, CREB1, EIF4A2, ETV6, FOXP1, HLF, HOXA11, JAZF1, KIT, LMO1, MYCN, NCOA2, NR4A3, PDGFRA, PHOX2B, PLAG1, RUNX1, and SS18L1 |
| MicroCosm Targets v5 | http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/ | 1,104 | NR6A1, TMEM64, TMED8, TMED4, TADA1L, ZNF17, ZNF487, SLC3A1, CARD11, DOCK7, SLC10A7, THBS4, E2F5, PRDX3, PLCL2, RGMA, DHX29, FAM58A, DMRT3, MAB21L1, C19orf59, TGFBRAP1, DEPDC6, CARM1, WDR45L, MDH1B, etc | AKAP9, BCL11A, C16orf75, CAGE1, CARD11, CSF3R, EIF4A2, EXT2, FANCE, FAS, FGFR2, FOS, FOXP1, FVT1, HOOK3, IDH1, IL2, LMO1, MLF1, MRAS, MYBL1, PALB2, PHOX2B, RAB38, RALA, RASGRP4, RASIP1, RASSF1, RASSF6, SSX1, SSX2, STAG2, TAF15, TSG101, TUSC3, WIF1, and ZNF521 |
miRanda-mirSVR predicted that hsa-miR-181a-5p could regulate 7,847 transcripts/targets (Table 3 and Table S3). These included ZNF527, ZNF439, ZNF781, ZNF559, ZNF204P, BAZ2B, ZNF844, C15orf29, FBXO34, C5orf41, KIAA0528, ZNF594, EIF4A2, ZNF833, GATM, ZNF440, MARK1, OSBPL3, AP1G1, GABRA1, DDX3X, etc. Most of these genes regulate a number of important cellular processes, such as cell proliferation, cell death, division, mitosis, metabolism of glucose, lipids, nuclear acids, and amino acids, and transport. Notably, 223 genes from these predicted targets were cancer genes (2.84%) (Table S4). These included ABI1, ABL1, ABL2, AKAP9, AKT2, APC, ARHGEF12, ARID2, ARNT, ATF1, ATM, ATP2B3, ATRX, BAP1, BCL2, BCL6, BCL7A, BCL9, BCOR, BCR, BRCA1, BTG1, C16orf75, CALR, CAMTA1, CANT1, CARD11, CASP8, CBFA2T3, CBL, CBLB, CCDC6, CCNE1, CD274, CDC73, CDH1, CDK6, CDX2, CEBPA, CHCHD7, CHN1, CLTC, CREB1, CREB3L2, CREBBP, CRTC3, CTNNB1, CYLD, DAXX, DDX10, DDX5, DEK, DICER1, DNM2, ECT2L, EGFR, EIF4A2, EML4, EPS15, ERG, ETV1, ETV6, EXT2, EZH2, FAM46C, FANCA, FANCD2, FANCF, FANCG, FAS, FBXO11, FGFR1, FGFR1OP, FGFR2, FGFR3, FLT3, FNBP1, FOXP1, FSTL3, FUS, GAS7, GATA2, GNAS, GOPC, H3F3B, HERPUD1, HEY1, HLF, HMGA2, HNRNPA2B1, HOOK3, HOXA11, HOXC11, HOXC13, IDH1, IGL@, IL2, IL21R, IL6ST, IL7R, IRF4, JAK2, JAZF1, JUN, KCNJ5, KDM5A, KDM5C, KDM6A, KDR, KIAA1549, KLK2, KRAS, LASP1, LCP1, LIFR, LMO1, LPP, MAF, MAFB, MALAT1, MALT1, MAP2K1, MAP2K4, MDM2, MDM4, MDS2, MET, MITF, MKL1, MLF1, MLH1, MLL, MLL3, MLLT10, MLLT3, MLLT4, MLLT6, MN1, MPL, MSH2, MSI2, MYB, MYCN, MYH11, NCOA1, NCOA2, NF1, NF2, NFE2L2, NFIB, NIN, NOTCH2, NPM1, NR4A3, NRAS, NSD1, NT5C2, NUP98, PAFAH1B2, PAX3, PAX5, PAX7, PBRM1, PBX1, PDE4DIP, PDGFRA, PDGFRB, PHF6, PHOX2B, PICALM, PIK3R1, PLAG1, PML, PMS1, PPARG, PRDM1, PTEN, PTPRC, RAC1, RAD21, RALGDS, RANBP17, RAP1GDS1, RB1, RNF43, RPL5, RUNX1, SDC4, SETBP1, SF3B1, SH2B3, SLC34A2, SMARCA4, SMARCE1, SRGAP3, SS18L1, STAT3, SUFU, SUZ12, SYK, TAF15, TAL2, TBL1XR1, TCF7L2, TCL6, TET2, TFRC, TMPRSS2, TOP1, TPM3, TRA@, TRIM27, TRIM33, TRRAP, TSC1, TSHR, U2AF1, UBR5, VTI1A, WHSC1, WIF1, YWHAE, and ZNF521 (Table S4).
miRDB predicted that 1,065 targets/transcripts were possibly regulated by hsa-miR-181a-5p (Table 3 and Table S5). These included PDE5A, ZNF439, PRTG, BRWD1, ZNF549, NFAT5, SH3TC2, OSBPL3, GFPT1, ZNF781, TNPO1, PAPD5, FIGN, S1PR1, TMEM87B, DDX3X, ZNF559, ZNF844, CLMN, GPR26, CTDSPL, ANKRD13C, FUT9, RAB3IP, DLGAP2, BIRC6, ZNF268, C5orf41, PAM, KIAA0528, ARHGEF3, etc. Many of the predicted targets play a role in the regulation of cell proliferation, cell cycle, apoptosis, energy, nuclear acid, and protein metabolism, signaling transduction, and transport. Among these targets, 43 targets were cancer genes (4.04%). These included ABI1, ATF1, ATM, BCL11A, CBLB, CREB1, EIF4A2, ETV6, FAS, FOXP1, GAS7, HLF, HOOK3, HOXA11, IL2, JAZF1, KDM5A, LIFR, LPP, MAP2K1, MAP2K4, MDM4, MET, MLL, MLL3, MLLT10, NCOA2, NFIB, NOTCH2, NR4A3, NRAS, PBX1, PDGFRA, PLAG1, RAD21, SH2B3, TCF7L2, TET2, TFRC, TMPRSS2, VHL, VTI1A, and WIF1 (Table 3 and Table S5).
Using RNA22 v2, 7,028 targets were predicted to be regulated by hsa-miR-181a-5p (Table 3 and Table S6). These included NIPAL3, PAX7, METTL13, FMO1, CLCN6, CLCA1, CLCA1, ATP1A2, KPNA6, SLAMF7, ZZZ3, USH2A, TRIT1, TPR, VPS13D, PER3, COL9A2, HHAT, TNFRSF9, KIF1B, ATP2B4, ZC3H11A, ZC3H11A, TMEM48, TARBP1, DDX20, GNAI3, SPEN, TIE1, IARS2, KCNAB2, TGFBR3, etc. Many of the predicted targets play a role in the regulation of cell proliferation, cell cycle, apoptosis, energy, nuclear acid and protein metabolism, signaling transduction, and transport. Among the predicted targets, 211 genes were cancer genes (3.00%) (Table S7). These included AKAP9, ARHGEF12, ARID2, ARNT, ASXL1, ATM, ATP1A1, ATRX, AXIN1, BAP1, BCL11A, BCL11B, BCL3, BCL6, BCOR, BCR, BLM, BMPR1A, BRAF, BRCA1, BRD4, BTG1, BUB1B, C15orf55, C2orf44, CAMTA1, CARD11, CARS, CBLB, CCND1, CD74, CDH1, CDK4, CHCHD7, CHEK2, CHN1, CIITA, CLTC, COL1A1, CREB1, CREB3L2, CRTC3, CTNNB1, CYLD, DDX5, DICER1, DNM2, DNMT3A, ECT2L, EGFR, ELF4, ELL, ERBB2, ERCC2, ETV5, EXT1, EZH2, FANCA, FANCD2, FBXO11, FBXW7, FGFR1, FGFR1OP, FGFR2, FHIT, FNBP1, FOXP1, GAS7, GATA2, GATA3, GNAQ, GNAS, GOLGA5, GPHN, HERPUD1, HIP1, HMGA1, HMGA2, HRAS, IDH1, IDH2, IL6ST, IL7R, ITK, JAK3, JAZF1, JUN, KCNJ5, KDM5A, KIAA1549, KIT, KRAS, LASP1, LCP1, LIFR, LPP, MAF, MALT1, MAML2, MAP2K1, MAX, MDM2, MDM4, MED12, MITF, MKL1, MLF1, MLH1, MLL, MLL3, MLLT10, MLLT3, MLLT4, MLLT6, MN1, MSH6, MSI2, MSN, MYD88, MYH11, MYST4, NACA, NCOA1, NDRG1, NF1, NF2, NIN, NONO, NOTCH1, NOTCH2, NT5C2, NTRK3, NUP214, NUP98, OLIG2, P2RY8, PAFAH1B2, PALB2, PAX5, PAX7, PBRM1, PBX1, PDE4DIP, PDGFRA, PDGFRB, PER1, PHF6, PHOX2B, PIK3CA, PIK3R1, PLAG1, PML, POU2AF1, PPARG, PRDM16, PTEN, PTPN11, PTPRC, RAD21, RAF1, RALGDS, RANBP17, RET, ROS1, RPN1, SBDS, SDC4, SDHD, SETBP1, SETD2, SFPQ, SLC45A3, SMO, SOX2, SRGAP3, SS18, SS18L1, SSX1, SSX2, SSX4, STAG2, STAT3, SUZ12, TAF15, TAL1, TBL1XR1, TCL1A, TERT, TET2, TFRC, THRAP3, TMPRSS2, TNFAIP3, TNFRSF14, TOP1, TRAF7, TRIM33, TRIP11, TRRAP, TSC1, TSC2, TSHR, UBR5, VHL, WHSC1, WHSC1L1, WRN, WWTR1, ZNF331, ZNF384, and ZNF521 (Table S7).
TargetMiner predicted that 108 targets were regulated by hsa-miR-181a-5p, including GPD2, THRB, DIO2, GABRA4, KITLG, PGR, SMAD5, LDLRAD4, MFAP3L, MTX3, CREB5, KCNMA1, RAB3IP, AP1G1, FOXK1, GK5, CREBZF, CHIC1, PAPD5, CYLD, KDM5A, ACVR2B, LOC124389, CALCR, AFF2, ITGA2, etc (Table 3 and Table S8). Among these predicted targets, five were cancer genes, including CYLD, KDM5A, KRAS, LIFR, and TET2 (4.63%) (Table 3 and Table S8).
TargetScan 6.2 only provided the predicted targets for the precursor hsa-miR-181a. It predicted that hsa-miR-181a could regulate 1,194 transcripts with conserved sites, with a total of 1,412 conserved sites and 626 poorly conserved sites (Table 3 and Table S9). Among these transcripts, 626 were functional genes. These included ZNF780A, PPIP5K2, NUDT12, HOXC8, MARK1, TOM1L1, CLVS1, ZNF563, S1PR1, ZNF568, FLT1, BTBD3, TCERG1, CTD-SPL, SLC25A37, DDX3Y, RPS6KB1, METAP1, FGD4, PBMUCL1, CDON, DDX3X, ZFP62, CLMN, TMEM165, PAPD5, ZFP82, CLIP1, SLC7A2, etc. Many of the targets were involved in the regulation of cell proliferation, cell cycle, apoptosis, energy, nuclear acid, and protein metabolism, signaling transduction, and transport. Among the predicted targets, 36 were cancer genes (5.75%). These included ABL2, ARID2, ATP2B3, ATXN1, BCL2, BCL9, BCR, CALR, CBFA2T3, CBLB, CCDC6, CDC73, CEBPA, CREBL2, CYLD, EIF4A2, ERG, FGFR3, H3F3B, HLF, HOXA11, KDM5A, KIAA1549, LMO1, LPP, MAP2K1, NCOA2, NFIB, NOTCH2, PAFAH1B2, PBX1, PRDM1, SH2B3, SS18L1, TCF7L2, and WHSC1 (Table 3 and Table S9).
PicTar only provided predicted targets for the precursor hsa-miR-181a. It predicted that hsa-miR-181a could regulate 510 transcripts with 399 genes, including KIAA0195, OSBPL3, CTDSPL, HIC2, GRIK2, ATXN1, ADAM11, ZBTB4, KIAA0802, FBXO33, PIP3AP, EYA3, CBX7, TARSH, CPEB4, LRRC5, MMP14, RLF, AKAP7, ZIC2, CLASP1, ATP2B2, SEMA4G, YTHDF3, FLJ23548, ALS2CR3, HOXC8, RSN, SOX6, TCERG1, COPEB, etc (Table 3 and Table S10). Many of the predicted targets play a role in the regulation of cell proliferation, cell cycle, apoptosis, energy, nuclear acid, and protein metabolism, signaling transduction, and transport. Among the predicted targets, 23 (5.76%) were cancer genes involved in the initiation, growth, and development and metastasis of cancer, including ATXN1, CARD11, CBFA2T3, CBLB, CHN1, COPEB, CREB1, EIF4A2, ETV6, FOXP1, HLF, HOXA11, JAZF1, KIT, LMO1, MYCN, NCOA2, NR4A3, PDGFRA, PHOX2B, PLAG1, RUNX1, and SS18L1 (Table 3 and Table S10).
MicroCosm Targets v5 predicted that 1,104 targets/transcripts were likely regulated by hsa-miR-181a-5p (Table 3 and Table S11). These included NR6A1, TMEM64, TMED8, TMED4, TADA1L, ZNF17, ZNF487, SLC3A1, CARD11, DOCK7, SLC10A7, THBS4, E2F5, PRDX3, PLCL2, RGMA, DHX29, FAM58A, DMRT3, MAB21L1, C19orf59, TGFBRAP1, DEPDC6, CARM1, WDR45L, MDH1B, etc. Among the predicted targets, 37 (3.35%) were cancer genes, including AKAP9, BCL11A, C16orf75, CAGE1, CARD11, CSF3R, EIF4A2, EXT2, FANCE, FAS, FGFR2, FOS, FOXP1, FVT1, HOOK3, IDH1, IL2, LMO1, MLF1, MRAS, MYBL1, PALB2, PHOX2B, RAB38, RALA, RASGRP4, RASIP1, RASSF1, RASSF6, SSX1, SSX2, STAG2, TAF15, TSG101, TUSC3, WIF1, and ZNF521 (Table 3 and Table S11).
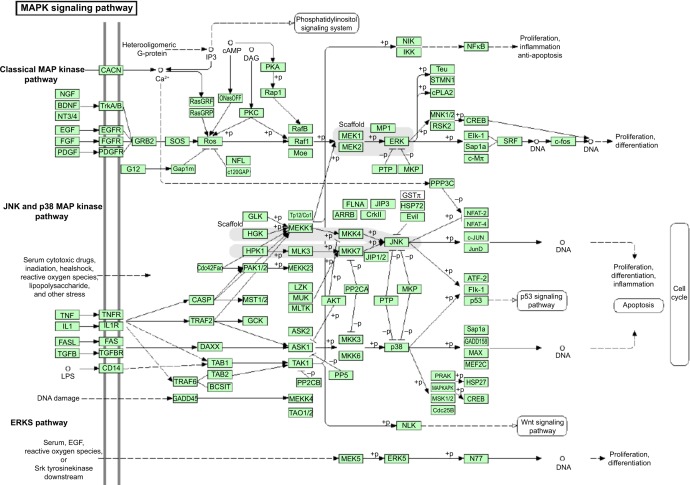
These results showed that the number of predicted targets of hsa-miR-181a-5p by the eight algorithms was very different, ranging from 108 to 7,847, with a mean of 2,424. Most of the predicted targets are involved in the regulation of cell proliferation, cell division, cell apoptosis, energy metabolism, amino acid, and nucleic acid metabolism, and transport, inflammation, redox homeostasis, and stress response. Many of the predicted targets are cancer genes, which participate in cancer initiation, development, growth, and metastasis. These cancer genes, including tumor suppressor genes and oncogenes, act as drivers or passengers in tumorigenesis. They are involved in various aspects of functions implicated in cancer initiation, development, and metastasis, including control of cell proliferation, apoptosis, signal transduction, transcription regulation, immunity, and defense.
Predicted targets of hsa-miR-181a-3p
Using DIANA microT v3.0, hsa-miR-181a-3p was predicted to regulate 249 transcripts/targets when the threshold was set at 0.45 (Table 4 and Table S12). These included ETV1, GGCT, ODZ1, NUB1, CPS1, AGPAT4, TRIO, LMO3, COL9A2, ELN, HEBP2, CYFIP2, MCOLN3, RC3H2, FLYWCH1, COL11A1, MRPS35, DGKA, LAPTM4A, IFT80, CLEC2D, AFF4, MARK2, SENP1, STX7, PDS5B, NKAIN1, PSMC5, SLC26A4, etc. Among these targets, 15 were cancer genes (6.12%), including ASPSCR1, BCL11A, CACNA1D, CCND2, ELN, ETV1, EZH2, GATA3, HIP1, HRAS, MSI2, PIM1, TSHR, USP6, and WIF1 (Table 4 and Table S12).
Table 4.
Predicted targets of hsa-miR-181a-3p by various predicting tools
| Tool | Website (URL) | Number of predicted transcripts | Examples of predicted targets | Cancer genes |
|---|---|---|---|---|
| DIANA microT v5.0 | http://diana.cslab.ece.ntua.gr/microT/ | 249 (threshold set at 0.45) | ETV1, GGCT, ODZ1, NUB1, CPS1, AGPAT4, TRIO, LMO3, COL9A2, ELN, HEBP2, CYFIP2, MCOLN3, RC3H2, FLYWCH1, COL11A1, MRPS35, DGKA, LAPTM4A, IFT80, CLEC2D, AFF4, MARK2, SENP1, STX7, PDS5B, NKAIN1, PSMC5, SLC26A4, etc | ASPSCR1, BCL11A, CACNA1D, CCND2, ELN, ETV1, EZH2, GATA3, HIP1, HRAS, MSI2, PIM1, TSHR, USP6, and WIF1 |
| miRanda- mirSVR | http://www.microrna.org/ | 1,873 | PMS2L2, COL27A1, SAE1, CNTNAP3B, FAM153B, GRIP2, NR4A1, RXRA, AES, POM121C, AFG3L1, XDH, XPA, MID1, AIRE, CTSK, HSD17B1, LOR, LTBP2, NEU1, AMPD3, AMT, APP, FUT2, etc | ABI2, ATM, AXIN1, BCL11A, BCL2, BTG1, CACNA1D, CASP8, CDH1, CDK12, CEBPA, CHCHD7, CREB1, CRTC3, DDB2, DDX10, DNM2, DUX4, EBF1, EIF4A2, ELL, ETV6, EZH2, FOXP1, GAS7, GATA3, GNA11, GNAS, HIP1, LMO1, LPP, MALAT1, MAX, MKL1, MLL3, MLLT1, MN1, MSI2, NF1, PAFAH1B2, PAX5, PAX7, PDE4DIP, PER1, PIM1, PML, PTEN, RANBP17, RPL10, SEPT6, SET, SETBP1, SMARCE1, SS18L1, TAL1, TBL1XR1, TCL6, TET2, TP53, TSHR, and TTL |
| miRDB | http://mirdb.org/miRDB/ | 22 | ALDH18A1, ATP13A4, ALDH6A1, C16orf57, NIPA2, RIBC1, SLC20A2, C14orf28, CFL2, ZNF3, RHOBTB1, H1F0, AP1S3, ARL4A, RBM22, MIER1, ACTR3, CLEC2D, AFF2, ACAP2, RABGEF1, and KCTD12 | None |
| RNA22 v2 | https://cm.jefferson.edu/rna22v2.0/ | 5,142 (2,718 genes) | CFH, TTC22, FMO1, ATP1A2, VPS13D, COL9A2, UTS2, LAMC2, PIGV, PTPRU, COL11A1, DDX20, WDR3, YBX1, ASPM, LRRC40, EPHA8, ARHGEF10L, RASAL2, PLXNA2, RAP1GAP, PPP1R12B, SDF4, TP73, NKAIN1, WDR47, OVGP1, SLC25A24, EPS15, POMGNT1, etc | ABL1, ABL2, AKAP9, AKT1, AKT2, ALDH2, ARID1A, ARID2, ASXL1, ATP1A1, AXIN1, BCL6, BCL7A, BCR, BRD3, CCNB1IP1, CCND2, CDK6, CHEK2, CREB1, CTNNB1, CYLD, DDB2, DNMT3A, ELK4, EPS15, FANCA, FANCC, FGFR2, FOXP1, FUS, GNAS, HIP1, HLF, HMGA2, HOXD11, IL7R, KCNJ5, KDR, KLF4, KTN1, LASP1, MAML2, MDM4, MED12, MET, MKL1, MLL3, MUTYH, MYB, MYH11, NACA, NCOA2, NFIB, NOTCH2, NSD1, NTRK3, PAX8, PBRM1, PDGFRB, POT1, POU2AF1, REL, RNF43, RPL10, RUNDC2A, RUNX1, SETBP1, SF3B1, SMARCA4, STAT3, SUFU, TAF15, TCEA1, TCF3, TFEB, TSC1, UBR5, USP6, VHL, WHSC1L1, and YWHAE |
| TargetMiner | http://www.isical.ac.in/~bioinfo_miu/targetminer20.htm | 13 | CD47, CELF2, CPNE3, FECH, FGF5, IKZF2, MIER1, NLGN1, NR2C2, RBM12B, SMAD2, SRSF8, and TLR4 | None |
| TargetScan 6.2 | http://www.targetscan.org/ | See Table 3 | ||
| PicTar | http://pictar.mdc-berlin.de/ | See Table 3 | ||
| MicroCosm Targets v5 | http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/ | 1,039 | JAG2, Q6ZWB7, PCDH11Y, KLF1, MAN1B1, TMED5, ONECUT1, OCIAD1, FAM84B, WDR69, ARFIP1, LRRC45, SLC1A7, ATP8A2, SPACA5, TTN, IER3, PPIL1, C12orf45, IL27, KRT33B, HYI, TSPAN32, TCHP, KDELR2, FOXA3, CPXM2, etc | AKT2, ALDH2, BCL2A1, BCL7C, CARS, CDK4, DAXX, DDB2, DLEU7, ERCC5, FUBP1, FUS, H3F3A, HOXA9, IRF4, JAK1, LCK, LMO1, MKL1, MSI2, NFIB, PAX3, RAC2, RAF1, RASIP1, SETBP1, SSX2, USP6, and WHSC1L1 |
miRanda-mirSVR predicted that hsa-miR-181-a-3p could regulate 1,873 targets (Table 4 and Table S13). These genes included PMS2L2, COL27A1, SAE1, CNTNAP3B, FAM153B, GRIP2, NR4A1, RXRA, AES, POM121C, AFG3L1, XDH, XPA, MID1, AIRE, CTSK, HSD17B1, LOR, LTBP2, NEU1, AMPD3, AMT, APP, FUT2, etc. Most of these genes regulate a number of important cellular processes, such as cell proliferation, cell death, division, mitosis, metabolism of glucose, lipids, nuclear acids, and amino acids, and transport. Among the predicted targets, 61 were cancer genes (3.26%). These included ABI2, ATM, AXIN1, BCL11A, BCL2, BTG1, CACNA1D, CASP8, CDH1, CDK12, CEBPA, CHCHD7, CREB1, CRTC3, DDB2, DDX10, DNM2, DUX4, EBF1, EIF4A2, ELL, ETV6, EZH2, FOXP1, GAS7, GATA3, GNA11, GNAS, HIP1, LMO1, LPP, MALAT1, MAX, MKL1, MLL3, MLLT1, MN1, MSI2, NF1, PAFAH1B2, PAX5, PAX7, PDE4DIP, PER1, PIM1, PML, PTEN, RANBP17, RPL10, SEPT6, SET, SETBP1, SMARCE1, SS18L1, TAL1, TBL1XR1, TCL6, TET2, TP53, TSHR, and TTL (Table 4 and Table S14).
miRDB predicted that 22 targets were possibly regulated by hsa-miR-181a-3p (Table 4 and Table S15). These included ALDH18A1, ATP13A4, ALDH6A1, C16orf57, NIPA2, RIBC1, SLC20A2, C14orf28, CFL2, ZNF3, RHOBTB1, H1F0, AP1S3, ARL4A, RBM22, MIER1, ACTR3, CLEC2D, AFF2, ACAP2, RABGEF1, and KCTD12. These targets play a role in the regulation of cell proliferation, cell cycle, apoptosis, energy, nuclear acid, and protein metabolism, signaling transduction, and transport. However, none of the predicted targets were cancer genes.
Using RNA22 v2, 5,142 transcripts/2,718 genes were predicted to be regulated by hsa-miR-181a-3p (Table 4 and Table S16). These included CFH, TTC22, FMO1, ATP1A2, VPS13D, COL9A2, UTS2, LAMC2, PIGV, PTPRU, COL11A1, DDX20, WDR3, YBX1, ASPM, LRRC40, EPHA8, ARHGEF10L, RASAL2, PLXNA2, RAP1GAP, PPP1R12B, SDF4, TP73, NKAIN1, WDR47, OVGP1, SLC25A24, EPS15, POMGNT1, etc. Many of the predicted targets play a role in the regulation of cell proliferation, cell cycle, apoptosis, energy, nuclear acid, and protein metabolism, signaling transduction, and transport. Among these targets, 82 were cancer genes (3.02%), including ABL1, ABL2, AKAP9, AKT1, AKT2, ALDH2, ARID1A, ARID2, ASXL1, ATP1A1, AXIN1, BCL6, BCL7A, BCR, BRD3, CCNB1IP1, CCND2, CDK6, CHEK2, CREB1, CTNNB1, CYLD, DDB2, DNMT3A, ELK4, EPS15, FANCA, FANCC, FGFR2, FOXP1, FUS, GNAS, HIP1, HLF, HMGA2, HOXD11, IL7R, KCNJ5, KDR, KLF4, KTN1, LASP1, MAML2, MDM4, MED12, MET, MKL1, MLL3, MUTYH, MYB, MYH11, NACA, NCOA2, NFIB, NOTCH2, NSD1, NTRK3, PAX8, PBRM1, PDGFRB, POT1, POU2AF1, REL, RNF43, RPL10, RUNDC2A, RUNX1, SETBP1, SF3B1, SMARCA4, STAT3, SUFU, TAF15, TCEA1, TCF3, TFEB, TSC1, UBR5, USP6, VHL, WHSC1L1, and YWHAE (Table 4 and Table S16).
TargetMiner predicted that 13 targets were regulated by hsa-miR-181a-3p, including CD47, CELF2, CPNE3, FECH, FGF5, IKZF2, MIER1, NLGN1, NR2C2, RBM12B, SMAD2, SRSF8, and TLR4 (Table 4 and Table S17). None of the predicted targets was a cancer gene.
TargetScan 6.2 predicted that hsa-miR-181a could regulate 626 targets (Tables 3 and 4). In PicTar, only hsa-miR-181a could be searched. It probably regulated 510 transcripts with 399 genes (Tables 3 and 4).
MicroCosm Targets v5 predicted that 1,039 targets were likely regulated by hsa-miR-181a-3p (Table 4 and Table S18). These included JAG2, Q6ZWB7, PCDH11Y, KLF1, MAN1B1, TMED5, ONECUT1, OCIAD1, FAM84B, WDR69, ARFIP1, LRRC45, SLC1A7, ATP8A2, SPACA5, TTN, IER3, PPIL1, C12orf45, IL27, KRT33B, HYI, TSPAN32, TCHP, KDELR2, FOXA3, CPXM2, etc. Among the predicted targets, 29 were cancer genes (2.79%), including AKT2, ALDH2, BCL2A1, BCL7C, CARS, CDK4, DAXX, DDB2, DLEU7, ERCC5, FUBP1, FUS, H3F3A, HOXA9, IRF4, JAK1, LCK, LMO1, MKL1, MSI2, NFIB, PAX3, RAC2, RAF1, RASIP1, SETBP1, SSX2, USP6, and WHSC1L1 (Table 4 and Table S18).
These results demonstrated that the number of predicted targets of hsa-miR-181a-3p by the eight tools was very different, ranging from 13 to 5,142, with a mean of 1,184. Most of the predicted targets are involved in the regulation of cell proliferation, cell division, cell apoptosis, energy metabolism, amino acid and nucleic acid metabolism, and transport, inflammation, redox homeostasis, and stress response. Many of the predicted targets are cancer genes which participate in cancer initiation, development, growth, and metastasis.
Predicted targets of hsa-miR-181a by miRWALK
miRWALK provides information on predicted targets from ten algorithms. When only miRWALK was chosen, where only one longest binding site was set per miRNA per mRNA, 3,762 transcripts were predicted to be regulated by hsa-miR-181a (Table S19). These included AASS, ABCB11, ACN9, ACTA2, ADAM28, BAG2, BCL11A, BCL6B, BTRC, CARD11, CCR3, CDK8, CDKN3, CIT, CNOT1, DAD1, DCN, DKC1, DR1, EHF, ESCO2, FBN2, FUT1, GALNT3, GCS1, GPC5, HK2, HSPB3, IL2, IL25, IVD, KCNG3, KIF2C, LACE1, MAEL, MAOA, MLL5, NCL, NOVA1, OCA2, PAG1, PROCR, RAD21, RFC3, RNF6, SELT, SIX2, TAF15, etc. Among these transcripts, 1,436 were functional genes (Table S20).
miRWALK also provided a summarized table that included all targets predicted to be regulated by hsa-miR-181a by the ten algorithms. In total, there were 17,395 transcripts that would be regulated by hsa-miR-181a (Table S21). Only 24 genes were predicted to be the targets of hsa-miR-181a by all the algorithms or at least nine algorithms. These included RNF145, TCERG1, SIRT1, SS18L1, NR6A1, FOXP1, GLS, HOXA11, SMAD7, MAP1B, INOC1, CDKN2AIP, LRRN1, TGFBI, YWHAG, C17orf39, CHD9, NAT13, C6orf62, ACVR2A, DCLK1, NMT2, NPEPPS, and KIAA0195. There were 102 targets that were predicted to be regulated by hsa-miR-181a by eight algorithms, including CDH13, PDIA6, IPO8, IGF2BP2, SLITRK1, SSX2IP, COL16A1, ADM, ZNF800, ADAMTS18, DPYSL2, E2F5, EIF4A2, EN2, ETV6, ACSL1, FBN2, C7orf41, FKBP1A, BTBD3, KIAA0423, HIC2, KANK1, HISPPD1, SYNE1, ZNF281, FOS, LEMD3, BRD1, OSBPL3, EPC2, GAPVD1, GATA6, NPTN, ATP11C, KLF15, LRP12, BAZ2B, HLF, HOXA1, ID4, JARID2, KPNA1, etc. There were 191, 778, 1,615, and 3,168 targets that were predicted to be regulated by hsa-miR-181a by seven, six, five, and four algorithms, respectively.
Validated targets of hsa-miR-181a-5p based on TarBase
Based on TarBase, 211 targets of hsa-miR-181a-5p have been validated with experimental evidence (Table 5). These included ACOT12, ALG10B, AMMECR1, ANKRD1, ANKRD13C, ARF6, ARHGAP11A, ARL6IP1, ARL6IP6, ATF7IP2, ATG10, ATM, ATP6V0E1, BAG2, BCL2, BDNF, BPGM, BRIX1, BRMS1L, BTBD3, C1orf109, C1orf43, C1QTNF9, C8A, CBX3, CCDC6, CCDC82, CCND1, CCNG1, etc. Among these validated targets, only eleven of them are cancer genes (5.21%), including ATM, BCL2, CCDC6, CCND1, CDX2, EP300, HOXA11, KRAS, PLAG1, TAF15, and TSHR.
Table 5.
Targets of hsa-miR-181a-5p with experimental evidence based on TarBase 6.0
| Gene symbol | Accession number | Full name | Alias | Function | Cancer gene |
|---|---|---|---|---|---|
| ACOT12 | NM_130767 | Acyl-CoA thioesterase 12 | CACH-1, Cach, STARD15, THEAL | Hydrolyzes acetyl-CoA to acetate and CoA | |
| ALG10B | NM_001013620 | α-1,2-Glucosyltransferase | ALG10, KCR1 | Transfers glucose from dolichyl phosphate glucose onto the lipid-linked oligosaccharide Glc(2) Man(9)GlcNAc(2)-PP-Dol | |
| AMMECR1 | NM_001025580 | Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 | RP13-360B22.1, AMMERC1 | ||
| ANKRD1 | NM_014391 | Ankyrin repeat domain 1 (cardiac muscle) | ALRP, C-193, CARP, CVARP, MCARP, bA320F15.2 | Plays an important role in endothelial cell activation | |
| ANKRD13C | NM_030816 | Ankyrin repeat domain 13C | RP4-677H15.5, dJ677H15.3 | ||
| ARF6 | NM_001663 | ADP-ribosylation factor 6 | Involved in protein trafficking | ||
| ARHGAP11A | NM_001286479 | Rho GTPase activating protein 11A | RP11-1000B6.5, GAP (1–12) | GTPase activator activity | |
| ARL6IP1 | NM_015161 | ADP-ribosylation factor-like 6 interacting protein 1 | AIP1, ARL6IP, ARMER, SPG61 | May be involved in protein transport, membrane trafficking, or cell signaling during hematopoietic maturation | |
| ARL6IP6 | NM_022989 | ADP-ribosylation factor-like 6 interacting protein 6 | RP23-265N10.1, 2310057C01Rik, 2610529A11Rik, Aip-6 | ||
| ATF7IP2 | NM_001256160 | Activating transcription factor 7 interacting protein 2 | MCAF2 | Recruiter that couples transcriptional factors to general transcription apparatus and thereby modulates transcription regulation and chromatin formation | |
| ATG10 | NM_001131028 | Autophagy related 10 | PP12616, APG10, APG10L, pp12616 | Plays a role in autophagy | |
| ATM | NM_000051 | ATM serine/threonine kinase | AT1, ATA, ATC, ATD, ATDC, ATE, TEL1, TELO1 | Serine/threonine protein kinase | Yes |
| ATP6V0E1 | NM_003945 | ATPase, H+ transporting, lysosomal 9 kDa, V0 subunit e1 | ATP6H, ATP6V0E, M9.2, Vma21, Vma21p | Vacuolar ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells | |
| BAG2 | NM_004282 | BCL2-associated athanogene 2 | RP3-496N17.2, BAG-2, dJ417I1.2 | Inhibits the chaperone activity of HSP70/HSC70 by promoting substrate release | |
| BCL2 | NM_000633 | B-cell CLL/lymphoma 2 | Bcl-2, PPP1R50 | Suppresses apoptosis | Yes |
| BDNF | NM_001143805 | Brain-derived neurotrophic factor | ANON2, BULN2 | Promotes the survival of neuronal populations | |
| BPGM | NM_001293085 | 2,3-Bisphosphoglycerate mutase | DPGM | Plays a major role in regulating hemoglobin oxygen affinity | |
| BRIX1 | NM_018321 | Biogenesis of ribosomes, homolog (S. cerevisiae) | BRIX, BXDC2 | Required for biogenesis of the 60S ribosomal subunit | |
| BRMS1L | NM_032352 | Breast cancer metastasis-suppressor 1-like | BRMS1 | Involved in the HDAC1-dependent transcriptional repression activity | |
| BTBD3 | NM_001282550 | BTB (POZ) domain containing 3 | RP4-742J24.3, dJ742J24.1 | Acts as a key regulator of dendritic field orientation during development of sensory cortex | |
| C1orf109 | NM_017850 | Chromosome 1 open reading frame 109 | |||
| C1orf43 | NM_001098616 | Chromosome 1 open reading frame 43 | HSPC012, NICE-3, NS5ATP4, S863-3 | ||
| C1QTNF9 | NM_183175 | C1q and tumor necrosis factor related protein 9 | 9130217G22Rik, CTRP9, Ciqtnf9 | Activates AMPK, AKT, and p44/42 MAPK signaling pathways | |
| C8A | NM_000562 | Complement component 8, α polypeptide | C8 is a constituent of the membrane attack complex | ||
| CBX3 | NM_007276 | Chromobox homolog 3 | HECH, HP1-GAMMA, HP1Hs-γ | Involved in transcriptional silencing in heterochromatin-like complexes | |
| CCDC6 | NM_005436 | Coiled-coil domain containing 6 | D10S170, H4, PTC, TPC, TST1 | Functions as a tumor suppressor | Yes |
| CCDC82 | NM_024725 | Coiled-coil domain containing 82 | HT025, HSPC048 | ||
| CCND1 | NM_053056 | Cyclin D1 | BCL1, D11S287E, PRAD1, U21B31 | Essential for the control of the cell cycle at the G1/S (start) transition | Yes |
| CCNG1 | NM_004060 | Cyclin G1 | CCNG | May play a role in growth regulation | |
| CD46 | NM_002389 | CD46 molecule, complement regulatory protein | AHUS2, MCP, MIC10, TLX, TRA2.10 | Acts as a cofactor for complement factor I | |
| CDKN1B | NM_004064 | Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | CDKN4, KIP1, MEN1B, MEN4, P27KIP1 | Important regulator of cell cycle progression | |
| CDX2 | NM_001265 | Caudal type homeobox 2 | CDX-3, CDX3 | Involved in the transcriptional regulation of multiple genes expressed in the intestinal epithelium | Yes |
| CEP97 | NM_024548 | Centrosomal protein 97 kDa | 2810403B08Rik, LRRIQ2 | Collaborates with cep110, being involved in the suppression of a cilia assembly program | |
| CFI | NM_000204 | Complement factor I | AHUS3, ARMD13, C3BINA, C3b-INA, FI, IF, KAF | Responsible for cleaving the α-chains of C4b and C3b in the presence of the cofactors C4-binding protein and factor H, respectively | |
| CHD1 | NM_001270 | Chromodomain helicase DNA binding protein 1 | Sequence-selective DNA-binding protein | ||
| CHL1 | NM_001253387 | Cell adhesion molecule L1-like | CALL, L1CAM2 | Plays a role in nervous system development and in synaptic plasticity | |
| CHRFAM7A | NM_139320 | CHRNA7 (cholinergic receptor, nicotinic, α 7, exons 5–10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion | CHRNA7, CHRNA7-DR1, D-10 | Extracellular ligand-gated ion channel activity | |
| CLUAP1 | NM_015041 | Clusterin associated protein 1 | CFAP22, FAP22 | May play a role in cell proliferation or apoptosis | |
| COL27A1 | NM_032888 | Collagen, type XXVII, α 1 | RP11-82I1.1 | Plays a role during the calcification of cartilage and the transition of cartilage to bone | |
| COPS2 | NM_001143887 | COP9 signalosome subunit 2 | ALIEN, CSN2, SGN2, TRIP15 | Involved in various cellular and developmental processes | |
| CST5 | NM_001900 | Cystatin D | Cysteine proteinase inhibitor | ||
| CXorf1 | NM_004709 | Transmembrane protein 257 | CXorf1 | ||
| D3R | NM_000796.5 | DRD3 | D3DR; ETM1; FET1 | Associated with cognitive, emotional, and endocrine functions | |
| DCP2 | NM_001242377 | Decapping mRNA 2 | NUDT20 | Necessary for the degradation of mRNAs | |
| DCST1 | NM_001143687 | DC-STAMP domain containing 1 | RP11-307C12.10-003 | Protein and zinc ion binding | |
| DDIT4 | NM_019058 | DNA-damage-inducible transcript 4 | RP11-442H21.1, Dig2, REDD-1, REDD1 | Inhibits cell growth by regulating the frap1 pathway upstream of the tsc1-tsc2 complex and downstream of Akt1 | |
| DNAJC7 | NM_001144766 | DnaJ (HSP40) homolog, subfamily C, member 7 | DJ11, DJC7, TPR2, TTC2 | Acts as co-chaperone regulating the molecular chaperones HSP70 and HSP90 in folding of steroid receptors | |
| DSCR8 | NM_032589 | Down syndrome critical region gene 8 | C21orf65, CT25.1a, CT25.1b, MMA-1, MMA-1a, MMA-1b, MMA1, MTAG2 | ||
| EIF1 | NM_005801 | Eukaryotic translation initiation factor 1 | A121, EIF-1A, ISO1, SUI1, EIF1 | Necessary for scanning and involved in initiation site selection | |
| EIF2C1 | NM_012199 | Argonaute RISC catalytic component 1 | RP4-789D17.1, EIF2C, AGO1, GERP95, Q99 | Required for RNA-mediated gene silencing | |
| EIF2C3 | NM_024852 | Argonaute RISC catalytic component 3 | AGO3 | Required for RNA-mediated gene silencing | |
| ELAVL1 | NM_001419 | ELAV like RNA binding protein 1 | ELAV1, HUR, Hua, MelG | Binds avidly to the AU-rich element in FOS and IL3/interleukin-3 mRNAs | |
| ENAH | NM_001008493 | Enabled homolog | RP11-496N12.7, ENA, MENA, NDPP1 | Ena/VASP proteins are actin-associated proteins involved in a range of processes dependent on cytoskeleton remodeling and cell polarity | |
| EP300 | NM_001429 | E1A binding protein p300 | RP1-85F18.1, KAT3B, RSTS2, p300 | Functions as HAT and regulates transcription via chromatin remodeling | Yes |
| EPHA5 | NM_001281765 | EPH receptor A5 | CEK7, EHK-1, EHK1, EK7, HEK7, TYRO4 | Receptor for members of the ephrin-A family | |
| ESR1 | NM_000125 | Estrogen receptor 1 | RP1-130E4.1, ER, ESR, ESRA, ESTRR, Era, NR3A1 | Nuclear hormone receptor | |
| EYA4 | NM_001301012 | EYA transcriptional coactivator and phosphatase 4 | RP11-704J17.4, CMD1J, DFNA10 | Tyrosine phosphatase that specifically dephosphorylates “Tyr-142” of histone H2AX (H2AXY142ph) | |
| FAM47B | NM_152631 | Family with sequence similarity 47, member B | RP13-520K9.1 | ||
| FBXO34 | NM_017943 | F-box protein 34 | CGI-301, Fbx34 | Substrate-recognition component of the SCF E3 ubiquitin ligase complex | |
| FKBP10 | NM_021939 | FK506 binding protein 10 | PSEC0056, FKBP65, OI11, OI6, PPIASE, hFKBP65 | PPIases accelerate the folding of proteins during protein synthesis | |
| FKBP4 | NM_002014 | FK506 binding protein 4 | FKBP51, FKBP52, FKBP59, HBI, Hsp56, PPIase, p52 | May play a role in the intracellular trafficking of heterooligomeric forms of steroid hormone receptors | |
| FKBP7 | NM_001135212 | FK506 binding protein 7 | UNQ670/PRO1304, FKBP23, PPIase | PPIases accelerate the folding of proteins during protein synthesis | |
| FRA10AC1 | NM_145246 | Fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 | PRO2972, C10orf4, F26C11.1-like, FRA10A | ||
| FSIP1 | NM_152597 | Fibrous sheath interacting protein 1 | HSD10 | ||
| FXYD6 | NM_001164831 | FXYD domain containing ion transport regulator 6 | UNQ521/PRO1056 | ||
| GADD45G | NM_006705 | Growth arrest and DNA-damage-inducible, γ | RP11-260L6.1, CR6, DDIT2, GADD45γ, GRP17 | Involved in the regulation of growth and apoptosis | |
| GATA6 | NM_005257 | GATA binding protein 6 | Regulates terminal differentiation and/or proliferation | ||
| GCNT1 | NM_001097633 | Glucosaminyl (N-acetyl) transferase 1, core 2 | RP11-214N16.1, C2GNT, C2GNT-L, C2GNT1, G6NT, NACGT2, NAGCT2 | Forms critical branches in O-glycans | |
| GNA13 | NM_001282425 | G protein, α 13 | G13 | Modulators or transducers in various transmembrane signaling systems | |
| GNB1 | NM_001282538 | G protein, β polypeptide 1 | RP1-283E3.7 | A modulator or transducer in various transmembrane signaling systems | |
| GPR137B | NM_003272 | G protein-coupled receptor 137B | RP5-985L19.1, TM7SF1 | ||
| GPR83 | NM_016540 | G protein-coupled receptor 83 | GIR, GPR72 | Orphan receptor. Could be a neuropeptide y receptor | |
| GSTM2 | NM_000848 | Glutathione S-transferase mu 2 (muscle) | GST4, GSTM-2, GTHMUS, GSTM2 | Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles | |
| H1F0 | NM_005318 | H1 histone family, member 0 | H10, H1FV | Histones H1 are necessary for the condensation of nucleosome chains into higher order structures | |
| HERC3 | NM_001271602 | HECT and RLD domain containing E3 ubiquitin protein ligase 3 | E3 ubiquitin-protein ligase | ||
| HEY2 | NM_012259 | Hes-related family bHLH transcription factor with YRPW motif 2 | RP1-293L8.3, CHF1, GRIDLOCK, GRL, HERP1, HESR2, HRT2, bHLHb32 | Downstream effector of Notch signaling which may be required for cardiovascular development | |
| HIPK2 | NM_001113239 | Homeodomain interacting protein kinase 2 | PRO0593 | Protein kinase acting as a corepressor of several transcription factors unwinds double-stranded DNA | |
| HMGB2 | NM_001130688 | High mobility group box 2 | HMG2 | Binds preferentially ssDNA and | |
| HNRPDL | NM_001207000 | Heterogeneous nuclear ribonucleoprotein D-like | HNRNP, HNRPDL, JKTBP, JKTBP2, laAUF1 | Acts as a transcriptional regulator | |
| HOXA10 | NM_018951 | Homeobox A10 | HOX1, HOX1.8, HOX1H, PL | Sequence-specific transcription factor | |
| HOXA11 | NM_005523 | Homeobox A11 | HOX1, HOX1I | Sequence-specific transcription factor | Yes |
| HSD17B3 | NM_000197 | Hydroxysteroid (17-β) dehydrogenase 3 | RP11-240L7.3, EDH17B3, SDR12C2 | Favors the reduction of androstenedione to testosterone | |
| HSP90B1 | NM_003299 | Heat-shock protein 90 kDa β (Grp94), member 1 | ECGP, GP96, GRP94, HEL-S-125m, HEL35, TRA1 | Molecular chaperone that functions in the processing and transport of secreted proteins | |
| HSPA1B | NM_005346 | Heat-shock 70 kDa protein 1B | DAAP-21F2.7, HSP70-1B, HSP70-2 | Stabilizes preexistent proteins against aggregation and mediates the folding of newly translated polypeptides in the cytosol as well as within organelles | |
| ICMT | NM_012405 | Isoprenylcysteine carboxyl methyltransferase | RP1-120G22.4, HSTE14, MST098, MSTP098, PCCMT, PCMT, PPMT | Catalyzes the posttranslational methylation of isoprenylated C-terminal cysteine residues | |
| IDS | NM_000202 | Iduronate 2-sulfatase | MPS2, SIDS | Required for the lysosomal degradation of heparan sulfate and dermatan sulfate | |
| INCENP | NM_001040694 | Inner centromere protein antigens | Component of the chromosomal passenger complex, a complex that acts as a key regulator of mitosis | ||
| IQCG | NM_001134435 | IQ motif containing G | CFAP122, DRC9 | ||
| KAT2B | NM_003884 | K(lysine) acetyltransferase 2B | CAF, P/CAF, PCAF | Functions as a HAT to promote transcriptional activation | |
| KBTBD3 | NM_152433 | Kelch repeat and BTB (POZ) domain containing 3 | BKLHD3 | ||
| KBTBD7 | NM_032138 | Kelch repeat and BTB (POZ) domain containing 7 | |||
| KCTD3 | NM_016121 | Potassium channel tetramerization domain containing 3 | RP11-5F19.1, NY-REN-45 | ||
| KIAA0101 | NM_001029989 | KIAA0101 | L5, NS5ATP9, OEATC, OEATC-1, OEATC1, PAF, PAF15, p15(PAF), p15/PAF, p15PAF | May be involved in protection of cells from UV-induced cell death | |
| KIAA2026 | NM_001017969 | KIAA2026 | |||
| KLHL15 | NM_030624 | Kelch-like family member 15 | HEL-S-305 | Probable substrate-specific adapter of an E3 ubiquitin-protein ligase complex which mediates the ubiq uitination and subsequent proteasomal degradation of target proteins | |
| KLRC4 | NM_013431 | Killer cell lectin-like receptor subfamily C, member 4 | NKG2-F, NKG2F | May play a role as a receptor for the recognition of MHC class I HLA-E molecules by NK cells | |
| KRAS | NM_004985 | Kirsten rat sarcoma viral oncogene homolog | C-K-RAS, CFC2, K-RAS2A, K-RAS2B, K-RAS4A, K-RAS4B, KI-RAS1, KRAS2, NS, NS3, RASK2, KRAS | Binds GDP/GTP and possesses intrinsic GTPase activity | Yes |
| LFNG | NM_001040167 | LFNG O-fucosylpeptide 3-β-N-acetylglucosaminyltransferase | SCDO3 | Glycosyltransferase | |
| LPGAT1 | NM_014873 | Lysophosphatidylglycerol acyltransferase 1 | FAM34A, FAM34A1, NET8 | Lysophoshatidylglycerol-specific acyltransferase | |
| LRRC17 | NM_001031692 | Leucine rich repeat containing 17 | UNQ3076/PRO9909, P37NB | Involved in bone homeostasis, acting as a negative regulator of RANKL-induced osteoclast precursor differentiation from bone marrow precursors | |
| LRRN3 | NM_001099658 | Leucine rich repeat neuronal 3 | Nbla10363, FIGLER5, NLRR-3, NLRR3 | ||
| LYSMD3 | NM_001286812 | LysM, putative peptidoglycan-binding, domain containing 3 | |||
| MAP1B | NM_005909 | Microtubule-associated protein 1B | FUTSCH, MAP5, PPP1R102 | May play a role in the cytoskeletal changes that accompany neurite extension | |
| METAP1 | NM_015143 | Methionyl aminopeptidase 1 | MAP1A, MetAP1A | Removes the amino-terminal methionine from nascent proteins | |
| MFAP3 | NM_001135037 | Microfibrillar-associated protein 3 | Component of the elastin-associated microfibrils | ||
| MIF | NM_002415 | Macrophage migration inhibitory factor (glycosylation-inhibiting factor) | GIF, GLIF, MMIF | The expression of MIF at sites of inflammation suggests a role for the mediator in regulating the function of macrophage in host defense. Also acts as a phenylpyruvate tautomerase | |
| MOB3B | NM_024761 | MOB kinase activator 3B | C9orf35, MOB1D, MOBKL2B | May regulate the activity of kinases | |
| MRPS14 | NM_022100 | Mitochondrial ribosomal protein S14 | DJ262D12.2, HSMRPS14, MRP-S14, S14mt | ||
| MTMR12 | NM_001040446 | Myotubularin-related protein 12 | 3-PAP, PIP3AP | Inactive phosphatase that plays a role as an adapter for the phosphatase myotubularin to regulate myotubularin intracellular location | |
| MTRR | NM_002454 | 5-Methyltetrahydrofolate-homocysteine methyltransferase reductase | MSR, cblE | Involved in the reductive regeneration of cob(I)alamin cofactor required for the maintenance of methionine synthase in a functional state | |
| MYO9A | NM_006901 | Myosin IXA | Myosins are actin-based motor molecules with ATPase activity Unconventional myosins serve in intracellular movements | ||
| NCAPG | NM_022346 | Non-SMC condensin I complex, subunit G | CAPG, CHCG, NY-MEL-3, YCG1 | Regulatory subunit of the condensin complex, a complex required for conversion of interphase chromatin into mitotic-like condense chromosomes | |
| NKX3-2 | NM_001189 | NK3 homeobox 2 | BAPX1, NKX3.2, NKX3B, SMMD | Transcriptional repressor that acts as a negative regulator of chondrocyte maturation | |
| NLK | NM_016231 | Nemo-like kinase | Role in cell fate determination, required for differentiation of bone marrow stromal cells | ||
| NMRK2 | NM_001289117 | Nicotinamide riboside kinase 2 | ITGB1BP3, MIBP, NRK2 | ||
| NOL4 | NM_001198546 | Nucleolar protein 4 | HRIHFB2255, CT125, NOLP | ||
| NUDT12 | NM_001300741 | Nudix-type motif 12 | Hydrolyzes NAD(P)H to NMNH and AMP (2′,5′-ADP), and diadenosine diphosphate to AMP | ||
| OAZ1 | NM_001301020 | Ornithine decarboxylase antizyme 1 | AZI, OAZ | Binds to and destabilizes ornithine decarboxylase, which is then degraded. Also inhibits cellular uptake of polyamines by inactivating the polyamine uptake transporter | |
| OFCC1 | NM_153003 | Orofacial cleft 1 candidate 1 | MRDS1 | ||
| OR11A1 | NM_013937 | Olfactory receptor, family 11, subfamily A, member 1 | DAAP-34I1.2, 6M1-18, OR11A2, dJ994E9.6, hs6M1-18 | Odorant receptor | |
| OTUD1 | NM_001145373 | OTU deubiquitinase 1 | DUBA7, OTDC1 | Deubiquitinating enzyme that specifically hydrolyzes ‘Lys-63′-linked polyubiquitin to monoubiquitin | |
| OTX2 | NM_001270523 | Orthodenticle homeobox 2 | CPHD6, MCOPS5 | Probably plays a role in the development of the brain and the sense organs | |
| PCAF | NM_003884 | K(lysine) acetyltransferase 2B | CAF, P/CAF, PCAF | Functions as a component of the PCAF complex | |
| PCDHB8 | NM_019120 | Protocadherin β 8 | PCDH-β8, PCDH3I | Potential calcium-dependent cell-adhesion protein | |
| PHOX2A | NM_005169 | Paired-like homeobox 2a | ARIX, CFEOM2, FEOM2, NCAM2, PMX2A | May be involved in regulating the specificity of expression of the catecholamine biosynthetic genes | |
| PIM3 | NM_001001852 | Pim-3 proto-oncogene, serine/threonine kinase | CITF22-49E9.1, pim-3 | May be involved in cell cycle progression and antiapoptotic process | |
| PLA2G4C | NM_001159322 | Phospholipase A2, group IVC (cytosolic, calcium-independent) | CPLA2-γ | Has a preference for arachidonic acid at the sn-2 position of phosphatidylcholine as compared with palmitic acid | |
| MTRR | NM_002454 | 5-Methyltetrahydrofolate-homocysteine methyltransferase reductase | MSR, cblE | Involved in the reductive regeneration of cob(I)alamin cofactor required for the maintenance of methionine synthase in a functional state | |
| MYO9A | NM_006901 | Myosin IXA | Myosins are actin-based motor molecules with ATPase activity Unconventional myosins serve in intracellular movements | ||
| NCAPG | NM_022346 | Non-SMC condensin I complex, subunit G | CAPG, CHCG, NY-MEL-3, YCG1 | Regulatory subunit of the condensin complex, a complex required for conversion of interphase chromatin into mitotic-like condense chromosomes | |
| NKX3-2 | NM_001189 | NK3 homeobox 2 | BAPX1, NKX3.2, NKX3B, SMMD | Transcriptional repressor that acts as a negative regulator of chondrocyte maturation | |
| NLK | NM_016231 | Nemo-like kinase | Role in cell fate determination, required for differentiation of bone marrow stromal cells | ||
| NMRK2 | NM_001289117 | Nicotinamide riboside kinase 2 | ITGB1BP3, MIBP, NRK2 | ||
| NOL4 | NM_001198546 | Nucleolar protein 4 | HRIHFB2255, CT125, NOLP | ||
| NUDT12 | NM_001300741 | Nudix-type motif 12 | Hydrolyzes NAD(P)H to NMNH and AMP (2′,5′-ADP), and diadenosine diphosphate to AMP | ||
| OAZ1 | NM_001301020 | Ornithine decarboxylase antizyme 1 | AZI, OAZ | Binds to and destabilizes ornithine decarboxylase, which is then degraded. Also inhibits cellular uptake of polyamines by inactivating the polyamine uptake transporter | |
| OFCC1 | NM_153003 | Orofacial cleft 1 candidate 1 | MRDS1 | ||
| OR11A1 | NM_013937 | Olfactory receptor, family 11, subfamily A, member 1 | DAAP-34I1.2, 6M1-18, OR11A2, dJ994E9.6, hs6M1-18 | Odorant receptor | |
| OTUD1 | NM_001145373 | OTU deubiquitinase 1 | DUBA7, OTDC1 | Deubiquitinating enzyme that specifically hydrolyzes ‘Lys-63′-linked polyubiquitin to monoubiquitin | |
| OTX2 | NM_001270523 | Orthodenticle homeobox 2 | CPHD6, MCOPS5 | Probably plays a role in the development of the brain and the sense organs | |
| PCAF | NM_003884 | K(lysine) acetyltransferase 2B | CAF, P/CAF, PCAF | Functions as a component of the PCAF complex | |
| PCDHB8 | NM_019120 | Protocadherin β 8 | PCDH-β8, PCDH3I | Potential calcium-dependent cell-adhesion protein | |
| PHOX2A | NM_005169 | Paired-like homeobox 2a | ARIX, CFEOM2, FEOM2, NCAM2, PMX2A | May be involved in regulating the specificity of expression of the catecholamine biosynthetic genes | |
| PIM3 | NM_001001852 | Pim-3 proto-oncogene, serine/threonine kinase | CITF22-49E9.1, pim-3 | May be involved in cell cycle progression and antiapoptotic process | |
| PLA2G4C | NM_001159322 | Phospholipase A2, group IVC (cytosolic, calcium-independent) | CPLA2-γ | Has a preference for arachidonic acid at the sn-2 position of phosphatidylcholine as compared with palmitic acid | |
| PLAG1 | NM_001114634 | Pleiomorphic adenoma gene 1 | PSA, SGPA, ZNF912 | Transcription factor whose activation results in upregulation of target genes, such as IGFII, leading to uncontrolled cell proliferation | Yes |
| PLCL2 | NM_001144382 | Phospholipase C-like 2 | PLCE2 | May play a role in the regulation of Ins(1,4,5)P3 around the endoplasmic reticulum | |
| PLXDC2 | NM_001282736 | Plexin domain containing 2 | UNQ2514/PRO6003, TEM7R | May play a role in tumor angiogenesis | |
| PNPT1 | NM_033109 | Polyribonucleotide nucleotidyltransferase 1 | COXPD13, DFNB70, OLD35, PNPASE, old-35 | Involved in mRNA degradation | |
| POLR2B | NM_000938 | Polymerase (RNA) II (DNA directed) polypeptide B | POL2RB, RPB2, hRPB140, hsRPB2 | DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucle oside triphosphates as substrates | |
| PPA1 | NM_021129 | Pyrophosphatase (inorganic) 1 | RP11-367H5.1, HEL-S-66p, IOPPP, PP, PP1, SID6-8061 | ||
| PPM1A | NM_021003 | Protein phosphatase, Mg2+/Mn2+ dependent, 1A | PP2C-ALPHA, PP2CA, PP2Cα | Enzyme with a broad specificity | |
| PPP2CA | NM_002715 | Protein phosphatase 2, catalytic subunit, α isozyme | PP2Ac, PP2CA, PP2Cα, RP-C | PP2A can modulate the activity of phosphorylase B kinase casein kinase 2, mitogen-stimulated S6 kinase, and MAP-2 kinase | |
| PPP2R5C | NM_001161725 | Protein phosphatase 2, regulatory subunit B′, γ | B56G, PR61G | The B regulatory subunit might modu late substrate selectivity and catalytic activity, and also might direct the localization of the catalytic enzyme to a particular subcellular compartment | |
| PRDX3 | NM_006793 | Peroxiredoxin 3 | AOP-1, AOP1, HBC189, MER5, PRO1748, SP-22, prx-III | Involved in redox regulation of the cell | |
| PRLR | NM_000949 | Prolactin receptor | HPRL, MFAB, hPRLrI | This is a receptor for the anterior pituitary hormone prolactin | |
| PROSC | NM_007198 | Proline synthetase co-transcribed homolog (bacterial) | |||
| PROX1 | NM_001270616 | Prospero homeobox 1 | May play a fundamental role in early development of central nervous system | ||
| PRR4 | NM_001098538 | Proline rich 4 (lacrimal) | LPRP, PROL4 | ||
| PTGS2 | NM_000963 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | May have a role as a major mediator of inflammation and/or a role for prostanoid signaling in activity-dependent plasticity | |
| PTPLAD1 | NM_016395 | Protein tyrosine phosphatase-like A domain containing 1 | B-IND1, HACD3, HSPC121 | Involved in Rac1-signaling pathways leading to the modulation of gene expression | |
| PTPRZ1 | NM_001206838 | Protein tyrosine phosphatase, receptor-type, Z polypeptide 1 | HPTPZ, HPTPζ, PTP-ζ, PTP18, PTPRZ, PTPZ, R-PTP-ζ-2, RPTPB, RPTPβ, phosphacan | May be involved in the regulation of specific developmental processes in the central nervous system | |
| RAB8B | NM_016530 | RAB8B, member RAS oncogene family | May be involved in vesicular trafficking and neurotransmitter release | ||
| RASSF6 | NM_001270391 | Ras association (RalGDS/AF-6) domain family member 6 | May act as a Ras effector protein | ||
| RBM15 | NM_001201545 | RNA binding motif protein 15 | OTT, OTT1, SPEN | May be implicated in HOX gene regulation | |
| RLF | NM_012421 | Rearranged L-myc fusion | RP1-39G22.1, ZN-15L, ZNF292L | May be involved in transcriptional regulation | |
| ROPN1L | NM_001201466 | Rhophilin associated tail protein 1-like | RP11-1C1.7, ASP, RSPH11 | ||
| RPS14 | NM_001025070 | Ribosomal protein S14 | PRO2640, EMTB, S14 | ||
| RTEL1-TNFRSF6B | NR_037882 | RTEL1-TNFRSF6B readthrough (NMD candidate) | |||
| S100A1 | NM_006271 | S100 calcium binding protein A1 | RP1-178F15.1, S100, S100-α, S100A | Weakly binds calcium but binds zinc very tightly-distinct binding sites with different affinities exist for both ions on each monomer | |
| SCAMP2 | NM_005697 | Secretory carrier membrane protein 2 | Functions in post-Golgi recycling pathways. Acts as a recycling carrier to the cell surface | ||
| SEPT2 | NM_001008491 | Septin 2 | DIFF6, NEDD-5, NEDD5, Pnutl3, hNedd5 | Required for normal progress through mitosis. Involved in cytokinesis | |
| SF3B3 | NM_012426 | Splicing factor 3b, subunit 3 | RSE1, SAP130, SF3b130, STAF13 | Subunit of the splicing factor SF3B required for ‘A’ complex assembly formed by the stable binding of U2 snRNP to the branch point sequence in pre-mRNA | |
| SH3BGRL | NM_003022 | SH3 domain binding glutamate-rich protein like | HEL-S-115, SH3BGR | ||
| SIX6 | NM_007374 | SIX homeobox 6 | MCOPCT2, OPTX2, Six9 | May be involved in eye development | |
| SLC37A3 | NM_001287498 | Solute carrier family 37, member 3 | |||
| SLC7A11 | NM_014331 | Solute carrier family 7 (anionic amino acid transporter light chain, xc-system), member 11 | CCBR1, xCT | Sodium-independent, high-affinity exchange of anionic amino acids with high specificity for anionic form of cystine and glutamate | |
| SLCO2A1 | NM_005630 | Solute carrier organic anion transporter family, member 2A1 | MATR1, OATP2A1, PGT, PHOAR2, SLC21A2 | May mediate the release of newly synthesized prostaglandins from cells, the transepithelial transport of prostaglandins, and the clearance of prostaglandins from the circulation | |
| SMAD5 | NM_001001419 | SMAD family member 5 | DWFC, JV5-1, MADH5 | Transcriptional modulator activated by BMP type 1 receptor kinase | |
| SMCHD1 | NM_015295 | Structural maintenance of chromosomes flexible hinge domain containing 1 | Required for maintenance of X inactivation in females and hypermethylation of CpG islands associated with inactive X | ||
| SNAI2 | NM_003068 | Snail family zinc finger 2 | SLUG, SLUGH1, SNAIL2, WS2D | Transcriptional repressor. Involved in the generation and migration of neural crest cells | |
| SRPK2 | NM_001278273 | SRSF protein kinase 2 | SFRSK2 | Phosphorylates RS domain-containing proteins | |
| TAAR6 | NM_175067 | Trace amine associated receptor 6 | RP11-295F4.3, TA4, TAR4, TAR6, TRAR4, taR-4, taR-6 | Orphan receptor. Could be a receptor for trace amines | |
| TAF15 | NM_003487 | TAF15 RNA polymerase II, TBP-associated factor | Npl3, RBP56, TAF2N, TAFII68 | RNA and ssDNA-binding protein that may play specific roles during transcription initiation at distinct promoters | Yes |
| TAF2 | NM_003184 | TAF2 RNA polymerase II, TBP-associated factor | CIF150, MRT40B, TAFII150, TAF2 | Transcription factor TFIID is one of the general factors required for accurate and regulated initiation by RNA polymerase II | |
| TAF6L | NM_006473 | TAF6-like RNA PCAF-associated factor | PAF65A | Functions as a component of the PCAF complex | |
| TBX4 | NM_018488 | T-box 4 | SPS | Involved in the transcriptional regulation of genes required for mesoderm differentiation | |
| TCF21 | NM_003206 | Transcription factor 21 | POD1, bHLHa23 | Involved in epithelial–mesenchymal interactions in kidney and lung morphogenesis that include epithelial differentiation and branching morphogenesis | |
| THUMPD1 | NM_017736 | THUMP domain containing 1 | |||
| TM9SF3 | NM_020123 | Transmembrane 9 superfamily member 3 | RP11-34E5.1, EP70-P-iso, SMBP | ||
| TMEM14A | NM_014051 | Transmembrane protein 14A | PTD011, C6orf73 | ||
| TMEM45A | NM_018004 | Transmembrane protein 45A | DERP7 | ||
| TMPO | NM_001032283 | Thymopoietin | CMD1T, LAP2, LEMD4, PRO0868, TP | May help direct the assembly of the nuclear lamina and thereby help maintain the structural organization of the nuclear envelope | |
| TMPRSS11A | NM_001114387 | Transmembrane protease, serine 11A | ECRG1 | Probable serine protease, which may play a role in cellular senescence | |
| TNPO1 | NM_002270 | Transportin 1 | IPO2, KPNB2, MIP, MIP1, TRN | Functions in nuclear protein import as nuclear transport receptor | |
| TNRC6C | NM_001142640 | Trinucleotide repeat containing 6C | Plays a role in RNA-mediated gene silencing by micro-RNAs | ||
| TRUB1 | NM_139169 | TruB pseudouridine (psi) synthase family member 1 | PUS4 | May be responsible for synthesis of psi from uracil in transfer RNAs | |
| TSG101 | NM_006292 | Tumor susceptibility 101 | TSG10, VPS23 | Component of the ESCRT-I complex, a regulator of vesicular trafficking process | |
| TSHR | NM_000369 | Thyroid stimulating hormone receptor | CHNG1, LGR3, hTSHR-I | Receptor for thyrothropin. Plays a central role in controlling thyroid cell metabolism | Yes |
| TUSC1 | NM_001004125 | Tumor suppressor candidate 1 | TSG-9, TSG9 | ||
| TWF1 | NM_001242397 | Twinfilin actin-binding protein 1 | A6, PTK9 | Actin-binding protein involved in motile and morphological processes | |
| UGT3A1 | NM_001171873 | UDP glycosyltransferase 3 family, polypeptide A1 | UDP-glucuronosyltransferases catalyze phase II biotransformation reactions | ||
| USP28 | NM_001301029 | Ubiquitin specific peptidase 28 | Deubiquitinase involved in DNA damage response checkpoint and MYC proto-oncogene stability | ||
| VBP1 | NM_003372 | Von Hippel-Lindau binding protein 1 | RP13-228J13.4, PFD3, PFDN3, VBP-1 | Binds specifically to c-CPN and transfers target proteins to it | |
| WDR33 | NM_001006622 | WD repeat domain 33 | NET14, WDC146 | Essential for both cleavage and polyadenylation of pre-mRNA 3′ ends | |
| WNT16 | NM_016087 | Wingless-type MMTV integration site family, member 16 | Ligand for members of the Frizzled family of seven transmembrane receptors | ||
| WNT2 | NM_003391 | Wingless-type MMTV integration site family member 2 | INT1L1, IRP | Ligand for members of the Frizzled family of seven transmembrane receptors | |
| WNT3A | NM_033131 | Wingless-type MMTV integration site family, member 3A | Ligand for members of the Frizzled family of seven transmembrane receptors | ||
| YY1 | NM_003403 | YY1 transcription factor | DELTA, INO80S, NF-E1, UCRBP, YIN-YANG-1 | May play an important role in development and differentiation | |
| ZIC2 | NM_007129 | Zinc family member 2 | HPE5 | Involved in cerebellar development | |
| ZNF12 | NM_006956 | Zinc finger protein 12 | GIOT-3, HZF11, KOX3, ZNF325 | May be involved in transcriptional regulation | |
| ZNF121 | NM_001008727 | Zinc finger protein 121 | D19S204, ZHC32, ZNF20 | May be involved in transcriptional regulation | |
| ZNF132 | NM_003433 | Zinc finger protein 132 | pHZ-12 | May be involved in transcriptional regulation | |
| ZNF180 | NM_001278508 | Zinc finger protein 180 | HHZ168 | May be involved in transcriptional regulation | |
| ZNF238 | NM_001278196 | Zinc finger and BTB domain containing 18 | C2H2-171, MRD22, RP58, TAZ-1, ZNF18 | Sequence-specific DNA-binding protein with transcriptional repression activity | |
| ZNF25 | NM_145011 | Zinc finger protein 25 | KOX19, Zfp9 | May be involved in transcriptional regulation | |
| ZNF30 | NM_001099437 | Zinc finger protein 30 | KOX28 | May be involved in transcriptional regulation | |
| ZNF426 | NM_001300883 | Zinc finger protein 426 | May be involved in transcriptional regulation | ||
| ZNF558 | NM_144693 | Zinc finger protein 558 | May be involved in transcriptional regulation | ||
| ZNF562 | NM_001130031 | Zinc finger protein 562 | May be involved in transcriptional regulation | ||
| ZNF564 | NM_144976 | Zinc finger protein 564 | May be involved in transcriptional regulation | ||
| ZNF594 | NM_032530 | Zinc finger protein 594 | hCG_1775942 | May be involved in transcriptional regulation | |
| ZNF644 | NM_016620 | Zinc finger protein 644 | BM-005, MYP21, NatF, ZEP-2 | May be involved in transcriptional regulation | |
| ZNF652 | NM_001145365 | Zinc finger protein 652 | Functions as a transcriptional repressor | ||
| ZNF700 | NM_001271848 | Zinc finger protein 700 | May be involved in transcriptional regulation | ||
| ZNF703 | NM_025069 | Zinc finger protein 703 | ZEPPO1, ZNF503L, ZPO1 | May function as a transcriptional repressor | |
| ZNF711 | NM_021998 | Zinc finger protein 711 | CMPX1, MRX97, ZNF4, ZNF5, ZNF6, Zfp711, dJ75N13.1 | May be involved in transcriptional regulation | |
| ZNF763 | NM_001012753 | Zinc finger protein 763 | ZNF, ZNF440L | May be involved in transcriptional regulation | |
| ZNF780A | NM_001010880 | Zinc finger protein 780A | ZNF780 | May be involved in transcriptional regulation | |
Abbreviations: CLL, chronic lymphocytic leukemia; HLA, human leukocyte antigen; HSP, heat shock protein; IGF, insulin-like growth factor; IL, interleukin; mRNA, messenger RNA; NK cells, natural killer cells; ssDNA, single-stranded DNA; UV, ultraviolet.
As shown in Table 6, our DAVID analysis showed that there were 16 functional clusters that were identified to be enriched with an enrichment score >1.0 in the target list of hsa-miR-181a-5p, based on TarBase. The functions of these clusters involved negative regulation of macromolecule biosynthetic process, negative regulation of the cellular bio-synthetic process, negative regulation of biosynthetic process, negative regulation of apoptosis, negative regulation of programmed cell death, negative regulation of cell death, negative regulation of transcription, negative regulation of nucleobase, nucleoside, nucleotide, and nucleic acid metabolic processes, negative regulation of nitrogen compound metabolic process, and lung and respiratory tube development, positive regulation of protein polymerization, positive regulation of protein complex assembly, positive regulation of protein polymerization, and positive regulation of protein complex assembly.
Table 6.
The top enriched clusters (enrich score >1) by DAVID for the targets of hsa-miR-181a-5p from TarBase 6.0
| Category | Term | Gene count | P-value | FDR |
|---|---|---|---|---|
| Annotation cluster 1 | Enrichment score: 4.3 | |||
| INTERPRO | Zinc finger, C2H2-type | 24 | 2.20E-05 | 4.60E-03 |
| INTERPRO | Zinc finger, C2H2-like | 24 | 2.90E-05 | 3.90E-03 |
| SMART | Zinc finger_C2H2 | 24 | 2.00E-04 | 1.90E-02 |
| Annotation cluster 2 | Enrichment score: 3.04 | |||
| GOTERM_BP_FAT | Negative regulation of macromolecule biosynthetic process | 17 | 7.10E-04 | 9.80E-02 |
| GOTERM_BP_FAT | Negative regulation of cellular biosynthetic process | 17 | 9.30E-04 | 1.20E-01 |
| GOTERM_BP_FAT | Negative regulation of biosynthetic process | 17 | 1.20E-03 | 9.50E-02 |
| Annotation cluster 3 | Enrichment score: 2.97 | |||
| GOTERM_BP_FAT | Negative regulation of apoptosis | 13 | 9.70E-04 | 1.10E-01 |
| GOTERM_BP_FAT | Negative regulation of programmed cell death | 13 | 1.10E-03 | 1.00E-01 |
| GOTERM_BP_FAT | Negative regulation of cell death | 13 | 1.10E-03 | 9.70E-02 |
| Annotation cluster 4 | Enrichment score: 2.67 | |||
| GOTERM_BP_FAT | Negative regulation of transcription | 15 | 1.00E-03 | 1.00E-01 |
| GOTERM_BP_FAT | Negative regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 15 | 2.90E-03 | 1.50E-01 |
| GOTERM_BP_FAT | Negative regulation of nitrogen compound metabolic process | 15 | 3.20E-03 | 1.50E-01 |
| Annotation cluster 5 | Enrichment score: 2.14 | |||
| GOTERM_BP_FAT | Lung development | 6 | 6.20E-03 | 2.30E-01 |
| GOTERM_BP_FAT | Respiratory tube development | 6 | 7.00E-03 | 2.40E-01 |
| GOTERM_BP_FAT | Respiratory system development | 6 | 8.90E-03 | 2.50E-01 |
| Annotation cluster 6 | Enrichment score: 1.75 | |||
| GOTERM_BP_FAT | Positive regulation of protein polymerization | 4 | 3.00E-03 | 1.50E-01 |
| GOTERM_BP_FAT | Positive regulation of protein complex assembly | 4 | 7.90E-03 | 2.60E-01 |
| GOTERM_BP_FAT | Regulation of protein polymerization | 4 | 4.60E-02 | 4.90E-01 |
| GOTERM_BP_FAT | Regulation of protein complex assembly | 4 | 9.00E-02 | 6.00E-01 |
| Annotation cluster 7 | Enrichment score: 1.59 | |||
| GOTERM_BP_FAT | Regulation of phosphorylation | 12 | 2.20E-02 | 3.50E-01 |
| GOTERM_BP_FAT | Regulation of phosphate metabolic process | 12 | 2.80E-02 | 3.90E-01 |
| GOTERM_BP_FAT | Regulation of phosphorus metabolic process | 12 | 2.80E-02 | 3.90E-01 |
| Annotation cluster 8 | Enrichment score: 1.57 | |||
| INTERPRO | Secreted growth factor Wnt protein, conserved site | 3 | 1.90E-02 | 7.90E-01 |
| INTERPRO | Secreted growth factor Wnt protein | 3 | 1.90E-02 | 7.90E-01 |
| INTERPRO | Wnt superfamily | 3 | 1.90E-02 | 7.90E-01 |
| PIR_SUPERFAMILY | PIRSF001784:int-1 transforming protein | 3 | 2.00E-02 | 8.60E-01 |
| SMART | Wnt1 | 3 | 2.50E-02 | 5.50E-01 |
| GOTERM_BP_FAT | Wnt receptor signaling pathway, calcium modulating pathway | 3 | 2.50E-02 | 3.70E-01 |
| KEGG_PATHWAY | Basal cell carcinoma | 3 | 1.40E-01 | 7.00E-01 |
| Annotation cluster 9 | Enrichment score: 1.56 | |||
| GOTERM_BP_FAT | Positive regulation of microtubule polymerization | 3 | 8.40E-03 | 2.50E-01 |
| GOTERM_BP_FAT | Regulation of microtubule polymerization | 3 | 9.80E-03 | 2.60E-01 |
| GOTERM_BP_FAT | Positive regulation of microtubule polymerization or depolymerization | 3 | 9.80E-03 | 2.60E-01 |
| GOTERM_BP_FAT | Regulation of microtubule polymerization or depolymerization | 3 | 5.10E-02 | 5.10E-01 |
| GOTERM_BP_FAT | Regulation of microtubule cytoskeleton organization | 3 | 8.70E-02 | 6.00E-01 |
| GOTERM_BP_FAT | Regulation of microtubule-based process | 3 | 1.10E-01 | 6.60E-01 |
| Annotation cluster 10 | Enrichment score: 1.49 | |||
| GOTERM_BP_FAT | Positive regulation of cytoskeleton organization | 5 | 1.90E-03 | 1.20E-01 |
| GOTERM_BP_FAT | Positive regulation of organelle organization | 5 | 1.60E-02 | 3.30E-01 |
| GOTERM_BP_FAT | Regulation of cytoskeleton organization | 5 | 7.60E-02 | 5.80E-01 |
| GOTERM_BP_FAT | Regulation of cellular component biogenesis | 5 | 8.60E-02 | 6.00E-01 |
| GOTERM_BP_FAT | Positive regulation of cellular component organization | 5 | 1.60E-01 | 7.30E-01 |
| Annotation cluster 11 | Enrichment score: 1.36 | |||
| UP_SEQ_FEATURE | DNA-binding region: Homeobox | 7 | 1.60E-02 | 4.70E-01 |
| INTERPRO | Homeobox, conserved site | 7 | 4.60E-02 | 9.10E-01 |
| INTERPRO | Homeobox | 7 | 4.90E-02 | 9.00E-01 |
| INTERPRO | Homeodomain-related | 7 | 5.10E-02 | 8.60E-01 |
| SMART | Hox | 7 | 8.50E-02 | 7.60E-01 |
| UP_SEQ_FEATURE | Domain: BTB | 6 | 1.80E-02 | 4.90E-01 |
| INTERPRO | BTB/POZ-like | 6 | 5.10E-02 | 8.80E-01 |
| INTERPRO | BTB/POZ fold | 6 | 5.30E-02 | 8.40E-01 |
| SMART | Btb | 6 | 8.40E-02 | 8.10E-01 |
| Annotation cluster 13 | Enrichment score: 1.31 | |||
| INTERPRO | Peptidyl-prolyl cis-trans isomerase, FKBP-type | 3 | 2.10E-02 | 7.60E-01 |
| SP_PIR_KEYWORDS | Rotamase | 3 | 5.90E-02 | 5.00E-01 |
| GOTERM_MF_FAT | Peptidyl-prolyl cis-trans isomerase activity | 3 | 6.50E-02 | 9.50E-01 |
| GOTERM_MF_FAT | cis-trans Isomerase activity | 3 | 7.10E-02 | 9.30E-01 |
| Annotation cluster 14 | Enrichment score: 1.27 | |||
| GOTERM_BP_FAT | Regulation of apoptosis | 16 | 5.10E-02 | 5.10E-01 |
| GOTERM_BP_FAT | Regulation of programmed cell death | 16 | 5.50E-02 | 5.20E-01 |
| GOTERM_BP_FAT | Regulation of cell death | 16 | 5.60E-02 | 5.20E-01 |
| Annotation cluster 15 | Enrichment score: 1.26 | |||
| GOTERM_BP_FAT | Positive regulation of protein kinase activity | 7 | 4.80E-02 | 5.00E-01 |
| GOTERM_BP_FAT | Positive regulation of kinase activity | 7 | 5.50E-02 | 5.20E-01 |
| GOTERM_BP_FAT | Positive regulation of transferase activity | 7 | 6.40E-02 | 5.40E-01 |
| Annotation cluster 16 | Enrichment score: 1.09 | |||
| SP_PIR_KEYWORDS | RNA-mediated gene silencing | 3 | 3.30E-02 | 4.10E-01 |
| GOTERM_BP_FAT | Gene silencing by RNA | 3 | 5.70E-02 | 5.20E-01 |
| GOTERM_BP_FAT | Gene silencing | 3 | 1.40E-01 | 6.90E-01 |
| SP_PIR_KEYWORDS | Translation regulation | 3 | 1.60E-01 | 7.60E-01 |
Abbreviations: DAVID, Database for Annotation, Visualization and Integrated Discovery; FDR, false discovery rate.
Furthermore, our DAVID analysis revealed that there were nine KEGG pathways significantly enriched in the target list of hsa-miR-181a-5p, based on TarBase (Table 7). These pathways included pathways in cancer pathways (Figure 1), the Wnt signaling pathway (Figure 2), prostate cancer, melanogenesis, cell cycle (Figure 3), hedgehog signaling pathway, p53 signaling pathway (Figure 4), small cell lung cancer, and thyroid cancer.
Table 7.
The KEGG pathways by DAVID, for the target list of hsa-miR-181a-5p based on TarBase 6.0
| Signaling pathway | Gene count | % | P-value | FDR |
|---|---|---|---|---|
| Pathways in cancer | 11 | 0.5 | 4.50E-03 | 1.30E-01 |
| Wnt signaling pathway | 8 | 0.4 | 1.80E-03 | 1.50E-01 |
| Prostate cancer | 6 | 0.3 | 3.80E-03 | 1.60E-01 |
| Melanogenesis | 5 | 0.2 | 2.90E-02 | 4.10E-01 |
| Cell cycle | 5 | 0.2 | 5.90E-02 | 5.00E-01 |
| Hedgehog signaling pathway | 4 | 0.2 | 2.80E-02 | 4.70E-01 |
| p53 signaling pathway | 4 | 0.2 | 4.60E-02 | 4.50E-01 |
| Small cell lung cancer | 4 | 0.2 | 7.60E-02 | 5.50E-01 |
| Thyroid cancer | 3 | 0.1 | 4.50E-02 | 5.00E-01 |
Abbreviations: DAVID, Database for Annotation, Visualization and Integrated Discovery; FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 1.
Cancer pathways in the target list of hsa-miR-181a-5p, based on TarBase 6.0.
Notes: Several important oncogenes and tumor suppressors are likely regulated by hsa-miR-181a-5p (marked with a red star), including Wnt, axin, CBP, Bcl-2, p27, cyclin D1, Ras, and HSP. These genes play an important role in the regulation of angiogenesis, cell proliferation, apoptosis, and metastasis.
Figure 2.
Wnt signaling pathway in the target list of hsa-miR-181a-5p, based on TarBase 6.0.
Notes: hsa-181a-5p is a regulator of Wnt. Three Wnt signaling pathways have been characterized: the canonical Wnt pathway, the noncanonical planar cell polarity pathway, and the noncanonical Wnt/calcium pathway.103 All three Wnt signaling pathways are activated by the binding of a Wnt-protein ligand to a Frizzled family receptor, which passes the biological signal to the protein Dishevelled inside the cell. The canonical Wnt pathway leads to regulation of gene transcription, the noncanonical planar cell polarity pathway regulates the cytoskeleton that is responsible for the shape of the cell, and the noncanonical Wnt/calcium pathway regulates calcium level inside the cell. Wnt signaling pathways are highly evolutionarily conserved. Wnt signaling has been implicated in the development of breast cancer, EC, and other types of cancer.103,104 Changes in CTNNB1 expression, which is the gene that encodes β-catenin, can be measured in, not just breast cancer but also, colorectal cancer, melanoma, prostate cancer, lung cancer, EC, and several other cancer types. Increased expression of Wnt ligand-proteins, such as Wnt 1, Wnt2, and Wnt7A, has been observed in the development of glioblastoma, esophageal cancer, EC, and ovarian cancer. There is clinical and experimental evidence that Wnt/β-catenin pathways are deregulated and play an important role in the initiation, development, growth, and metastasis of EC.105–109 Targets of hsa-miR-181a-5p are marked with a red star.
Abbreviation: EC, endometrial cancer.
Figure 3.
Cell cycle pathway in the target list of hsa-miR-181a-5p, based on TarBase 6.0.
Notes: hsa-miR-181a-5p can regulate a number of important proteins that control the cell cycle. Control of eukaryotic cell growth and division involves molecular circuits known as “checkpoints” that ensure proper timing of cellular events. Passage through a checkpoint from one cell cycle phase to the next needs a coordinated set of proteins that monitor cell growth and DNA integrity. Uncontrolled cell division or propagation of damaged DNA can contribute to genomic instability and tumorigenesis.110 The G1/S checkpoint controls progression of cells through the restriction point (R) into the DNA synthesis S phase. During G1, the tumor suppressor Rb binds and inhibits transcription factor E2F. Phosphorylation of Rb by CDKs in late G1 induces dissociation of Rb and permits E2F-mediated transcription of S phase-promoting genes. Responding to upstream signals, INK4 and Kip/Cip family inhibitors control CDK activity and prevent entry into S phase.110 DNA damage activates response pathways through ATM/ATR and Chk1/2 kinases to block CDK activity, leading to cell cycle arrest and DNA repair or cell death. The G2/M checkpoint prevents cells containing damaged DNA from entering mitosis. Activated CDK1/Cdc2 bound to cyclin B promotes entry into M phase. Wee1 and Myt1 kinases and Cdc25 phosphatase competitively regulate CDK1 activity; Wee1 and Myt1 inhibit CDK1 and prevent entry into M phase, while Cdc25 removes inhibitory phosphates. DNA damage activates multiple kinases that phosphorylate kinases Chk1/2 and tumor suppressor protein p53. Chk1/2 kinases stimulate Wee1 activity and inhibit Cdc25C, preventing entry into M phase. Phosphorylation of p53 promotes dissociation between p53 and MDM2 and allows binding of the transcription factor to DNA. In addition, the spindle checkpoint ensures proper chromatid attachment prior to progression from metaphase to anaphase.110 The SCF and APC/C protein complexes play prominent roles, with APC-Cdc20 initiating the entry into anaphase by promoting ubiquitin-mediated degradation of multiple substrates, including cyclin B and the regulatory protein securing. Targets of hsa-miR-181a-5p are marked with a red star.
Figure 4.
p53 pathway in the target list of hsa-miR-181a-5p, based on TarBase 6.0.
Notes: hsa-miR-181a-5p can regulate the p53 signaling pathway. p53 is situated at the crossroads of a network of signaling pathways that are essential for cell growth regulation and apoptosis induced by genotoxic and nongenotoxic stresses. In normal unstressed cells, the level of p53 protein is downregulated via the binding of proteins such as MDM2, COP1, PIRH2, or JNK, which promote p53 degradation via the ubiquitin/proteasome pathway. As most of these genes are upregulated by p53, this leads to a regulation loop that will keep p53 level very low in normal cells. After exposure to genotoxic or nongenotoxic stresses, p53 protein level is increased via the inhibition of its interaction with MDM2 and the other negative regulators. A series of modulators, such as kinases and acetylases, will activate p53 transcriptional activity. Regardless of the type of stress, the final outcome of p53 activation is either cell cycle arrest and DNA repair or apoptosis. Dysfunctional p53 due to mutations will lead to tumorigenesis. p53 mutations can be found in 50% of human cancers including EC, but their penetrance is highly heterogeneous. Mutations in various pathways upstream of p53 (eg, ATM, p19ARF, or Hcdk2 gene) can be observed in various types of cancer, including EC. Targets of hsa-miR-181a-5p are marked with a red star.
Abbreviation: EC, endometrial cancer.
Validated targets of hsa-miR-181a-5p based on miRTarBase
Based on miRTarBase, 241 targets of hsa-miR-181a-5p have been validated with experimental evidence (Table 8). These included ACOT12, AFTPH, AKAP12, AMMECR1, ANKRD1, ANKRD13C, AP1M1, ARF6, ARHGAP12, ARL6IP6, ATF7IP2, ATG10, ATM, ATP6V0E1, ATP8A1, BAG2, BCL2, BCL2L11, BDNF, BPGM, BRCA1, BRMS1L, BTBD3, C1orf109, C1QTNF9, C8A, CCDC6, CCNG1, CD46, and CDKN1B. Among these validated targets, only 18 are cancer genes (7.47%), including ATM, BCL2, CCDC6, CDX2, FBXO11, H3F3B, HOOK3, HOXA11, HRAS, KRAS, MAP2K1, NOTCH1, NOTCH2, PLAG1, PTPN11, STAG2, TAF15, and TSHR (Table 8). Only half of these cancer genes have been included in TarBase.
Table 8.
Targets of hsa-miR-181a-5p with experimental evidence based on miRTarBase 4.0
| Gene symbol | Accession | Full name | Alias | Function | Cancer gene |
|---|---|---|---|---|---|
| ACOT12 | NM_130767 | Acyl-CoA thioesterase 12 | CACH-1, Cach, STARD15, THEAL | Hydrolyzes acetyl-CoA to acetate and CoA | |
| AFTPH | NM_001002243 | Aftiphilin | Nbla10388 | May play a role in membrane trafficking | |
| AKAP12 | NM_005100 | A kinase anchor protein 12 | AKAP250, SSeCKS | Anchoring protein that mediates the subcellular compartmentation of PKA and PKC | |
| AMMECR1 | NM_001025580 | Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 | RP13-360B22.1, AMMERC1 | ||
| ANKRD1 | NM_014391 | Ankyrin repeat domain 1 (cardiac muscle) | ALRP, C-193, CARP, CVARP, MCARP, bA320F15.2 | Plays an important role in endothelial cell activation | |
| ANKRD13C | NM_030816 | Ankyrin repeat domain 13C | RP4-677H15.5, dJ677H15.3 | ||
| AP1M1 | NM_001130524 | Adaptor-related protein complex 1, mu 1 subunit | AP47, CLAPM2, CLTNM, MU-1A | Subunit of clathrin-associated adaptor protein complex 1 that plays a role in protein sorting in the trans-Golgi network and endosomes | |
| ARF6 | NM_001663 | ADP-ribosylation factor 6 | Involved in protein trafficking | ||
| ARHGAP12 | NM_001270695 | Rho GTPase activating protein 12 | GTPase activator for the Rho-type GTPases by converting them to an inactive GDP-bound state | ||
| ARL6IP6 | NM_022989 | ADP-ribosylation factor-like 6 interacting protein 6 | RP23-265N10.1, 2310057C01Rik, 2610529A11Rik, Aip-6 | May be involved in protein transport, membrane trafficking, or cell signaling during hematopoietic maturation | |
| ATF7IP2 | NM_001256160 | Activating transcription factor 7 interacting protein 2 | MCAF2 | Recruiter that couples transcriptional factors to general transcription apparatus and thereby modulates transcription regulation and chromatin formation | |
| ATG10 | NM_001131028 | Autophagy related 10 | PP12616, APG10, APG10L, pp12616 | Plays a role in autophagy | |
| ATM | NM_000051 | ATM serine/threonine kinase | AT1, ATA, ATC, ATD, ATDC, ATE, TEL1, TELO1 | Serine/threonine protein kinase | Yes |
| ATP6V0E1 | NM_003945 | ATPase, H+ transporting, lysosomal 9 kDa, V0 subunit e1 | ATP6H, ATP6V0E, M9.2, Vma21, Vma21p | Vacuolar ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells | |
| ATP8A1 | NM_001105529 | ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 | ATPASEII, ATPIA, ATPP2 | May play a role in the transport of aminophospholipids from the outer to the inner leaflet of various membranes and the maintenance of asymmetric distribution of phospholipids | |
| BAG2 | NM_004282 | BCL2-associated athanogene 2 | RP3-496N17.2, BAG-2, dJ417I1.2 | Inhibits the chaperone activity of HSP70/HSC70 by promoting substrate release | |
| BCL2 | NM_000633 | B-cell CLL/lymphoma 2 | Bcl-2, PPP1R50 | Suppresses apoptosis | Yes |
| BCL2L11 | NM_001204106 | BCL2-like 11 | BAM, BIM, BOD | Induces apoptosis | |
| BDNF | NM_001143805 | Brain-derived neurotrophic factor | ANON2, BULN2 | Promotes the survival of neuronal populations | |
| BPGM | NM_001293085 | 2,3-bisphosphoglycerate mutase | DPGM | Plays a major role in regulating hemoglobin oxygen affinity | |
| BRCA1 | NM_007294 | Breast cancer 1, early onset | BRCAI, BRCC1, BROVCA1, IRIS, PNCA4, PPP1R53, PSCP, RNF53 | Plays a central role in DNA repair by facilitating cellular response to DNA repair | |
| BRMS1L | NM_032352 | Breast cancer metastasis-suppressor 1-like | BRMS1 | Involved in the HDAC1-dependent transcriptional repression activity | |
| BTBD3 | NM_001282550 | BTB (POZ) domain containing 3 | RP4-742J24.3, dJ742J24.1 | Acts as a key regulator of dendritic field orientation during development of sensory cortex | |
| C1orf109 | NM_017850 | Chromosome 1 open reading frame 109 | |||
| C1QTNF9 | NM_183175 | C1q and tumor necrosis factor related protein 9 | 9130217G22Rik, CTRP9, Ciqtnf9 | Activates AMPK, AKT, and p44/42 MAPK signaling pathways | |
| C8A | NM_000562 | Complement component 8, α polypeptide | C8 is a constituent of the membrane attack complex | ||
| CCDC6 | NM_005436 | Coiled-coil domain containing 6 | D10S170, H4, PTC, TPC, TST1 | A tumor suppressor | Yes |
| CCNG1 | NM_004060 | Cyclin G1 | CCNG | May play a role in growth regulation | |
| CD46 | NM_002389 | CD46 molecule, complement regulatory protein | AHUS2, MCP, MIC10, TLX, TRA2.10 | Acts as a cofactor for complement factor I | |
| CDKN1B | NM_004064 | Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | CDKN4, KIP1, MEN1B, MEN4, P27KIP1 | Important regulator of cell cycle progression | |
| CDX2 | NM_001265 | Caudal type homeobox 2 | CDX-3, CDX3 | Involved in the transcriptional regulation of multiple genes expressed in the intestinal epithelium | Yes |
| CFI | NM_000204 | Complement factor I | AHUS3, ARMD13, C3BINA, C3b-INA, FI, IF, KAF | Responsible for cleaving the α-chains of C4b and C3b in the presence of the cofactors C4-binding protein and factor H, respectively | |
| CHL1 | NM_001253387 | Cell adhesion molecule L1-like | CALL, L1CAM2 | Plays a role in nervous system development and in synaptic plasticity | |
| CHRFAM7A | NM_139320 | CHRNA7 (cholinergic receptor, nicotinic, α 7, exons 5–10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion | CHRNA7, CHRNA7-DR1, D-10 | Extracellular ligand-gated ion channel activity | |
| CLUAP1 | NM_015041 | Clusterin associated protein 1 | CFAP22, FAP22 | May play a role in cell proliferation or apoptosis | |
| COL27A1 | NM_032888 | Collagen, type XXVII, α 1 | RP11-82I1.1 | Plays a role during the calcification of cartilage and the transition of cartilage to bone | |
| COPS2 | NM_001143887 | COP9 signalosome subunit 2 | ALIEN, CSN2, SGN2, TRIP15 | Involved in various cellular and developmental processes | |
| CST5 | NM_001900 | Cystatin D | Cysteine proteinase inhibitor | ||
| DCST1 | NM_001143687 | DC-STAMP domain containing 1 | RP11-307C12.10-003 | Protein and zinc ion binding | |
| DDIT4 | NM_019058 | DNA-damage-inducible transcript 4 | RP11-442H21.1, Dig2, REDD-1, REDD1 | Inhibits cell growth by regulating the frap1 pathway upstream of the tsc1-tsc2 complex and downstream of Akt1 | |
| DDX27 | NM_017895 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 | HSPC259, DRS1, Drs1p, PP3241, RHLP, dJ686N3.1 | Probable ATP-dependent RNA helicase | |
| DDX3X | NM_001193416 | DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked | DBX, DDX14, DDX3, HLP2 | ATP-dependent RNA helicase | |
| DNAJC7 | NM_001144766 | DnaJ (HSP40) homolog, subfamily C, member 7 | DJ11, DJC7, TPR2, TTC2 | Acts as co-chaperone regulating the molecular chaperones HSP70 and HSP90 in folding of steroid receptors | |
| DSCR8 | NM_032589 | Down syndrome critical region gene 8 | C21orf65, CT25.1a, CT25.1b, MMA-1, MMA-1a, MMA-1b, MMA1, MTAG2 | ||
| DUSP5 | NM_004419 | Dual specificity phosphatase 5 | DUSP, HVH3 | Displays phosphatase activity toward several substrates | |
| DUSP6 | NM_001946 | Dual specificity phosphatase 6 | HH19, MKP3, PYST1 | Inactivates MAP kinases | |
| ENAH | NM_001008493 | Enabled homolog | RP11-496N12.7, ENA, MENA, NDPP1 | Ena/VASP proteins are actin-associated proteins involved in a range of processes dependent on cytoskeleton remodeling and cell polarity | |
| EPHA5 | NM_001281765 | EPH receptor A5 | CEK7, EHK-1, EHK1, EK7, HEK7, TYRO4 | Receptor for members of the ephrin-A family | |
| ESR1 | NM_000125 | Estrogen receptor 1 | RP1-130E4.1, ER, ESR, ESRA, ESTRR, Era, NR3A1 | Nuclear hormone receptor | |
| EYA4 | NM_001301012 | EYA transcriptional coactivator and phosphatase 4 | RP11-704J17.4, CMD1J, DFNA10 | Tyrosine phosphatase that specifically dephosphorylates ‘Tyr-142’ of histone H2AX (H2AXY142ph) | |
| FAM160A2 | NM_001098794 | Family with sequence similarity 160, member A2 | C11orf56 | ||
| FAM222B | NM_001077498 | Family with sequence similarity 222, member B | C17orf63 | ||
| FAM47B | NM_152631 | Family with sequence similarity 47, member B | RP13-520K9.1 | ||
| FAT1 | NM_005245 | FAT atypical cadherin 1 | CDHF7, CDHR8, FAT, ME5, hFat1 | Could function as a cell-adhesion protein | |
| FBXO11 | NM_001190274 | F-box protein 11 | UG063H01, FBX11, PRMT9, UBR6, VIT1 | Substrate recognition component of the SCF E3 ubiquitin-protein ligase complex | Yes |
| FBXO28 | NM_001136115 | F-box protein 28 | CENP-30, Fbx28 | Probably recognizes and binds to some phosphorylated proteins and promotes their ubiquitination and degradation | |
| FBXO33 | NM_203301 | F-box protein 33 | BMND12, Fbx33, c14_5247 | Substrate recognition component of the SCF E3 ubiquitin-protein ligase complex, which mediates the ubiquitination and subsequent proteasomal degradation of target proteins | |
| FBXO34 | NM_017943 | F-box protein 34 | CGI-301, Fbx34 | Substrate recognition component of the SCF E3 ubiquitin-protein ligase complex, which mediates the ubiquitination and subsequent proteasomal degradation of target proteins | |
| FKBP10 | NM_021939 | FK506 binding protein 10 | PSEC0056, FKBP65, OI11, OI6, PPIASE, hFKBP65 | PPIases accelerate the folding of proteins during protein synthesis | |
| FKBP7 | NM_001135212 | FK506 binding protein 7 | UNQ670/PRO1304, FKBP23, PPIase | PPIases accelerate the folding of proteins during protein synthesis | |
| FOS | NM_005252 | FBJ murine osteosarcoma viral oncogene homolog | AP-1, C-FOS, p55 | Nuclear phosphoprotein, which forms a tight but noncovalently linked complex with the JUN/AP-1 transcription factor | |
| FRA10AC1 | NM_145246 | Fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 | PRO2972, C10orf4, F26C11.1-like, FRA10A | ||
| FSIP1 | NM_152597 | Fibrous sheath interacting protein 1 | HSD10 | ||
| FXYD6 | NM_001164831 | FXYD domain containing ion transport regulator 6 | UNQ521/PRO1056 | ||
| GADD45G | NM_006705 | Growth arrest and DNA-damage-inducible, γ | RP11-260L6.1, CR6, DDIT2, GADD45 γ, GRP17 | Involved in the regulation of growth and apoptosis | |
| GANAB | NM_001278192 | Glucosidase, α; neutral AB | G2AN, GLUII | Cleaves sequentially the 2 innermost α-1,3-linked glucose residues from the Glc(2) Man(9)GlcNAc(2) oligosaccharide precursor of immature glycoproteins | |
| GATA6 | NM_005257 | GATA binding protein 6 | Regulates terminal differentiation and/or proliferation | ||
| GATAD2B | NM_020699 | GATA zinc finger domain containing 2B | RP11-216N14.6, MRD18, P66β, p68 | Has transcriptional repressor activity | |
| GCNT1 | NM_001097633 | Glucosaminyl (N-acetyl) transferase 1, core 2 | RP11-214N16.1, C2GNT, C2GNT-L, C2GNT1, G6NT, NACGT2, NAGCT2 | Forms critical branches in O-glycans | |
| GIGYF1 | NM_022574 | GRB10 interacting GYF protein 1 | PP3360, GYF1, PERQ1 | May act cooperatively with GRB10 to regulate tyrosine kinase receptor signaling | |
| GNAI3 | NM_006496 | G protein, α inhibiting activity polypeptide 3 | RP5-1160K1.2, 87U6, ARCND1 | G proteins are involved as modulators or transducers in various transmembrane signaling systems | |
| GPR137B | NM_003272 | G protein-coupled receptor 137B | RP5-985L19.1, TM7SF1 | ||
| GPR78 | NM_080819 | G protein-coupled receptor 78 | UNQ5925/PRO19818 | Orphan receptor | |
| GPR83 | NM_016540 | G protein-coupled receptor 83 | GIR, GPR72 | Orphan receptor. Could be a neuropeptide y receptor | |
| GPRIN3 | NM_198281 | GPRIN family member 3 | GRIN3 | May be involved in neurite outgrowth | |
| GSTM2 | NM_000848 | Glutathione S-transferase mu 2 (muscle) | GST4, GSTM-2, GTHMUS, GSTM2 | Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles | |
| H1F0 | NM_005318 | H1 histone family, member 0 | H10, H1FV | Histones H1 are necessary for the condensation of nucleosome chains into higher order structures | |
| H2AFY | NM_001040158 | H2A histone family, member Y | H2A.y, H2A/y, H2AF12M, H2AFJ, MACROH2A1.1, mH2A1, macroH2A1.2 | Plays a central role in transcription regulation, DNA repair, DNA replication, and chromosomal stability | |
| H3F3B | NM_005324 | H3 histone, family 3B | H3.3B | Plays a central role in transcription regulation, DNA repair, DNA replication, and chromosomal stability | Yes |
| HDAC6 | NM_006044 | Histone deacetylase 6 | JM21, CPBHM, HD6, PPP1R90 | Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3, and H4) | |
| HERC3 | NM_001271602 | HECT and RLD domain containing E3 ubiquitin protein ligase 3 | E3 ubiquitin-protein ligase | ||
| HEY2 | NM_012259 | Hes-related family bHLH transcription factor with YRPW motif 2 | RP1-293L8.3, CHF1, GRIDLOCK, GRL, HERP1, HESR2, HRT2, bHLHb32 | Downstream effector of Notch signaling, which may be required for cardiovascular development | |
| HIPK2 | NM_001113239 | Homeodomain interacting protein kinase 2 | PRO0593 | Protein kinase acting as a corepressor of several transcription factors | |
| HMGB2 | NM_001130688 | High mobility group box 2 | HMG2 | Binds preferentially ssDNA and unwinds double-stranded DNA | |
| HNRNPAB | NM_004499 | Heterogeneous nuclear ribonucleoprotein A/B | ABBP1, HNRPAB | Binds single-stranded RNA | |
| HOOK3 | NM_032410 | Hook microtubule-tethering protein 3 | HK3 | Probable cytoskeletal linker protein, involved in tethering the G olgi complex to the cytoskeleton | Yes |
| HOXA11 | NM_005523 | Homeobox A11 | HOX1, HOX1I | Sequence-specific transcription factor | Yes |
| HRAS | NM_001130442 | Harvey rat sarcoma viral oncogene homolog | C-BAS/HAS, C-H-RAS, C-HA-RAS1, CTLO, H-RASIDX, HAMSV1, RASH1, p21ras, HRAS | Ras proteins bind GDP/GTP and possess intrinsic GTPase activity | Yes |
| HSD17B3 | NM_000197 | Hydroxysteroid (17-β) dehydrogenase 3 | RP11-240L7.3, EDH17B3, SDR12C2 | Favors the reduction of androstenedione to testosterone | |
| HSPA13 | NM_006948 | Heat shock protein 70 kDa family, member 13 | STCH | Has peptide-independent ATPase activity | |
| HUWE1 | NM_031407 | HECT, UBA and WWE domain containing 1 | RP3-339A18.4, ARF-BP1, HECTH9, HSPC272, Ib772, LASU1, MULE, URE-B1, UREB1 | E3 ubiquitin-protein ligase mediating ubiquitination and subsequent proteasomal degradation of target proteins | |
| IDS | NM_000202 | Iduronate 2-sulfatase | MPS2, SIDS | Required for the lysosomal degradation of heparan sulfate and dermatan sulfate | |
| INCENP | NM_001040694 | Inner centromere protein antigens | Component of the chromosomal passenger complex, a complex that acts as a key regulator of mitosis | ||
| IQCG | NM_001134435 | IQ motif containing G | CFAP122, DRC9 | ||
| KAT2B | NM_003884 | K(lysine) acetyltransferase 2B | CAF, P/CAF, PCAF | Functions as a HAT to promote transcriptional activation | |
| KBTBD3 | NM_152433 | Kelch repeat and BTB (POZ) domain containing 3 | BKLHD3 | ||
| KCTD2 | NM_015353 | Potassium channel tetramerization domain containing 2 | |||
| KCTD3 | NM_016121 | Potassium channel tetramerization domain containing 3 | RP11-5F19.1, NY-REN-45 | ||
| KIAA0100 | NM_014680 | KIAA0100 | BCOX, BCOX1, CT101 | ||
| KIAA0101 | NM_001029989 | KIAA0101 | L5, NS5ATP9, OEATC, OEATC-1, OEATC1, PAF, PAF15, p15(PAF), p15/PAF, p15PAF | May be involved in protection of cells from UV-induced cell death | |
| KIAA1462 | NM_020848 | KIAA1462 | JCAD | ||
| KLF6 | NM_001160124 | Kruppel-like factor 6 | RP11-184A2.1, BCD1, CBA1, COPEB, CPBP, GBF, PAC1, ST12, ZF9 | Plays a role in B-cell growth and development | |
| KLHL15 | NM_030624 | Kelch-like family, member 15 | HEL-S-305 | Probable substrate-specific adapter of an E3 ubiquitin-protein ligase complex, which mediates the ubiquitination and subsequent proteasomal degradation of target proteins | |
| KLHL42 | NM_020782 | Kelch-like family, member 42 | Ctb9, KLHDC5 | Substrate-specific adapter of a BCR (BTB-CUL3-RBX1) E3 ubiquitin-protein ligase complex required for mitotic progression and cytokinesis | |
| KLRC4 | NM_013431 | Killer cell lectin-like receptor subfamily C, member 4 | NKG2-F, NKG2F | May play a role as a receptor for the recognition of MHC class I HLA-E molecules by NK cells | |
| KRAS | NM_004985 | Kirsten rat sarcoma viral oncogene homolog | C-K-RAS, CFC2, K-RAS2A, K-RAS2B, K-RAS4A, K-RAS4B, KI-RAS1, KRAS2, NS, NS3, RASK2, KRAS | Binds GDP/GTP and possesses intrinsic GTPase activity | Yes |
| LAMA3 | NM_000227 | Laminin, α 3 | BM600, E170, LAMNA, LOCS, lama3a | Binding to cells via a high-affinity receptor, mediating the attachment, migration, and organization of cells into tissues | |
| LBR | NM_002296 | Lamin B receptor | PRO0650, DHCR14B, LMN2R, PHA, TDRD18 | Anchors the lamina and the heterochromatin to the inner nuclear membrane | |
| LCLAT1 | NM_001002257 | Lysocardiolipin acyltransferase 1 | UNQ1849/PRO3579, 1AGPAT8, AGPAT8, ALCAT1, HSRG1849, LYCAT, UNQ1849 | Acyl-CoA: lysocardiolipin acyltransferase | |
| LFNG | NM_001040167 | LFNG O-fucosylpeptide 3-β-N-acetylglucosaminyltransferase | SCDO3 | Glycosyltransferase | |
| LGALSL | NM_014181 | Lectin, galactoside-binding-like | HSPC159, GRP | Does not bind lactose and may not bind carbohydrates | |
| LPGAT1 | NM_014873 | Lysophosphatidylglycerol acyltransferase 1 | FAM34A, FAM34A1, NET8 | Lysophoshatidylglycerol-specific acyltransferase | |
| LRRC17 | NM_001031692 | Leucine rich repeat containing 17 | UNQ3076/PRO9909, P37NB | Involved in bone homeostasis, acting as a negative regulator of RANKL-induced osteoclast precursor differentiation from bone marrow precursors | |
| LRRN3 | NM_001099658 | Leucine rich repeat neuronal 3 | Nbla10363, FIGLER5, NLRR-3, NLRR3 | ||
| LYSMD3 | NM_001286812 | LysM, putative peptidoglycan-binding, domain containing 3 | |||
| MAP2K1 | NM_002755 | Mitogen-activated protein kinase kinase 1 | CFC3, MAPKK1, MEK1, MKK1, PRKMK1 | Catalyzes the concomitant phosphorylation of a threonine and a tyrosine residue in a Thr-Glu-Tyr sequence located in MAP kinases | Yes |
| MAZ | NM_001042539 | MYC-associated zinc finger protein (purine-binding transcription factor) | PUR1, Pur-1, SAF-1, SAF-2, SAF-3, ZF87, ZNF801, Zif87 | May function as a transcription factor with dual roles in transcription initiation and termination | |
| MCL1 | NM_001197320 | Myeloid cell leukemia 1 | BCL2L3, EAT-ES, MCL1L, MCL1S, Mcl-1, TM, bcl2-L-3, mcl1/EAT, MCL1 | Involved in the regulation of apoptosis versus cell survival, and in the maintenance of viability but not of proliferation | |
| METAP1 | NM_015143 | Methionyl aminopeptidase 1 | MAP1A, MetAP1A | Removes the amino terminal methionine from nascent proteins | |
| MGAT5 | NM_002410 | Mannosyl (α-1,6-)-glycoprotein β-1,6-N-acetyl-glucosaminyltransferase | GNT-V, GNT-VA | Catalyzes the addition of N-acetylglucosamine in β 1–6 linkage to the α-linked mannose of biantennary N-linked oligosaccharides | |
| MOB1A | NM_018221 | MOB kinase activator 1A | C2orf6, MATS1, MOB1, MOBK1B, MOBKL1B, Mob4B | Activator of LATS1/2 in the Hippo signaling pathway | |
| MOB1B | NM_001244766 | MOB kinase activator 1B | MATS2, MOB4A, MOBKL1A | Activator of LATS1/2 in the Hippo signaling pathway | |
| MOB3B | NM_024761 | MOB kinase activator 3B | C9orf35, MOB1D, MOBKL2B | May regulate the activity of kinases | |
| MRPS14 | NM_022100 | Mitochondrial ribosomal protein S14 | DJ262D12.2, HSMRPS14, MRP-S14, S14mt | ||
| MTMR12 | NM_001040446 | Myotubularin related protein 12 | 3-PAP, PIP3AP | Inactive phosphatase that plays a role as an adapter for the phosphatase myotubularin, to regulate myotubularin intracellular location | |
| MTMR3 | NM_021090 | Myotubularin related protein 3 | hCG_2011013, FYVE-DSP1, ZFYVE10 | Phosphatase that acts on lipids with a phosphoinositol head group | |
| ND2 | NADH dehydrogenase subunit 2 | Core subunit of the mitochondrial membrane re-spiratory chain NADH dehydrogenase (Complex I) | |||
| NFYB | NM_006166 | Nuclear transcription factor Yβ | CBF-A, CBF-B, HAP3, NF-YB | Stimulates the transcription of various genes by recognizing and binding to a CCAAT motif in promoters | |
| NKX3-2 | NM_001189 | NK3 homeobox 2 | BAPX1, NKX3.2, NKX3B, SMMD | Transcriptional repressor that acts as a negative regulator of chondrocyte maturation | |
| NLK | NM_016231 | Nemo-like kinase | Role in cell fate determination, required for differentiation of bone marrow stromal cells | ||
| NMRK2 | NM_001289117 | Nicotinamide riboside kinase 2 | ITGB1BP3, MIBP, NRK2 | ||
| NOL4 | NM_001198546 | Nucleolar protein 4 | HRIHFB2255, CT125, NOLP | ||
| NOTCH1 | NM_017617 | Notch 1 | TAN1, hN1 | Functions as a receptor for membrane-bound ligands Jagged1, Jagged2, and Delta1 to regulate cell fate determination | Yes |
| NOTCH2 | NM_001200001 | Notch 2 | AGS2, HJCYS, hN2 | Functions as a receptor for membrane-bound ligands Jagged1, Jagged2, and Delta1 to regulate cell fate determination | Yes |
| NR6A1 | NM_001278546 | Nuclear receptor subfamily 6, group A, member 1 | CT150, GCNF, GCNF1, NR61, RTR, hGCNF, hRTR | Orphan nuclear receptor. May be involved in the regulation of gene expression in germ cell development during gametogenesis | |
| NRP1 | NM_001024628 | Neuropilin 1 | RP11-342D11.1, BDCA4, CD304, NP1, NRP, VEGF165R | The membrane-bound isoform 1 is a receptor involved in the development of the cardiovascular system, in angiogenesis, in the formation of certain neuronal circuits, and in organogenesis outside the nervous system | |
| NUDT12 | NM_001300741 | Nudix-type motif 12 | Hydrolyzes NAD(P)H to NMNH and AMP (2′,5′-ADP), and diadenosine diphosphate to AMP | ||
| NUPL1 | NM_001008564 | Nucleoporin like 1 | RP11-206I15.1, PRO2463 | Component of the nuclear pore complex, a complex required for the trafficking across the nuclear membrane | |
| OCA2 | NM_000275 | Oculocutaneous albinism II | BEY, BEY1, BEY2, BOCA, D15S12, EYCL, EYCL2, EYCL3, HCL3, P, PED, SHEP1 | Could be involved in the transport of tyrosine | |
| OFCC1 | NM_153003 | Orofacial cleft 1 candidate 1 | MRDS1 | ||
| OR11A1 | NM_013937 | Olfactory receptor, family 11, subfamily A, member 1 | DAAP-34I1.2, 6M1-18, OR11A2, dJ994E9.6, hs6M1-18 | Odorant receptor | |
| OTUD1 | NM_001145373 | OTU deubiquitinase 1 | DUBA7, OTDC1 | Deubiquitinating enzyme that specifically hydrolyzes ‘Lys-63’-linked polyubiquitin to monoubiquitin | |
| OTX2 | NM_001270523 | Orthodenticle homeobox 2 | CPHD6, MCOPS5 | Probably plays a role in the development of the brain and the sense organs | |
| PABPC1 | NM_002568 | Poly(A) binding protein, cytoplasmic 1 | PAB1, PABP, PABP1, PABPC2, PABPL1 | Binds the poly(A) tail of mRNA | |
| PCDHB8 | NM_019120 | Protocadherin β8 | PCDH-β8, PCDH3I | Potential calcium-dependent cell-adhesion protein | |
| PFKFB2 | NM_001018053 | 6-phosphofructo-2-kinase/fructose-2, 6-biphosphatase 2 | RP11-164O23.2, PFK-2/FBPase-2 | Synthesis and degradation of fructose 2,6-bisphosphate | |
| PGD | NM_002631 | Phosphogluconate dehydrogenase | 6PGD | Catalyzes the oxidative decarboxylation of 6-phosphogluconate to ribulose 5-phosphate and CO2, with concomitant reduction of NADP to NADPH | |
| PHOX2A | NM_005169 | Paired-like homeobox 2a | ARIX, CFEOM2, FEOM2, NCAM2, PMX2A | May be involved in regulating the specificity of expression of the catecholamine biosynthetic genes | |
| PHPT1 | NM_001135861 | Phosphohistidine phosphatase 1 | RP11-216L13.10-005, CGI-202, HEL-S-132P, HSPC141, PHP14 | Exhibits phosphohistidine phosphatase activity | |
| PITPNB | NM_001284277 | Phosphatidylinositol transfer protein β | RP3-353E16.2, PI-TP-β, PtdInsTP, VIB1B | Catalyzes the transfer of PtdIns and phosphatidylcholine between membranes | |
| PLA2G4C | NM_001159322 | Phospholipase A2, group IVC | CPLA2-γ | Has a preference for arachidonic acid at the sn-2 position of phosphatidylcholine as compared with palmitic acid | |
| PLAG1 | NM_001114634 | Pleiomorphic adenoma gene 1 | PSA, SGPA, ZNF912 | Transcription factor whose activation results Yes in upregulation of target genes, such as IGFII, leading to uncontrolled cell proliferation | |
| PLCL2 | NM_001144382 | Phospholipase C-like 2 | PLCE2 | May play a role in the regulation of Ins(1,4,5)P3 around the endoplasmic reticulum | |
| PLXDC2 | NM_001282736 | Plexin domain containing 2 | UNQ2514/PRO6003, TEM7R | May play a role in tumor angiogenesis | |
| PPP1R9A | NM_001166160 | protein phosphatase 1, regulatory subunit 9A | NRB1, NRBI, Neurabin-I | Binds to actin filaments (F-actin) and shows crosslinking activity | |
| PRAP1 | NM_001145201 | Proline-rich acidic protein 1 | RP11-122K13.6, PRO1195, UPA | May play an important role in maintaining normal growth homeostasis in epithelial cells | |
| PRDX3 | NM_006793 | Peroxiredoxin 3 | AOP-1, AOP1, HBC189, MER5, PRO1748, SP-22, prx-III | Involved in redox regulation of the cell | |
| PRLR | NM_000949 | Prolactin receptor | HPRL, MAB, hPRLrI | This is a receptor for the anterior pituitary hormone prolactin | |
| PROSC | NM_007198 | Proline synthetase co-transcribed homolog | |||
| PROX1 | NM_001270616 | Prospero homeobox 1 | May play a fundamental role in early development of the central nervous system | ||
| PRR4 | NM_001098538 | Proline rich 4 | LPRP, PROL4 | ||
| PRRC2B | NM_013318 | Proline-rich coiled-coil 2B | RP11-334J6.1, BAT2L, BAT2L1, KIAA0515, LQFBS-1 | ||
| PTGS2 | NM_000963 | Prostaglandin-endoperoxide synthase 2 | COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | May have a role as a major mediator of inflammation and/or a role for prostanoid signaling in activity-dependent plasticity | |
| PTPLAD1 | NM_016395 | Protein tyrosine phosphatase-like A domain containing 1 | B-IND1, HACD3, HSPC121 | Involved in Rac1-signaling pathways leading to the modulation of gene expression | |
| PTPN11 | NM_002834 | Protein tyrosine phosphatase, non-receptor type 11 | BPTP3, CFC, NS1, PTP-1D, PTP2C, SH-PTP2, SH-PTP3, SHP2 | Acts downstream of various receptor and Yes cytoplasmic protein tyrosine kinases to participate in the signal transduction from the cell surface to the nucleus | |
| PTPN22 | NM_001193431 | Protein tyrosine phosphatase, non-receptor type 22 | LYP, LYP1, LYP2, PEP, PTPN8 | Seems to act on Cbl | |
| PTPRZ1 | NM_001206838 | Protein tyrosine phosphatase, receptor-type, Z polypeptide 1 | HPTPZ, HPTPζ, PTP-ζ, PTP18, PTPRZ, PTPZ, R-PTP-ζ-2, RPTPB, RPTPβ, phosphacan | May be involved in the regulation of specific developmental processes in the central nervous system | |
| PUM1 | NM_001020658 | Pumilio RNA-binding family member 1 | RP1-65J11.4, HSPUM, PUMH, PUMH1, PUML1 | Sequence-specific RNA-binding protein that regulates translation and mRNA stability by binding the 3′-UTR of mRNA targets | |
| RALA | NM_005402 | V-ral simian leukemia viral oncogene homolog A | RAL | Multifunctional GTPase involved in a variety of cellular processes, including gene expression, cell migration, cell proliferation, oncogenic transformation, and membrane trafficking | |
| RASSF6 | NM_001270391 | Ras association (RalGDS/AF-6) domain family member 6 | May act as a Ras effector protein | ||
| RLF | NM_012421 | Rearranged L-myc fusion | RP1-39G22.1, ZN-15L, ZNF292L | May be involved in transcriptional regulation. | |
| RNF2 | NM_007212 | Ring finger protein 2 | GS1-120K12.1, BAP-1, BAP1, DING, HIPI3, RING1B, RING2 | E3 ubiquitin-protein ligase that mediates mono-ubiquitination of “Lys-119” of histone H2A, playing a central role in histone code and gene regulation | |
| RNF34 | NM_001256858 | Ring finger protein 34 | CARP-1, CARP1, RFI, RIF, RIFF, hRFI | Has E3 ubiquitin-protein ligase activity. Regulates the levels of CASP8 and CASP10 by targeting them for proteasomal degradation | |
| ROPN1L | NM_001201466 | Rhophilin associated tail protein 1-like | RP11-1C1.7, ASP, RSPH11 | ||
| RPL14 | NM_001034996 | Ribosomal protein L14 | CAG-ISL-7, CTG-B33, L14, RL14, hRL14 | ||
| RPS8 | NM_001012 | Ribosomal protein S8 | RP11-269F19.3, S8 | ||
| S100A1 | NM_006271 | S100 calcium binding protein A1 | RP1-178F15.1, S100, S100-α, S100A | Weakly binds calcium but binds zinc very tightly – distinct binding sites with different affinities exist for both ions on each monomer | |
| SCD | NM_005063 | Stearoyl-CoA desaturase | PRO1933, FADS5, MSTP0081, SCDOS, SCD | Terminal component of the liver microsomal stearyl-CoA desaturase system | |
| SH3BGRL | NM_003022 | SH3 domain binding glutamate-rich protein like | HEL-S-115, SH3BGR | ||
| SIK2 | NM_015191 | Salt-inducible kinase 2 | LOH11CR1I, QIK, SNF1LK2 | Phosphorylates Ser794 of IRS1 in insulin-stimulated adipocytes | |
| SIRT1 | NM_001142498 | Sirtuin 1 | RP11-57G10.3, SIR2L1 | NAD-dependent deacetylase, which regulates processes such as apoptosis and muscle differentiation by deacetylating key proteins | |
| SIX6 | NM_007374 | SIX homeobox 6 | MCOPCT2, OPTX2, Six9 | May be involved in eye development | |
| SLC35B4 | NM_032826 | Solute carrier family 35 (UDP-xylose/UDP-N-acetylglucosamine transporter), member B4 | PSEC0055, YEA, YEA4 | Sugar transporter that specifically mediates the transport of UDP-Xyl and UDP-GlcNAc from cytosol into Golgi | |
| SLC37A3 | NM_001287498 | Solute carrier family 37, member 3 | |||
| SLC7A11 | NM_014331 | Solute carrier family 7 (anionic amino acid transporter light chain, xc-system), member 11 | CCBR1, xCT | Sodium-independent, high-affinity exchange of anionic amino acids with high specificity for anionic form of cystine and glutamate | |
| SLCO2A1 | NM_005630 | Solute carrier organic anion transporter family, member 2A1 | MATR1, OATP2A1, PGT, PHOAR2, SLC21A2 | May mediate the release of newly synthesized prostaglandins from cells, the transepithelial transport of prostaglandins, and the clearance of prostaglandins from the circulation | |
| SMCHD1 | NM_015295 | Structural maintenance of chromosomes flexible hinge domain containing 1 | Required for maintenance of X inactivation in females and hypermethylation of CpG islands associated with inactive X | ||
| SMG1 | NM_015092 | SMG1 phosphatidylinositol 3-kinase-related kinase | 61E3.4, ATX, LIP | Serine/threonine protein kinase involved in both mRNA surveillance and genotoxic stress response pathways | |
| SNAI2 | NM_003068 | Snail family zinc finger 2 | SLUG, SLUGH1, SNAIL2, WS2D | Transcriptional repressor. Involved in the generation and migration of neural crest cells | |
| SOGA2 | NM_015210 | Microtubule crosslinking factor 1 | CCDC165, KIAA0802, MTCL1 | ||
| SPRY2 | NM_005842 | Sprouty homolog 2 | hSPRY2 | May function as an antagonist of fibroblast growth factor pathways and may negatively modulate respiratory organogenesis | |
| SRPK2 | NM_001278273 | SRSF protein kinase 2 | SFRSK2 | Phosphorylates RS domain-containing proteins | |
| SRSF7 | NM_001031684 | Serine/arginine-rich splicing factor 7 | 9G8, AAG3, SFRS7 | Required for pre-mRNA splicing | |
| STAG2 | NM_001042749 | Stromal antigen 2 | RP11-517O1.1, SA-2, SA2, SCC3B, bA517O1.1 | Component of cohesion complex, a complex required for the cohesion of sister chromatids after DNA replication | Yes |
| TAAR6 | NM_175067 | Trace amine associated receptor 6 | RP11-295F4.3, TA4, TAR4, TAR6, TRAR4, taR-4, taR-6 | Orphan receptor. Could be a receptor for trace amines | |
| TAB2 | NM_001292034 | TGF-β activated kinase 1/MAP3K7 binding protein 2 | CHTD2, MAP3K7IP2, TAB-2 | Adapter linking MAP3K7/TAK1 and TRAF6, and mediator of MAP3K7 activation in the IL1 signaling pathway | |
| TAF15 | NM_003487 | TAF15 RNA polymerase II, TBP-associated factor | Npl3, RBP56, TAF2N, TAFII68 | RNA and ssDNA-binding protein that may play specific roles during transcription initiation at distinct promoters | Yes |
| TAF2 | NM_003184 | TAF2 RNA polymerase II, TBP-associated factor | CIF150, MRT40B, TAFII150, TAF2 | Transcription factor TFIID is one of the general factors required for accurate and regulated initiation by RNA polymerase II | |
| TAF6L | NM_006473 | TAF6-like RNA PCAF-associated factor | PAF65A | Functions as a component of the PCAF complex | |
| TBX4 | NM_018488 | T-box 4 | SPS | Involved in the transcriptional regulation of genes required for mesoderm differentiation | |
| TCF21 | NM_003206 | Transcription factor 21 | POD1, bHLHa23 | Involved in epithelial–mesenchymal interactions in kidney and lung morphogenesis that include epi thelial differentiation and branching morphogenesis | |
| TEAD4 | NM_003213 | TEA domain family, member 4 | EFTR-2, RTEF1, TCF13L1, TEF-3, TEF3, TEFR-1, hRTEF-1B | Binds specifically and noncooperatively to the Sph and GT-IIC “enhansons” (5′GTGGAATGT-3′) and activates transcription | |
| TGFBR3 | NM_001195683 | Transforming growth factor β receptor III | BGCAN, β-glycan | Binds to TGF-β | |
| TGIF2 | NM_001199513 | TGFB-induced factor homeobox 2 | Transcriptional repressor. Probably represses tran scription via the recruitment of HDAC proteins | ||
| TIAL1 | NM_001033925 | TIA1 cytotoxic granule-associated RNA binding protein-like 1 | TCBP, TIAR | RNA-binding protein. Possesses nucleolytic activity against cytotoxic lymphocyte target cells May be involved in apoptosis | |
| TM9SF3 | NM_020123 | Transmembrane 9 superfamily member 3 | RP11-34E5.1, EP70-P-iso, SMBP | ||
| TMED4 | NM_182547 | Transmembrane emp24 protein transport domain containing 4 | ERS25, HNLF | Involved in endoplasmic reticulum stress response. May play a role in the regulation of heat-shock response and apoptosis | |
| TMEM132B | NM_001286219 | Transmembrane protein 132B | |||
| TMEM14A | NM_014051 | Transmembrane protein 14A | PTD011, C6orf73 | ||
| TMEM192 | NM_001100389 | Transmembrane protein 192 | |||
| TMEM257 | NM_004709 | Transmembrane protein 257 | CXorf1 | ||
| TMEM45A | NM_018004 | Transmembrane protein 45A | DERP7 | ||
| TMEM64 | NM_001008495 | Transmembrane protein 64 | |||
| TMPRSS11A | NM_001114387 | Transmembrane protease, serine 11A | ECRG1 | Probable serine protease, which may play a role in cellular senescence | |
| TNIP1 | NM_001252385 | TNFAIP3 interacting protein 1 | ABIN-1, NAF1, VAN, nip40-1 | Interacts with zinc finger protein A20/TNFAIP3 and inhibits TNF-induced NF-κB-dependent gene expression by interfering with an RIP-or TRAF2-mediated transactivation signal | |
| TRIM2 | NM_001130067 | Tripartite motif containing 2 | CMT2R, RNF86 | May contribute to the alteration of neural cellular mechanisms | |
| TSHR | NM_000369 | Thyroid stimulating hormone receptor | CHNG1, LGR3, hTSHR-I | Receptor for thyrothropin. Plays a central role in controlling thyroid cell metabolism | Yes |
| TUSC1 | NM_001004125 | Tumor suppressor candidate 1 | TSG-9, TSG9 | ||
| UBA2 | NM_005499 | Ubiquitin-like modifier activating enzyme 2 | HRIHFB2115, ARX, SAE2 | The dimeric enzyme acts as a E1 ligase for SUMO1, SUMO2, SUMO3, and probably SUMO4 | |
| UCHL1 | NM_004181 | Ubiquitin carboxyl-terminal esterase L1 | HEL-117, NDGOA, PARK5, PGP 9.5, PGP9.5, PGP95, Uch-L1 | Ubiquitin-protein hydrolase involved both in the processing of ubiquitin precursors and of ubiquitinated proteins | |
| UGT3A1 | NM_001171873 | UDP glycosyltransferase 3 family, polypeptide A1 | UDP-glucuronosyltransferases catalyze phase II biotransformation reactions | ||
| USP28 | NM_001301029 | Ubiquitin specific peptidase 28 | Deubiquitinase involved in DNA damage response checkpoint and MYC proto-oncogene stability | ||
| WNT16 | NM_016087 | Wingless-type MMTV integration site family, member 16 | Ligand for members of the Frizzled family of seven transmembrane receptors | ||
| WNT2 | NM_003391 | Wingless-type MMTV integration site family, member 2 | INT1L1, IRP | Ligand for members of the Frizzled family of seven transmembrane receptors | |
| WNT3A | NM_033131 | Wingless-type MMTV integration site family, member 3A | Ligand for members of the Frizzled family of seven transmembrane receptors | ||
| XIAP | NM_001167 | X-linked inhibitor of apoptosis | RP1-315G1.5, API3, BIRC4, IAP-3, ILP1, MIHA, XLP2, hIAP-3, hIAP3 | Apoptotic suppressor | |
| YOD1 | NM_001276320 | YOD1 deubiquitinase | RP11-164O23.1, DUBA8, OTUD2, PRO0907 | May play an important regulatory role at the level of protein turnover, by preventing degradation | |
| ZEB2 | NM_001171653 | Zinc finger E-box binding homeobox 2 | HRIHFB2411, HSPC082, SIP-1, SIP1, SMADIP1, ZFHX1B | Transcriptional inhibitor that binds to DNA sequence 5′-CACCT-3′ in different promoters Represses transcription of E-cadherin | |
| ZFP36L2 | NM_006887 | Zinc finger protein 36 homolog | BRF2, ERF-2, ERF2, RNF162C, TIS11D | Probable regulatory protein involved in regulating the response to growth factors | |
| ZNF12 | NM_006956 | Zinc finger protein 12 | GIOT-3, HZF11, KOX3, ZNF325 | May be involved in transcriptional regulation | |
| ZNF148 | NM_021964 | Zinc finger protein 148 | BERF-1, BFCOL1, HT-β, ZBP-89, ZFP148, pHZ-52 | Involved in transcriptional regulation | |
| ZNF25 | NM_145011 | Zinc finger protein 25 | KOX19, Zfp9 | May be involved in transcriptional regulation | |
| ZNF35 | NM_003420 | Zinc finger protein 35 | HF.10, HF10, Zfp105 | May be involved in transcriptional regulation | |
| ZNF350 | NM_021632 | Zinc finger protein 350 | ZBRK1, ZFQR | Transcriptional repressor | |
| ZNF426 | NM_001300883 | Zinc finger protein 426 | May be involved in transcriptional regulation | ||
| ZNF445 | NM_181489 | Zinc finger protein 445 | ZKSCAN15, ZNF168, ZSCAN47 | May be involved in transcriptional regulation | |
| ZNF558 | NM_144693 | Zinc finger protein 558 | May be involved in transcriptional regulation | ||
| ZNF562 | NM_001130031 | Zinc finger protein 562 | May be involved in transcriptional regulation | ||
| ZNF594 | NM_032530 | Zinc finger protein 594 | hCG_1775942 | May be involved in transcriptional regulation | |
| ZNF652 | NM_001145365 | Zinc finger protein 652 | Functions as a transcriptional repressor | ||
| ZNF763 | NM_001012753 | Zinc finger protein 763 | ZNF, ZNF440L | May be involved in transcriptional regulation | |
Abbreviations: CLL, chronic lymphocytic leukemia; HLA, human leukocyte antigen; IGF, insulin-like growth factor; IL, interleukin; mRNA, messenger RNA; NK cells, natural killer cells; ssDNA, single-stranded DNA; TGF, transforming growth factor; TNF, tumor necrosis factor; UV, ultraviolet.
As shown in Table 9, our DAVID analysis showed that there were 20 functional clusters that were identified to be enriched with an enrichment score >1.0 in the target list of hsa-miR-181a-5p, based on miRTarBase. The functions of these clusters involved negative regulation of transcription, negative regulation of gene expression, negative regulation of nucleobase, nucleoside, nucleotide, and nucleic acid metabolic processes, negative regulation of nitrogen compound metabolic process, negative regulation of macromolecule biosynthetic process, negative regulation of cellular biosynthetic process, negative regulation of bio-synthetic process, regulation of phosphorylation, regulation of phosphate metabolic process, lung development, respiratory tube development, positive regulation of transcription, positive regulation of gene expression, mesenchymal cell differentiation and development, negative regulation of apoptosis and programmed cell death, the insulin-like growth factor (IGF)-1 signaling pathway, interleukin (IL)-6 signaling pathway, insulin signaling pathway, Ubl conjugation pathway, modification-dependent macromolecule catabolic process, and modification-dependent protein catabolic process.
Table 9.
The top enriched clusters (enrich score >1) by DAVID for the targets of hsa-miR-181a-5p from miRTarBase 4.0
| Category | Term | Gene count | P-value | FDR |
|---|---|---|---|---|
| Annotation cluster 1 | Enrichment score: 2.9 | |||
| GOTERM_BP_FAT | Negative regulation of transcription | 17 | 3.10E-04 | 7.80E-02 |
| GOTERM_BP_FAT | Negative regulation of gene expression | 17 | 8.50E-04 | 9.90E-02 |
| GOTERM_BP_FAT | Negative regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 17 | 1.00E-03 | 1.00E-01 |
| GOTERM_BP_FAT | Negative regulation of nitrogen compound metabolic process | 17 | 1.20E-03 | 1.00E-01 |
| GOTERM_BP_FAT | Negative regulation of macromolecule biosynthetic process | 17 | 2.00E-03 | 1.20E-01 |
| GOTERM_BP_FAT | Negative regulation of cellular biosynthetic process | 17 | 2.50E-03 | 1.20E-01 |
| GOTERM_BP_FAT | Negative regulation of biosynthetic process | 17 | 3.10E-03 | 1.20E-01 |
| Annotation cluster 2 | Enrichment score: 2.86 | |||
| GOTERM_BP_FAT | Regulation of phosphorylation | 16 | 1.10E-03 | 1.00E-01 |
| GOTERM_BP_FAT | Regulation of phosphorus metabolic process | 16 | 1.60E-03 | 1.10E-01 |
| GOTERM_BP_FAT | Regulation of phosphate metabolic process | 16 | 1.60E-03 | 1.10E-01 |
| Annotation cluster 3 | Enrichment score: 2.66 | |||
| GOTERM_BP_FAT | Lung development | 7 | 1.80E-03 | 1.10E-01 |
| GOTERM_BP_FAT | Respiratory tube development | 7 | 2.10E-03 | 1.20E-01 |
| GOTERM_BP_FAT | Respiratory system development | 7 | 2.80E-03 | 1.20E-01 |
| Annotation cluster 4 | Enrichment score: 2.38 | |||
| GOTERM_BP_FAT | Positive regulation of transcription, DNA-dependent | 16 | 1.30E-03 | 1.00E-01 |
| GOTERM_BP_FAT | Positive regulation of transcription | 16 | 6.50E-03 | 1.70E-01 |
| GOTERM_BP_FAT | Positive regulation of gene expression | 16 | 8.50E-03 | 1.80E-01 |
| Annotation cluster 5 | Enrichment score: 2.37 | |||
| GOTERM_BP_FAT | Mesenchymal cell differentiation | 5 | 4.20E-03 | 1.50E-01 |
| GOTERM_BP_FAT | Mesenchymal cell development | 5 | 4.20E-03 | 1.50E-01 |
| GOTERM_BP_FAT | Mesenchyme development | 5 | 4.50E-03 | 1.50E-01 |
| Annotation cluster 6 | Enrichment score: 2.16 | |||
| GOTERM_BP_FAT | Negative regulation of apoptosis | 12 | 6.40E-03 | 1.70E-01 |
| GOTERM_BP_FAT | Negative regulation of programmed cell death | 12 | 7.10E-03 | 1.70E-01 |
| GOTERM_BP_FAT | Negative regulation of cell death | 12 | 7.20E-03 | 1.70E-01 |
| Annotation cluster 7 | Enrichment score: 2.12 | |||
| BIOCARTA | IGF-1 signaling pathway | 4 | 7.60E-03 | 3.80E-01 |
| BIOCARTA | IL-6 signaling pathway | 4 | 7.60E-03 | 3.80E-01 |
| BIOCARTA | Insulin signaling pathway | 4 | 7.60E-03 | 3.80E-01 |
| Annotation cluster 8 | Enrichment score: 2.03 | |||
| SP_PIR_KEYWORDS | Ubl conjugation pathway | 15 | 2.80E-03 | 9.30E-02 |
| GOTERM_BP_FAT | Modification-dependent macromolecule catabolic process | 15 | 1.70E-02 | 2.40E-01 |
| GOTERM_BP_FAT | Modification-dependent protein catabolic process | 15 | 1.70E-02 | 2.40E-01 |
| Annotation cluster 9 | Enrichment score: 2 | |||
| GOTERM_CC_FAT | Intracellular organelle lumen | 33 | 7.40E-03 | 3.90E-01 |
| GOTERM_CC_FAT | Organelle lumen | 33 | 1.00E-02 | 4.20E-01 |
| GOTERM_CC_FAT | Membrane-enclosed lumen | 33 | 1.30E-02 | 4.50E-01 |
| Annotation cluster 10 | Enrichment score: 1.9 | |||
| GOTERM_BP_FAT | Proteolysis involved in cellular protein catabolic process | 16 | 1.10E-02 | 2.00E-01 |
| GOTERM_BP_FAT | Cellular protein catabolic process | 16 | 1.20E-02 | 2.00E-01 |
| GOTERM_BP_FAT | Protein catabolic process | 16 | 1.50E-02 | 2.20E-01 |
| Annotation cluster 11 | Enrichment score: 1.86 | |||
| INTERPRO | Zinc finger, C2H2-type | 19 | 7.50E-03 | 6.70E-01 |
| INTERPRO | Zinc finger, C2H2-like | 19 | 8.80E-03 | 6.30E-01 |
| SMART | Zinc finger_C2H2 | 19 | 4.00E-02 | 9.00E-01 |
| Annotation cluster 12 | Enrichment score: 1.76 | |||
| GOTERM_BP_FAT | Regulation of apoptosis | 19 | 1.60E-02 | 2.30E-01 |
| GOTERM_BP_FAT | Regulation of programmed cell death | 19 | 1.80E-02 | 2.40E-01 |
| GOTERM_BP_FAT | Regulation of cell death | 19 | 1.80E-02 | 2.40E-01 |
| Annotation cluster 13 | Enrichment score: 1.75 | |||
| GOTERM_BP_FAT | Determination of left/right symmetry | 4 | 1.70E-02 | 2.40E-01 |
| GOTERM_BP_FAT | Determination of symmetry | 4 | 1.80E-02 | 2.40E-01 |
| GOTERM_BP_FAT | Determination of bilateral symmetry | 4 | 1.80E-02 | 2.40E-01 |
| Annotation cluster 14 | Enrichment score: 1.73 | |||
| GOTERM_BP_FAT | Neuron projection morphogenesis | 9 | 6.50E-03 | 1.70E-01 |
| GOTERM_BP_FAT | Cell projection morphogenesis | 9 | 1.40E-02 | 2.20E-01 |
| GOTERM_BP_FAT | Cell part morphogenesis | 9 | 1.80E-02 | 2.40E-01 |
| GOTERM_BP_FAT | Neuron projection development | 9 | 1.80E-02 | 2.40E-01 |
| GOTERM_BP_FAT | Neuron development | 9 | 7.40E-02 | 4.90E-01 |
| Annotation cluster 15 | Enrichment score: 1.49 | |||
| INTERPRO | Homeobox, conserved site | 8 | 2.40E-02 | 7.40E-01 |
| INTERPRO | Homeobox | 8 | 2.50E-02 | 7.20E-01 |
| SMART | HOX | 8 | 5.50E-02 | 7.30E-01 |
| Annotation cluster 16 | Enrichment score: 1.38 | |||
| INTERPRO | Wnt superfamily | 3 | 2.20E-02 | 7.60E-01 |
| INTERPRO | Secreted growth factor Wnt protein | 3 | 2.20E-02 | 7.60E-01 |
| INTERPRO | Secreted growth factor Wnt protein, conserved site | 3 | 2.20E-02 | 7.60E-01 |
| PIR_SUPERFAMILY | PIRSF001784: int-1 transforming protein | 3 | 2.60E-02 | 7.60E-01 |
| GOTERM_BP_FAT | Wnt receptor signaling pathway, calcium modulating pathway | 3 | 3.00E-02 | 3.10E-01 |
| SMART | WNT1 | 3 | 3.10E-02 | 9.70E-01 |
| KEGG_PATHWAY | Basal cell carcinoma | 3 | 1.90E-01 | 6.00E-01 |
| KEGG_PATHWAY | Hedgehog signaling pathway | 3 | 2.00E-01 | 6.00E-01 |
| Annotation cluster 17 | Enrichment score: 1.26 | |||
| GOTERM_BP_FAT | Positive regulation of apoptosis | 11 | 5.30E-02 | 4.30E-01 |
| GOTERM_BP_FAT | Positive regulation of programmed cell death | 11 | 5.50E-02 | 4.40E-01 |
| GOTERM_BP_FAT | Positive regulation of cell death | 11 | 5.60E-02 | 4.40E-01 |
| Annotation cluster 18 | Enrichment Score: 1.22 | |||
| UP_SEQ_FEATURE | Domain: F-box | 4 | 5.30E-02 | 9.50E-01 |
| INTERPRO | Cyclin-like F-box | 4 | 5.40E-02 | 8.70E-01 |
| SMART | FBOX | 4 | 7.90E-02 | 8.00E-01 |
| Annotation cluster 19 | Enrichment score: 1.05 | |||
| BIOCARTA | Cadmium induces DNA synthesis and proliferation in macrophages | 3 | 3.20E-02 | 7.40E-01 |
| BIOCARTA | IL-3 signaling pathway | 3 | 3.70E-02 | 6.90E-01 |
| BIOCARTA | NGF pathway | 3 | 5.30E-02 | 7.40E-01 |
| BIOCARTA | EPO signaling pathway | 3 | 5.90E-02 | 7.10E-01 |
| BIOCARTA | Inhibition of cellular proliferation by Gleevec | 3 | 6.50E-02 | 6.90E-01 |
| BIOCARTA | TPO signaling pathway | 3 | 7.10E-02 | 6.80E-01 |
| BIOCARTA | Signaling pathway from G-protein families | 3 | 7.80E-02 | 6.70E-01 |
| BIOCARTA | IL-2 signaling pathway | 3 | 7.80E-02 | 6.70E-01 |
| BIOCARTA | PDGF signaling pathway | 3 | 1.00E-01 | 7.40E-01 |
| BIOCARTA | BCR signaling pathway | 3 | 1.10E-01 | 7.40E-01 |
| BIOCARTA | EGF signaling pathway | 3 | 1.10E-01 | 7.40E-01 |
| BIOCARTA | Fc epsilon receptor I signaling in mast cells | 3 | 1.40E-01 | 7.70E-01 |
| BIOCARTA | T-cell receptor signaling pathway | 3 | 1.70E-01 | 7.90E-01 |
| BIOCARTA | MAP kinase signaling pathway | 3 | 5.40E-01 | 1.00E+00 |
| Annotation cluster 20 | Enrichment score: 1.03 | |||
| UP_SEQ_FEATURE | Short sequence motif: BH3 | 3 | 1.70E-02 | 7.10E-01 |
| INTERPRO | Apoptosis regulator Bcl-2, BH | 3 | 2.90E-02 | 7.30E-01 |
| SMART | Bcl | 3 | 4.00E-02 | 7.90E-01 |
| GOTERM_CC_FAT | Mitochondrial outer membrane | 3 | 2.80E-01 | 9.50E-01 |
| GOTERM_CC_FAT | Organelle outer membrane | 3 | 3.40E-01 | 9.60E-01 |
| GOTERM_CC_FAT | Outer membrane | 3 | 3.60E-01 | 9.60E-01 |
Abbreviations: Bcl, B-cell lymphoma; BCR, B-cell receptor; DAVID, Database for Annotation, Visualization and Integrated Discovery; EGF, epidermal growth factor; EPO, erythropoietin; FDR, false discovery rate; IGF, insulin-like growth factor; IL, interleukin; NGF, nerve growth factor; PDGF, platelet-derived growth factor; TPO, thrombopoietin.
Furthermore, our DAVID analysis revealed that there were 14 KEGG pathways significantly enriched in the target list of hsa-miR-181a-5p, based on miRTarBase (Table 10). These pathways included pathways in cancer pathways (Figure 5), the MAPK signaling pathway (Figure 6), melanogenesis, chronic myeloid leukemia, small cell lung cancer, prostate cancer, dorsoventral axis formation, thyroid cancer, the Notch signaling pathway (Figure 7), long-term depression, renal cell carcinoma, the B cell receptor signaling pathway, vascular endothelial growth factor (VEGF) signaling pathway (Figure 8), and prion diseases.
Table 10.
The KEGG pathways by DAVID for the target list of hsa-miR-181a-5p based on miRTarBase
| Signaling pathway | Gene count | % | P-value | FDR |
|---|---|---|---|---|
| Pathways in cancer | 14 | 6.2 | 8.70E-04 | 8.20E-02 |
| MAPK signaling pathway | 9 | 4 | 4.20E-02 | 3.70E-01 |
| Melanogenesis | 7 | 3.1 | 3.20E-03 | 1.40E-01 |
| Chronic myeloid leukemia | 5 | 2.2 | 2.40E-02 | 3.80E-01 |
| Small cell lung cancer | 5 | 2.2 | 3.50E-02 | 3.90E-01 |
| Prostate cancer | 5 | 2.2 | 4.20E-02 | 4.10E-01 |
| Dorsoventral axis formation | 4 | 1.8 | 5.60E-03 | 1.70E-01 |
| Thyroid cancer | 4 | 1.8 | 8.60E-03 | 1.90E-01 |
| Notch signaling pathway | 4 | 1.8 | 3.10E-02 | 4.10E-01 |
| Long-term depression | 4 | 1.8 | 8.10E-02 | 5.60E-01 |
| Renal cell carcinoma | 4 | 1.8 | 8.40E-02 | 5.40E-01 |
| B-cell receptor signaling pathway | 4 | 1.8 | 9.80E-02 | 5.40E-01 |
| VEGF signaling pathway | 4 | 1.8 | 9.80E-02 | 5.40E-01 |
| Prion diseases | 3 | 1.3 | 9.40E-02 | 5.50E-01 |
Abbreviations: DAVID, Database for Annotation, Visualization and Integrated Discovery; FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes; VEGF, vascular endothelial growth factor.
Figure 5.
Cancer pathways in the target list of hsa-miR-181a-5p based on miRTarBase 4.0.
Notes: Several important oncogenes and tumor suppressors are likely regulated by hsa-miR-181a-5p (marked with a red star), including ATM, p300, p27, GADD45, and cyclin D1. These genes play an important role in the regulation of angiogenesis, cell proliferation, apoptosis, and metastasis.
Figure 6.
MAPK signaling pathway in the target list of hsa-miR-181a-5p based on miRTarBase 4.0.
Notes: hsa-miR-181a-5p can regulate MAPK signaling pathways. The MAPK/Erk signaling cascade is activated by a wide variety of receptors involved in growth and differentiation, including receptor tyrosine kinases, integrins, and ion channels.111 The specific components of the cascade vary greatly among different stimuli, but the architecture of the pathway usually includes a set of adaptors (Shc, GRB2, Crk, etc) linking the receptor to a guanine nucleotide exchange factor (SOS, C3G, etc) transducing the signal to small GTP-binding proteins (Ras, Rap1), which in turn activate the core unit of the cascade composed of a MAPKKK (Raf), a MAPKK (MEK1/2), and MAPK (Erk). An activated Erk dimer can regulate targets in the cytosol and also translocates to the nucleus, where it phosphorylates a variety of transcription factors regulating gene expression. p38 MAPKs (α, β, γ, and δ) are members of the MAPK family that are activated by a variety of environmental stresses and inflammatory cytokines. As with other MAPK cascades, the membrane-proximal component is a MAPKKK, typically a MEKK or a mixed lineage kinase (MLK). The MAPKKK phosphorylates and activates MKK3/6, the p38 MAPK kinases.111 MKK3/6 can also be activated directly by ASK1, which is stimulated by apoptotic stimuli. p38 MAPK is involved in regulation of HSP27, MAPKAPK-2 (MK2), MAPKAPK-3 (MK3), and several transcription factors, including ATF-2, Stat1, the Max/Myc complex, MEF-2, Elk-1, and indirectly, CREB via activation of MSK1.
Figure 7.
Notch signaling pathway in the target list of hsa-miR-181a-5p based on miRTarBase 4.0.
Notes: hsa-miR-181a-5p can regulate the function of the Notch signaling pathway. Notch signaling is an evolutionarily conserved pathway in multicellular organisms that regulates cell fate determination during development and maintains adult tissue homeostasis. In mammalian signal-sending cells, members of the Delta-like (DLL1, DLL3 & DLL4) and the Jagged (JAG1 & JAG2) families serve as ligands for Notch signaling receptors.112 Upon ligand binding, the NECD is cleaved away (S2 cleavage) from the TM-NICD domain by TACE (TNF-α ADAM metalloprotease converting enzyme). The NECD remains bound to the ligand, and this complex undergoes endocytosis/recycling within the signal-sending cell in a manner dependent on ubiquitination by Mib. In the signal-receiving cell, γ-secretase (also involved in Alzheimer’s disease) releases the NICD from the TM (S3 cleavage), which allows for nuclear translocation where it associates with the CSL (CBF1/Su(H)/Lag-1) transcription factor complex, resulting in subsequent activation of the canonical Notch target genes, including Myc, p21, and the HES-family members. Abnormal expression of Notch and related proteins has been observed in EC, and the Notch signaling pathway may play a role in the development, growth, and metastasis of EC.113–116 Targets of hsa-miR-181a-5p are marked with a red star.
Abbreviations: EC, endometrial cancer; TNF, tumor necrosis factor.
Figure 8.
VEGF signaling pathway in the target list of hsa-miR-181a-5p based on miRTarBase 4.0.
Notes: hsa-miR-181a-5p can regulate the VEGF signaling pathway. VEGF is an important signaling protein involved in both vasculogenesis and angiogenesis. All members of the VEGF family stimulate cellular responses by binding to tyrosine kinase receptors (the VEGFRs) on the cell surface, causing them to dimerize and become activated through transphosphorylation. This triggers a signaling cascade that activates several signaling pathways, such as PI3K/Akt, Erk1/2, Smad, and Notch, and results in endothelial cell proliferation and migration. A number of studies have shown that VEGF and its associated proteins are aberrant in EC.117–122 These proteins represent useful targets in the treatment of EC.
Abbreviations: EC, endometrial cancer; VEGF, vascular endothelial growth factor.
Combined validated targets of hsa-miR-181a-5p, based on both TarBase 6.0 and miRTarBase 4.0
When we combined the target lists of hsa-miR-181a-5p with experimental evidence from both TarBase 6.0 and miRTarBase 4.0, there were 313 validated targets for hsa-miR-181a-5p (Table 11). As shown in Table 12, our DAVID analysis showed that there were 26 functional clusters that were identified to be enriched with an enrichment score >1.0 in the combined target list of hsa-miR-181a-5p, based on both TarBase 6.0 and miRTarBase 4.0. The functions of these clusters involved response to hormone stimulus, response to endogenous stimulus, response to organic substance, negative regulation of apoptosis, negative and positive regulation of programmed cell death, negative and positive regulation of cell death, the DNA damage checkpoint, DNA integrity checkpoint, DNA damage response and signal transduction, the cell cycle checkpoint, response to DNA damage stimulus, bladder cancer, endometrial cancer (Figure 9), non-small-cell lung cancer, acute myeloid leukemia, glioma, melanoma, developmental growth, cell fate commitment, tissue morphogenesis, positive regulation of macromolecule biosynthetic process, positive regulation of cellular biosynthetic process, positive regulation of biosynthetic process, regulation of phosphorylation, regulation of phosphate metabolic process, regulation of phosphorus metabolic process, positive regulation of transcription, positive regulation of gene expression, positive regulation of protein kinase activity, positive regulation of kinase activity, positive regulation of transferase activity, regulation of protein kinase activity, regulation of kinase activity, positive regulation of cellular protein metabolic process, positive regulation of protein metabolic process, branching morphogenesis of a tube, positive regulation of cell development, morphogenesis of a branching structure, tube morphogenesis, regulation of cell development, neuron projection morphogenesis, cell projection morphogenesis, neuron projection development, cell part morphogenesis, neuron development, cell morphogenesis, cell projection organization, cellular component morphogenesis, neuron differentiation, IGF-1 signaling pathway, IL6 signaling pathway, insulin signaling pathway, signaling of hepatocyte growth factor receptor, embryonic appendage morphogenesis, embryonic limb morphogenesis, limb morphogenesis, appendage morphogenesis, limb development, appendage development, embryonic morphogenesis, response to ethanol, response to metal ion, response to inorganic substance, response to drug, response to estrogen stimulus, positive regulation of protein modification process, regulation of protein modification process, protein amino acid phosphorylation, phosphorylation, phosphate metabolic process, phosphorus metabolic process, cell aging, negative regulation of neuron apoptosis, aging, actin cytoskeleton organization, actin filament-based process, membrane organization, membrane insoluble fraction, Ras protein signal transduction, long-term depression, the B cell receptor signaling pathway, VEGF signaling pathway, Fc epsilon RI signaling pathway, ErbB signaling pathway, gap junction, gonadotropin-releasing hormone (GnRH) signaling pathway, T cell receptor signaling pathway, insulin signaling pathway, small GTPase-mediated signal transduction, chemokine signaling pathway, regulation of actin cytoskeleton, MAPK signaling pathway, axonogenesis, cell morphogenesis involved in neuron differentiation, cell morphogenesis involved in differentiation, nucleoplasm, nuclear lumen, intracellular organelle lumen, organelle lumen, membrane-enclosed lumen, hemopoiesis, hemopoietic or lymphoid organ development, immune system development, and transcription regulation (Table 12).
Table 11.
Combined targets of hsa-miR-181a-5p with experimental evidence based on both TarBase and miRTarBase 4.0
| Gene symbol | Full name | Alias | Function |
|---|---|---|---|
| ACOT12 | Acyl-CoA thioesterase 12 | CACH-1, Cach, STARD15, THEAL | Hydrolyzes acetyl-CoA to acetate and CoA |
| AFTPH | Aftiphilin | Nbla10388 | May play a role in membrane trafficking |
| AKAP12 | A kinase anchor protein 12 | AKAP250, SSeCKS | Anchoring protein that mediates the subcellular compartmentation of PKA and PKC |
| ALG10B | α-1,2-Glucosyltransferase | ALG10, KCR1 | Transfers glucose from dolichyl phosphate glucose onto the lipid-linked oligosaccharide Glc(2)Man(9)GlcNAc(2)-PP-Dol |
| AMMECR1 | Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 | RP13-360B22.1, AMMERC1 | |
| ANKRD1 | Ankyrin repeat domain 1 (cardiac muscle) | ALRP, C-193, CARP, CVARP, MCARP, bA320F15.2 | Plays an important role in endothelial cell activation |
| ANKRD13C | Ankyrin repeat domain 13C | RP4-677H15.5, dJ677H15.3 | |
| AP1M1 | Adaptor-related protein complex 1, mu 1 subunit | AP47, CLAPM2, CLTNM, MU-1A | Subunit of clathrin-associated adaptor protein complex 1 that plays a role in protein sorting in the trans-Golgi network and endosomes |
| ARF6 | ADP-ribosylation factor 6 | Involved in protein trafficking | |
| ARHGAP11A | Rho GTPase activating protein 11A | RP11-1000B6.5, GAP (1–12) | GTPase activator activity |
| ARHGAP12 | Rho GTPase activating protein 12 | GTPase activator for the Rho-type GTPases, by converting them to an inactive GDP-bound state | |
| ARL6IP1 | ADP-ribosylation factor-like 6 interacting protein 1 | AIP1, ARL6IP, ARMER, SPG61 | May be involved in protein transport, membrane trafficking, or cell signaling during hematopoietic maturation |
| ARL6IP6 | ADP-ribosylation factor-like 6 interacting protein 6 | RP23-265N10.1, 2310057C01Rik, 2610529A11Rik, Aip-6 | May be involved in protein transport, membrane trafficking, or cell signaling during hematopoietic maturation |
| ATF7IP2 | Activating transcription factor 7 interacting protein 2 | MCAF2 | Recruiter that couples transcriptional factors to general transcription apparatus and thereby modulates transcription regulation and chromatin formation |
| ATG10 | Autophagy related 10 | PP12616, APG10, APG10L, pp12616 | Plays a role in autophagy |
| ATM | ATM serine/threonine kinase | AT1, ATA, ATC, ATD, ATDC, ATE, TEL1, TELO1 | Serine/threonine protein kinase |
| ATP6V0E1 | ATPase, H+ transporting, lysosomal 9 kDa, V0 subunit e1 | ATP6H, ATP6V0E, M9.2, Vma21, Vma21p | Vacuolar ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells |
| ATP8A1 | ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 | ATPASEII, ATPIA, ATPP2 | May play a role in the transport of aminophospholipids from the outer to the inner leaflet of various membranes and the maintenance of asymmetric distribution of phospholipids |
| BAG2 | BCL2-associated athanogene 2 | RP3-496N17.2, BAG-2, dJ417I1.2 | Inhibits the chaperone activity of HSP70/HSC70 by promoting substrate release |
| BCL2 | B-cell CLL/lymphoma 2 | Bcl-2, PPP1R50 | Suppresses apoptosis |
| BCL2L11 | BCL2-like 11 | BAM, BIM, BOD | Induces apoptosis |
| BDNF | Brain-derived neurotrophic factor | ANON2, BULN2 | Promotes the survival of neuronal populations |
| BPGM | 2,3-bisphosphoglycerate mutase | DPGM | Plays a major role in regulating hemoglobin oxygen affinity |
| BRCA1 | Breast cancer 1, early onset | BRCAI, BRCC1, BROVCA1, IRIS, PNCA4, PPP1R53, PSCP, RNF53 | Plays a central role in DNA repair by facilitating cellular response to DNA repair |
| BRIX1 | Biogenesis of ribosomes, homolog (S. cerevisiae) | BRIX, BXDC2 | Required for biogenesis of the 60S ribosomal subunit |
| BRMS1L | Breast cancer metastasis-suppressor 1-like | BRMS1 | Involved in the HDAC1-dependent transcriptional repression activity |
| BTBD3 | BTB (POZ) domain containing 3 | RP4-742J24.3, dJ742J24.1 | Acts as a key regulator of dendritic field orientation during the development of sensory cortex |
| C1orf109 | Chromosome 1 open reading frame 109 | ||
| C1orf43 | Chromosome 1 open reading frame 43 | HSPC012, NICE-3, NS5ATP4, S863-3 | |
| C1QTNF9 | C1q and tumor necrosis factor related protein 9 | 9130217G22Rik, CTRP9, Ciqtnf9 | Activates AMPK, AKT, and p44/42 MAPK signaling pathways |
| C8A | Complement component 8, α polypeptide | C8 is a constituent of the membrane attack complex | |
| CBX3 | Chromobox homolog 3 | HECH, HP1-GAMMA, HP1Hs-γ | Involved in transcriptional silencing in heterochromatin-like complexes |
| CCDC6 | Coiled-coil domain containing 6 | D10S170, H4, PTC, TPC, TST1 | Functions as a tumor suppressor |
| CCDC82 | Coiled-coil domain containing 82 | HT025, HSPC048 | |
| CCND1 | Cyclin D1 | BCL1, D11S287E, PRAD1, U21B31 | Essential for the control of the cell cycle at the G1/S (start) transition |
| CCNG1 | Cyclin G1 | CCNG | May play a role in growth regulation |
| CD46 | CD46 molecule, complement regulatory protein | AHUS2, MCP, MIC10, TLX, TRA2.10 | Acts as a cofactor for complement factor I |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | CDKN4, KIP1, MEN1B, MEN4, P27KIP1 | Important regulator of cell cycle progression |
| CDX2 | Caudal type homeobox 2 | CDX-3, CDX3 | Involved in the transcriptional regulation of multiple genes expressed in the intestinal epithelium |
| CEP97 | Centrosomal protein 97 kDa | 2810403B08Rik, LRRIQ2 | Collaborates with cep110, being involved in the suppression of a cilia assembly program |
| CFI | Complement factor I | AHUS3, ARMD13, C3BINA, C3b-INA, FI, IF, KAF | Responsible for cleaving the α-chains of C4b and C3b, in the presence of the cofactors C4-binding protein and factor H, respectively |
| CHD1 | Chromodomain helicase DNA binding protein 1 | Sequence-selective DNA-binding protein | |
| CHL1 | Cell adhesion molecule L1-like | CALL, L1CAM2 | Plays a role in nervous system development and in synaptic plasticity |
| CHRFAM7A | CHRNA7 (cholinergic receptor, nicotinic, α 7, exons 5–10) and FAM7A (family with sequence similarity 7A, exons A–E) fusion | CHRNA7, CHRNA7-DR1, D-10 | Extracellular ligand-gated ion channel activity |
| CLUAP1 | Clusterin associated protein 1 | CFAP22, FAP22 | May play a role in cell proliferation or apoptosis |
| COL27A1 | Collagen, type XXVII, α 1 | RP11-82I1.1 | Plays a role during the calcification of cartilage and the transition of cartilage to bone |
| COPS2 | COP9 signalosome subunit 2 | ALIEN, CSN2, SGN2, TRIP15 | Involved in various cellular and developmental processes |
| CST5 | Cystatin D | Cysteine proteinase inhibitor | |
| CXorf1 | Transmembrane protein 257 | CXorf1 | |
| D3R | Dopamine receptor D3 | DRD3; D3DR; ETM1; FET1 | Inhibits adenylyl cyclase through inhibitory G-proteins, plays a role in cognitive and emotional functions |
| DCP2 | Decapping mRNA 2 | NUDT20 | Necessary for the degradation of mRNAs |
| DCST1 | DC-STAMP domain containing 1 | RP11-307C12.10-003 | Protein and zinc ion binding |
| DDIT4 | DNA-damage-inducible transcript 4 | RP11-442H21.1, Dig2, REDD-1, REDD1 | Inhibits cell growth by regulating the frap1 pathway upstream of the tsc1-tsc2 complex and downstream of Akt1 |
| DDX27 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 | HSPC259, DRS1, Drs1p, PP3241, RHLP, dJ686N3.1 | Probable ATP-dependent RNA helicase |
| DDX3X | DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked | DBX, DDX14, DDX3, HLP2 | ATP-dependent RNA helicase |
| DNAJC7 | DnaJ (HSP40) homolog, subfamily C, member 7 | DJ11, DJC7, TPR2, TTC2 | Acts as co-chaperone regulating the molecular chaperones HSP70 and HSP90 in folding of steroid receptors |
| DSCR8 | Down syndrome critical region gene 8 | C21orf65, CT25.1a, CT25.1b, MMA-1, MMA-1a, MMA-1b, MMA1, MTAG2 | |
| DUSP5 | Dual specificity phosphatase 5 | DUSP, HVH3 | Displays phosphatase activity toward several substrates |
| DUSP6 | Dual specificity phosphatase 6 | HH19, MKP3, PYST1 | Inactivates MAP kinases |
| EIF1 | Eukaryotic translation initiation factor 1 | A121, EIF-1A, ISO1, SUI1, EIF1 | Necessary for scanning and involved in initiation site selection |
| EIF2C1 | Argonaute RISC catalytic component 1 | RP4-789D17.1, EIF2C, AGO1, GERP95, Q99 | Required for RNA-mediated gene silencing |
| EIF2C3 | Argonaute RISC catalytic component 3 | AGO3 | Required for RNA-mediated gene silencing |
| ELAVL1 | ELAV like RNA binding protein 1 | ELAV1, HUR, Hua, MelG | Binds avidly to the AU-rich element in FOS and IL3 mRNAs |
| ENAH | Enabled homolog | RP11-496N12.7, ENA, MENA, NDPP1 | Ena/VASP proteins are actin-associated proteins involved in a range of processes dependent on cytoskeleton remodeling and cell polarity |
| EP300 | E1A binding protein p300 | RP1-85F18.1, KAT3B, RSTS2, p300 | Functions as HAT and regulates transcription via chromatin remodeling |
| EPHA5 | EPH receptor A5 | CEK7, EHK-1, EHK1, EK7, HEK7, TYRO4 | Receptor for members of the ephrin-A family |
| ESR1 | Estrogen receptor 1 | RP1-130E4.1, ER, ESR, ESRA, ESTRR, Era, NR3A1 | Nuclear hormone receptor |
| EYA4 | EYA transcriptional coactivator and phosphatase 4 | RP11-704J17.4, CMD1J, DFNA10 | Tyrosine phosphatase that specifically dephosphorylates “Tyr-142” of histone H2AX (H2AXY142ph) |
| FAM160A2 | Family with sequence similarity 160, member A2 | C11orf56 | |
| FAM222B | Family with sequence similarity 222, member B | C17orf63 | |
| FAM47B | Family with sequence similarity 47, member B | RP13-520K9.1 | |
| FAT1 | FAT atypical cadherin 1 | CDHF7, CDHR8, FAT, ME5, hFat1 | Could function as a cell-adhesion protein |
| FBXO11 | F-box protein 11 | UG063H01, FBX11, PRMT9, UBR6, VIT1 | Substrate recognition component of the SCF E3 ubiquitin-protein ligase complex |
| FBXO28 | F-box protein 28 | CENP-30, Fbx28 | Probably recognizes and binds to some phosphorylated proteins and promotes their ubiquitination and degradation |
| FBXO33 | F-box protein 33 | BMND12, Fbx33, c14_5247 | Substrate recognition component of the SCF E3 ubiquitin-protein ligase complex, which mediates the ubiquitination and subsequent proteasomal degradation of target proteins |
| FBXO34 | F-box protein 34 | CGI-301, Fbx34 | Substrate-recognition component of the SCF E3 ubiquitin ligase complex |
| FKBP10 | FK506 binding protein 10 | PSEC0056, FKBP65, OI11, OI6, PPIASE, hFKBP65 | PPIases accelerate the folding of proteins during protein synthesis |
| FKBP4 | FK506 binding protein 4 | FKBP51, FKBP52, FKBP59, HBI, Hsp56, PPIase, p52 | May play a role in the intracellular trafficking of heterooligomeric forms of steroid hormone receptors |
| FKBP7 | FK506 binding protein 7 | UNQ670/PRO1304, FKBP23, PPIase | PPIases accelerate the folding of proteins during protein synthesis |
| FOS | FBJ murine osteosarcoma viral oncogene homolog | AP-1, C-FOS, p55 | Nuclear phosphoprotein, which forms a tight but noncovalently linked complex with the JUN/AP-1 transcription factor |
| FRA10AC1 | Fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 | PRO2972, C10orf4, F26C11.1-like, FRA10A | |
| FSIP1 | Fibrous sheath interacting protein 1 | HSD10 | |
| FXYD6 | FXYD domain containing ion transport regulator 6 | UNQ521/PRO1056 | |
| GADD45G | Growth arrest and DNA-damage-inducible, γ | RP11-260L6.1, CR6, DDIT2, GADD45γ, GRP17 | Involved in the regulation of growth and apoptosis |
| GANAB | Glucosidase, α; neutral AB | G2AN, GLUII | Cleaves sequentially the 2 innermost α-1,3-linked glucose residues from the Glc(2)Man(9)GlcNAc(2) oligosaccharide precursor of immature glycoproteins |
| GATA6 | GATA binding protein 6 | Regulates terminal differentiation and/or proliferation | |
| GATAD2B | GATA zinc finger domain containing 2B | RP11-216N14.6, MRD18, P66β, p68 | Has transcriptional repressor activity |
| GCNT1 | Glucosaminyl (N-acetyl) transferase 1, core 2 | RP11-214N16.1, C2GNT, C2GNT-L, C2GNT1, G6NT, NACGT2, NAGCT2 | Forms critical branches in O-glycans |
| GIGYF1 | GRB10 interacting GYF protein 1 | PP3360, GYF1, PERQ1 | May act cooperatively with GRB10 to regulate tyrosine kinase receptor signaling |
| GNA13 | G protein, α 13 | G13 | Modulators or transducers in various transmembrane signaling systems |
| GNAI3 | G protein, α inhibiting activity polypeptide 3 | RP5-1160K1.2, 87U6, ARCND1 | G proteins are involved as modulators or transducers in various transmembrane signaling systems |
| GNB1 | G protein, β polypeptide 1 | RP1-283E3.7 | A modulator or transducer in various transmembrane signaling systems |
| GPR137B | G protein-coupled receptor 137B | RP5-985L19.1, TM7SF1 | |
| GPR78 | G protein-coupled receptor 78 | UNQ5925/PRO19818 | Orphan receptor |
| GPR83 | G protein-coupled receptor 83 | GIR, GPR72 | Orphan receptor. Could be a neuropeptide y receptor |
| GPRIN3 | GPRIN family member 3 | GRIN3 | May be involved in neurite outgrowth |
| GSTM2 | Glutathione S-transferase mu 2 (muscle) | GST4, GSTM-2, GTHMUS, GSTM2 | Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles |
| H1F0 | H1 histone family, member 0 | H10, H1FV | Histones H1 are necessary for the condensation of nucleosome chains into higher-order structures |
| H2AFY | H2A histone family, member Y | H2A.y, H2A/y, H2AF12M, H2AFJ, MACROH2A1.1, mH2A1, macroH2A1.2 | Plays a central role in transcription regulation, DNA repair, DNA replication, and chromosomal stability |
| H3F3B | H3 histone, family 3B | H3.3B | Plays a central role in transcription regulation, DNA repair, DNA replication, and chromosomal stability |
| HDAC6 | Histone deacetylase 6 | JM21, CPBHM, HD6, PPP1R90 | Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4) |
| HERC3 | HECT and RLD domain containing E3 ubiquitin protein ligase 3 | E3 ubiquitin-protein ligase | |
| HEY2 | Hes-related family bHLH transcription factor with YRPW motif 2 | RP1-293L8.3, CHF1, GRIDLOCK, GRL, HERP1, HESR2, HRT2, bHLHb32 | Downstream effector of Notch signaling, which may be required for cardiovascular development |
| HIPK2 | Homeodomain interacting protein kinase 2 | PRO0593 | Protein kinase acting as a corepressor of several transcription factors |
| HMGB2 | High mobility group box 2 | HMG2 | Binds preferentially ssDNA and unwinds double stranded DNA |
| HNRNPAB | Heterogeneous nuclear ribonucleoprotein A/B | ABBP1, HNRPAB | Binds single-stranded RNA |
| HNRPDL | Heterogeneous nuclear ribonucleoprotein D-like | HNRNP, HNRPDL, JKTBP, JKTBP2, laAUF1 | Acts as a transcriptional regulator |
| HOOK3 | Hook microtubule-tethering protein 3 | HK3 | Probable cytoskeletal linker protein, involved in tethering the Golgi complex to the cytoskeleton |
| HOXA10 | Homeobox A10 | HOX1, HOX1.8, HOX1H, PL | Sequence-specific transcription factor |
| HOXA11 | Homeobox A11 | HOX1, HOX1I | Sequence-specific transcription factor |
| HRAS | Harvey rat sarcoma viral oncogene homolog | C-BAS/HAS, C-H-RAS, C-HA-RAS1, CTLO, H-RASIDX, HAMSV1, RASH1, p21ras, HRAS | Ras proteins bind GDP/GTP and possess intrinsic GTPase activity |
| HSD17B3 | Hydroxysteroid (17-β) dehydrogenase 3 | RP11-240L7.3, EDH17B3, SDR12C2 | Favors the reduction of androstenedione to testosterone |
| HSP90B1 | Heat-shock protein 90 kDa β (Grp94), member 1 | ECGP, GP96, GRP94, HEL-S-125m, HEL35, TRA1 | Molecular chaperone that functions in the processing and transport of secreted proteins |
| HSPA13 | Heat-shock protein 70 kDa family, member 13 | STCH | Has peptide-independent ATPase activity |
| HSPA1B | Heat-shock 70 kDa protein 1B | DAAP-21F2.7, HSP70-1B, HSP70-2 | Stabilizes preexistent proteins against aggregation and mediates the folding of newly translated polypeptides in the cytosol as well as within organelles |
| HUWE1 | HECT, UBA and WWE domain containing 1 | RP3-339A18.4, ARF-BP1, HECTH9, HSPC272, Ib772, LASU1, MULE, URE-B1, UREB1 | E3 ubiquitin-protein ligase mediating ubiquitination and subsequent proteasomal degradation of target proteins. |
| ICMT | Isoprenylcysteine carboxyl methyltransferase | RP1-120G22.4, HSTE14, MST098, MSTP098, PCCMT, PCMT, PPMT | Catalyzes the posttranslational methylation of isoprenylated C-terminal cysteine residues |
| IDS | Iduronate 2-sulfatase | MPS2, SIDS | Required for the lysosomal degradation of heparan sulfate and dermatan sulfate |
| INCENP | Inner centromere protein antigens | Component of the chromosomal passenger complex, a complex that acts as a key regulator of mitosis | |
| IQCG | IQ motif containing G | CFAP122, DRC9 | |
| KAT2B | K(lysine) acetyltransferase 2B | CAF, P/CAF, PCAF | Functions as a HAT to promote transcriptional activation |
| KBTBD3 | Kelch repeat and BTB (POZ) domain containing 3 | BKLHD3 | |
| KBTBD7 | Kelch repeat and BTB (POZ) domain containing 7 | ||
| KCTD2 | Potassium channel tetramerization domain containing 2 | ||
| KCTD3 | Potassium channel tetramerization domain containing 3 | RP11-5F19.1, NY-REN-45 | |
| KIAA0100 | KIAA0100 | BCOX, BCOX1, CT101 | |
| KIAA0101 | KIAA0101 | L5, NS5ATP9, OEATC, OEATC-1, OEATC1, PAF, PAF15, p15(PAF), p15/PAF, p15PAF | May be involved in protection of cells from UV-induced cell death |
| KIAA1462 | KIAA1462 | JCAD | |
| KIAA2026 | KIAA2026 | ||
| KLF6 | Kruppel-like factor 6 | RP11-184A2.1, BCD1, CBA1, COPEB, CPBP, GBF, PAC1, ST12, ZF9 | Plays a role in B-cell growth and development |
| KLHL15 | Kelch-like family member 15 | HEL-S-305 | Probable substrate-specific adapter of an E3 ubiquitin-protein ligase complex, which mediates the ubiquitination and subsequent proteasomal degradation of target proteins |
| KLHL42 | Kelch-like family member 42 | Ctb9, KLHDC5 | Substrate-specific adapter of a BCR (BTB-CUL3-RBX1) E3 ubiquitin-protein ligase complex required for mitotic progression and cytokinesis |
| KLRC4 | Killer cell lectin-like receptor subfamily C, member 4 | NKG2-F, NKG2F | May play a role as a receptor for the recognition of MHC class I HLA-E molecules by NK cells |
| KRAS | Kirsten rat sarcoma viral oncogene homolog | C-K-RAS, CFC2, K-RAS2A, K-RAS2B, K-RAS4A, K-RAS4B, KI-RAS1, KRAS2, NS, NS3, RASK2, KRAS | Binds GDP/GTP and possesses intrinsic GTPase activity |
| LAMA3 | Laminin, α3 | BM600, E170, LAMNA, LOCS, lama3a | Binding to cells via a high-affinity receptor, mediating the attachment, migration, and organization of cells into tissues |
| LBR | Lamin B receptor | PRO0650, DHCR14B, LMN2R, PHA, TDRD18 | Anchors the lamina and the heterochromatin to the inner nuclear membrane. |
| LCLAT1 | Lysocardiolipin acyltransferase 1 | UNQ1849/PRO3579, 1AGPAT8, AGPAT8, ALCAT1, HSRG1849, LYCAT, UNQ1849 | Acyl-CoA: lysocardiolipin acyltransferase |
| LFNG | LFNG O-fucosylpeptide 3-β-N- acetylglucosaminyltransferase | SCDO3 | Glycosyltransferase |
| LGALSL | Lectin, galactoside-binding-like | HSPC159, GRP | Does not bind lactose and may not bind carbohydrates |
| LPGAT1 | Lysophosphatidylglycerol acyltransferase 1 | FAM34A, FAM34A1, NET8 | Lysophoshatidylglycerol-specific acyltransferase |
| LRRC17 | Leucine rich repeat containing 17 | UNQ3076/PRO9909, P37NB | Involved in bone homeostasis, acting as a negative regulator of RANKL-induced osteoclast precursor differentiation from bone marrow precursors |
| LRRN3 | Leucine rich repeat neuronal 3 | Nbla10363, FIGLER5, NLRR-3, NLRR3 | |
| LYSMD3 | LysM, putative peptidoglycan-binding, domain containing 3 | ||
| MAP1B | Microtubule-associated protein 1B | FUTSCH, MAP5, PPP1R102 | May play a role in the cytoskeletal changes that accompany neurite extension |
| MAP2K1 | Mitogen-activated protein kinase kinase 1 | CFC3, MAPKK1, MEK1, MKK1, PRKMK1 | Catalyzes the concomitant phosphorylation of a threonine and a tyrosine residue in a Thr-Glu-Tyr sequence located in MAP kinases |
| MAZ | MYC-associated zinc finger protein (purine-binding transcription factor) | PUR1, Pur-1, SAF-1, SAF-2, SAF-3, ZF87, ZNF801, Zif87 | May function as a transcription factor, with dual roles in transcription initiation and termination |
| MCL1 | Myeloid cell leukemia 1 | BCL2L3, EAT-ES, MCL1L, MCL1S, Mcl-1, TM, bcl2-L-3, mcl1/EAT, MCL1 | Involved in the regulation of apoptosis versus cell survival, and in the maintenance of viability but not of proliferation |
| METAP1 | Methionyl aminopeptidase 1 | MAP1A, MetAP1A | Removes the amino-terminal methionine from nascent proteins |
| MFAP3 | Microfibrillar-associated protein 3 | Component of the elastin-associated microfibrils | |
| MGAT5 | Mannosyl (α-1,6-)-glycoprotein β1,6-N-acetyl- glucosaminyltransferase | GNT-V, GNT-VA | Catalyzes the addition of N-acetylglucosamine in β1-6 linkage to the α-linked mannose of biantennary N-linked oligosaccharides |
| MIF | Macrophage migration inhibitory factor (glycosylation-inhibiting factor) | GIF, GLIF, MMIF | The expression of MIF at sites of inflammation suggests a role for the mediator in regulating the function of macrophage in host defense. Also acts as a phenylpyruvate tautomerase |
| MOB1A | MOB kinase activator 1A | C2orf6, MATS1, MOB1, MOBK1B, MOBKL1B, Mob4B | Activator of LATS1/2 in the Hippo signaling pathway |
| MOB1B | MOB kinase activator 1B | MATS2, MOB4A, MOBKL1A | Activator of LATS1/2 in the Hippo signaling pathway |
| MOB3B | MOB kinase activator 3B | C9orf35, MOB1D, MOBKL2B | May regulate the activity of kinases |
| MRPS14 | Mitochondrial ribosomal protein S14 | DJ262D12.2, HSMRPS14, MRP-S14, S14mt | |
| MTMR12 | Myotubularin related protein 12 | 3-PAP, PIP3AP | Inactive phosphatase that plays a role as an adapter for the phosphatase myotubularin to regulate myotubularin intracellular location |
| MTMR3 | Myotubularin related protein 3 | hCG_2011013, FYVE-DSP1, ZFYVE10 | Phosphatase that acts on lipids with a phosphoinositol head group |
| MTRR | 5-methyltetrahydrofolate-homocysteine methyltransferase reductase | MSR, cblE | Involved in the reductive regeneration of cob(I)alamin cofactor required for the maintenance of methionine synthase in a functional state |
| MYO9A | Myosin IXA | Myosins are actin-based motor molecules with ATPase activity Unconventional myosins serve in intracellular movements |
|
| NCAPG | Non-SMC condensin I complex, subunit G | CAPG, CHCG, NY-MEL-3, YCG1 | Regulatory subunit of the condensin complex, a complex required for conversion of interphase chromatin into mitotic-like condense chromosomes |
| ND2 | NADH dehydrogenase subunit 2 | Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) | |
| NFYB | Nuclear transcription factor Y β | CBF-A, CBF-B, HAP3, NF-YB | Stimulates the transcription of various genes by recognizing and binding to a CCAAT motif in promoters |
| NKX3-2 | NK3 homeobox 2 | BAPX1, NKX3.2, NKX3B, SMMD | Transcriptional repressor that acts as a negative regulator of chondrocyte maturation |
| NLK | Nemo-like kinase | Role in cell fate determination, required for differentiation of bone marrow stromal cells | |
| NMRK2 | Nicotinamide riboside kinase 2 | ITGB1BP3, MIBP, NRK2 | |
| NOL4 | Nucleolar protein 4 | HRIHFB2255, CT125, NOLP | |
| NOTCH1 | Notch 1 | TAN1, hN1 | Functions as a receptor for membrane-bound ligands Jagged1, Jagged2 and Delta1 to regulate cell fate determination |
| NOTCH2 | Notch 2 | AGS2, HJCYS, hN2 | Functions as a receptor for membrane-bound ligands Jagged1, Jagged2, and Delta1 to regulate cell fate determination |
| NR6A1 | Nuclear receptor subfamily 6, group A, member 1 | CT150, GCNF, GCNF1, NR61, RTR, hGCNF, hRTR | Orphan nuclear receptor. May be involved in the regulation of gene expression in germ cell development during gametogenesis |
| NRP1 | Neuropilin 1 | RP11-342D11.1, BDCA4, CD304, NP1, NRP, VEGF165R | The membrane-bound isoform 1 is a receptor involved in the development of the cardiovascular system, in angiogenesis, in the formation of certain neuronal circuits, and in organogenesis outside the nervous system |
| NUDT12 | Nudix-type motif 12 | Hydrolyzes NAD(P)H to NMNH and AMP (2′,5′-ADP), and diadenosine diphosphate to AMP | |
| NUPL1 | Nucleoporin like 1 | RP11-206I15.1, PRO2463 | Component of the nuclear pore complex, a complex required for the trafficking across the nuclear membrane |
| OAZ1 | Ornithine decarboxylase antizyme 1 | AZI, OAZ | Binds to and destabilizes ornithine decarboxylase, which is then degraded. Also inhibits cellular uptake of polyamines by inactivating the polyamine uptake transporter |
| OCA2 | Oculocutaneous albinism II | BEY, BEY1, BEY2, BOCA, D15S12, EYCL, EYCL2, EYCL3, HCL3, P, PED, SHEP1 | Could be involved in the transport of tyrosine |
| OFCC1 | Orofacial cleft 1 candidate 1 | MRDS1 | |
| OR11A1 | Olfactory receptor, family 11, subfamily A, member 1 | DAAP-34I1.2, 6M1-18, OR11A2, dJ994E9.6, hs6M1-18 | Odorant receptor |
| OTUD1 | OTU deubiquitinase 1 | DUBA7, OTDC1 | Deubiquitinating enzyme that specifically hydrolyzes “Lys-63”- linked polyubiquitin to monoubiquitin |
| OTX2 | Orthodenticle homeobox 2 | CPHD6, MCOPS5 | Probably plays a role in the development of the brain and the sense organs |
| PABPC1 | Poly(A) binding protein, cytoplasmic 1 | PAB1, PABP, PABP1, PABPC2, PABPL1 | Binds the poly(A) tail of mRNA |
| PCAF | K(lysine) acetyltransferase 2B | CAF, P/CAF, PCAF | Functions as a component of the PCAF complex |
| PCDHB8 | Protocadherin β 8 | PCDH-β8, PCDH3I | Potential calcium-dependent cell-adhesion protein |
| PFKFB2 | 6-Phosphofructo-2-kinase/fructose-2, 6-biphosphatase 2 | RP11-164O23.2, PFK-2/FBPase-2 | Synthesis and degradation of fructose 2,6-bisphosphate |
| PGD | Phosphogluconate dehydrogenase | 6PGD | Catalyzes the oxidative decarboxylation of 6-phosphogluconate to ribulose 5-phosphate and CO2, with concomitant reduction of NADP to NADPH |
| PHOX2A | Paired-like homeobox 2a | ARIX, CFEOM2, FEOM2, NCAM2, PMX2A | May be involved in regulating the specificity of expression of the catecholamine biosynthetic genes |
| PHPT1 | Phosphohistidine phosphatase 1 | RP11-216L13.10-005, CGI-202, HEL-S-132P, HSPC141, PHP14 | Exhibits phosphohistidine phosphatase activity |
| PIM3 | Pim-3 proto-oncogene, serine/threonine kinase | CITF22-49E9.1, pim-3 | May be involved in cell cycle progression and antiapoptotic process |
| PITPNB | Phosphatidylinositol transfer protein β | RP3-353E16.2, PI-TP-β, PtdInsTP, VIB1B | Catalyzes the transfer of PtdIns and phosphatidylcholine between membranes |
| PLA2G4C | Phospholipase A2, group IVC | CPLA2-γ | Has a preference for arachidonic acid at the sn-2 position of phosphatidylcholine as compared with palmitic acid |
| PLAG1 | Pleiomorphic adenoma gene 1 | PSA, SGPA, ZNF912 | Transcription factor whose activation results in upregulation of target genes, such as IGFII, leading to uncontrolled cell proliferation |
| PLCL2 | Phospholipase C-like 2 | PLCE2 | May play an role in the regulation of Ins(1,4,5)P3 around the endoplasmic reticulum |
| PLXDC2 | Plexin domain containing 2 | UNQ2514/PRO6003, TEM7R | May play a role in tumor angiogenesis |
| PNPT1 | Polyribonucleotide nucleotidyltransferase 1 | COXPD13, DFNB70, OLD35, PNPASE, old-35 | Involved in mRNA degradation |
| POLR2B | Polymerase (RNA) II (DNA directed) polypeptide B | POL2RB, RPB2, hRPB140, hsRPB2 | DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA, using the four ribonucleoside triphosphates as substrates |
| PPA1 | Pyrophosphatase (inorganic) 1 | RP11-367H5.1, HEL-S-66p, IOPPP, PP, PP1, SID6-8061 | |
| PPM1A | Protein phosphatase, Mg2+/Mn2+ dependent, 1A | PP2C-ALPHA, PP2CA, PP2Cα | Enzyme with a broad specificity |
| PPP1R9A | Protein phosphatase 1, regulatory subunit 9A | NRB1, NRBI, Neurabin-I | Binds to actin filaments (F-actin) and shows crosslinking activity |
| PPP2CA | Protein phosphatase 2, catalytic subunit, α isozyme | PP2Ac, PP2CA, PP2Cα, RP-C | PP2A can modulate the activity of phosphorylase B kinase casein kinase 2, mitogen-stimulated S6 kinase, and MAP-2 kinase |
| PPP2R5C | Protein phosphatase 2, regulatory subunit B’, γ | B56G, PR61G | The B regulatory subunit might modulate substrate selectivity and catalytic activity, and also might direct the localization of the catalytic enzyme to a particular subcellular compartment |
| PRAP1 | Proline-rich acidic protein 1 | RP11-122K13.6, PRO1195, UPA | May play an important role in maintaining normal growth homeostasis in epithelial cells |
| PRDX3 | Peroxiredoxin 3 | AOP-1, AOP1, HBC189, MER5, PRO1748, SP-22, prx-III | Involved in redox regulation of the cell |
| PRLR | Prolactin receptor | HPRL, MFAB, hPRLrI | This is a receptor for the anterior pituitary hormone prolactin |
| PROSC | Proline synthetase co-transcribed homolog | ||
| PROX1 | Prospero homeobox 1 | May play a fundamental role in early development of the central nervous system | |
| PRR4 | Proline rich 4 | LPRP, PROL4 | |
| PRRC2B | Proline-rich coiled-coil 2B | RP11-334J6.1, BAT2L, BAT2L1, KIAA0515, LQFBS-1 | |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | COX-2, COX2, GRIPGHS, PGG/HS, PGHS-2, PHS-2, hCox-2 | May have a role as a major mediator of inflammation and/or a role for prostanoid signaling in activity-dependent plasticity |
| PTPLAD1 | Protein tyrosine phosphatase-like A domain containing 1 | B-IND1, HACD3, HSPC121 | Involved in Rac1-signaling pathways leading to the modulation of gene expression |
| PTPN11 | Protein tyrosine phosphatase, non-receptor type 11 | BPTP3, CFC, NS1, PTP-1D, PTP2C, SH-PTP2, SH-PTP3, SHP2 | Acts downstream of various receptor and cytoplasmic protein tyrosine kinases to participate in the signal transduction from the cell surface to the nucleus |
| PTPN22 | Protein tyrosine phosphatase, non-receptor type 22 | LYP, LYP1, LYP2, PEP, PTPN8 | Seems to act on casitas B-lineage lymphoma (Cbl) |
| PTPRZ1 | Protein tyrosine phosphatase, receptor-type, Z polypeptide 1 | HPTPZ, HPTPζ, PTP-ζ, PTP18, PTPRZ, PTPZ, R-PTP-ζ-2, RPTPB, RPTPβ, phosphacan | May be involved in the regulation of specific developmental processes in the central nervous system |
| PUM1 | Pumilio RNA-binding family member 1 | RP1-65J11.4, HSPUM, PUMH, PUMH1, PUML1 | Sequence-specific RNA-binding protein that regulates translation and mRNA stability by binding the 3′-UTR of mRNA targets |
| RAB8B | RAB8B, member RAS oncogene family | May be involved in vesicular trafficking and neurotransmitter release | |
| RALA | V-ral simian leukemia viral oncogene homolog A | RAL | Multifunctional GTPase involved in a variety of cellular processes, including gene expression, cell migration, cell proliferation, oncogenic transformation, and membrane trafficking |
| RASSF6 | Ras association (RalGDS/AF-6) domain family member 6 | May act as a Ras effector protein | |
| RBM15 | RNA binding motif protein 15 | OTT, OTT1, SPEN | May be implicated in HOX gene regulation |
| RLF | Rearranged L-myc fusion | RP1-39G22.1, ZN-15L, ZNF292L | May be involved in transcriptional regulation |
| RNF2 | Ring finger protein 2 | GS1-120K12.1, BAP-1, BAP1, DING, HIPI3, RING1B, RING2 | E3 ubiquitin-protein ligase that mediates monoubiquitination of “Lys-119” of histone H2A, playing a central role in histone code and gene regulation |
| RNF34 | Ring finger protein 34 | CARP-1, CARP1, RFI, RIF, RIFF, hRFI | Has E3 ubiquitin-protein ligase activity. Regulates the levels of CASP8 and CASP10 by targeting them for proteasomal degradation |
| ROPN1L | Rhophilin associated tail protein 1-like | RP11-1C1.7, ASP, RSPH11 | |
| RPL14 | Ribosomal protein L14 | CAG-ISL-7, CTG-B33, L14, RL14, hRL14 | |
| RPS14 | Ribosomal protein S14 | PRO2640, EMTB, S14 | |
| RPS8 | Ribosomal protein S8 | RP11-269F19.3, S8 | |
| RTEL1-TNFRSF6B | RTEL1-TNFRSF6B readthrough (NMD candidate) | ||
| S100A1 | S100 calcium binding protein A1 | RP1-178F15.1, S100, S100-α, S100A | Weakly binds calcium but binds zinc very tightly – distinct binding sites with different affinities exist for both ions on each monomer |
| SCAMP2 | Secretory carrier membrane protein 2 | Functions in post-Golgi recycling pathways. Acts as a recycling carrier to the cell surface | |
| SCD | Stearoyl-CoA desaturase | PRO1933, FADS5, MSTP0081, SCDOS, SCD | Terminal component of the liver microsomal stearyl-CoA desaturase system |
| SEPT2 | Septin 2 | DIFF6, NEDD-5, NEDD5, Pnutl3, hNedd5 | Required for normal progress through mitosis. Involved in cytokinesis |
| SF3B3 | Splicing factor 3b, subunit 3 | RSE1, SAP130, SF3b130, STAF13 | Subunit of the splicing factor SF3B required for “A” complex assembly formed by the stable binding of U2 snRNP to the branch point sequence in pre-mRNA |
| SH3BGRL | SH3 domain binding glutamate-rich protein like | HEL-S-115, SH3BGR | |
| SIK2 | Salt-inducible kinase 2 | LOH11CR1I, QIK, SNF1LK2 | Phosphorylates “Ser-794” of IRS1 in insulin-stimulated adipocytes |
| SIRT1 | Sirtuin 1 | RP11-57G10.3, SIR2L1 | NAD-dependent deacetylase, which regulates processes such as apoptosis and muscle differentiation by deacetylating key proteins |
| SIX6 | SIX homeobox 6 | MCOPCT2, OPTX2, Six9 | May be involved in eye development |
| SLC35B4 | Solute carrier family 35 (UDP-xylose/UDP-N-acetylglucosamine transporter), member B4 | PSEC0055, YEA, YEA4 | Sugar transporter that specifically mediates the transport of UDP-Xyl and UDP-GlcNAc from cytosol into Golgi |
| SLC37A3 | Solute carrier family 37, member 3 | ||
| SLC7A11 | Solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 | CCBR1, xCT | Sodium-independent, high-affinity exchange of anionic amino acids with high specificity for anionic form of cystine and glutamate |
| SLCO2A1 | Solute carrier organic anion transporter family, member 2A1 | MATR1, OATP2A1, PGT, PHOAR2, SLC21A2 | May mediate the release of newly synthesized prostaglandins from cells, the transepithelial transport of prostaglandins, and the clearance of prostaglandins from the circulation |
| SMAD5 | SMAD family member 5 | DWFC, JV5-1, MADH5 | Transcriptional modulator activated by BMP type 1 receptor kinase |
| SMCHD1 | Structural maintenance of chromosomes flexible hinge domain containing 1 | Required for maintenance of X inactivation in females and hypermethylation of CpG islands associated with inactive X | |
| SMG1 | SMG1 phosphatidylinositol 3-kinase-related kinase | 61E3.4, ATX, LIP | Serine/threonine protein kinase involved in both mRNA surveillance and genotoxic stress response pathways |
| SNAI2 | Snail family zinc finger 2 | SLUG, SLUGH1, SNAIL2, WS2D | Transcriptional repressor. Involved in the generation and migration of neural crest cells |
| SOGA2 | Microtubule crosslinking factor 1 | CCDC165, KIAA0802, MTCL1 | |
| SPRY2 | Sprouty homolog 2 | hSPRY2 | May function as an antagonist of FGF pathways and may negatively modulate respiratory organogenesis |
| SRPK2 | SRSF protein kinase 2 | SFRSK2 | Phosphorylates RS domain-containing proteins |
| SRSF7 | Serine/arginine-rich splicing factor 7 | 9G8, AAG3, SFRS7 | Required for pre-mRNA splicing |
| STAG2 | Stromal antigen 2 | RP11-517O1.1, SA-2, SA2, SCC3B, bA517O1.1 | Component of cohesin complex, a complex required for the cohesion of sister chromatids after DNA replication |
| TAAR6 | Trace amine associated receptor 6 | RP11-295F4.3, TA4, TAR4, TAR6, TRAR4, taR-4, taR-6 | Orphan receptor. Could be a receptor for trace amines |
| TAB2 | TGF-β activated kinase 1/MAP3K7 binding protein 2 | CHTD2, MAP3K7IP2, TAB-2 | Adapter linking MAP3K7/TAK1 and TRAF6 and mediator of MAP3K7 activation in the IL1 signaling pathway |
| TAF15 | TAF15 RNA polymerase II, TBP-associated factor | Npl3, RBP56, TAF2N, TAFII68 | RNA and ssDNA-binding protein that may play specific roles during transcription initiation at distinct promoters |
| TAF2 | TAF2 RNA polymerase II, TBP-associated factor | CIF150, MRT40B, TAFII150, TAF2 | Transcription factor TFIID is one of the general factors required for accurate and regulated initiation by RNA polymerase II |
| TAF6L | TAF6-like RNA polymerase II, PCAF-associated factor | PAF65A | Functions as a component of the PCAF complex |
| TBX4 | T-box 4 | SPS | Involved in the transcriptional regulation of genes required for mesoderm differentiation |
| TCF21 | Transcription factor 21 | POD1, bHLHa23 | Involved in epithelial–mesenchymal interactions in kidney and lung morphogenesis that include epithelial differentiation and branching morphogenesis |
| TEAD4 | TEA domain family member 4 | EFTR-2, RTEF1, TCF13L1, TEF-3, TEF3, TEFR-1, hRTEF-1B | Binds specifically and noncooperatively to the Sph and GT-IIC “enhansons” (5′-GTGGAATGT-3′) and activates transcription |
| TGFBR3 | Transforming growth factor β receptor III | BGCAN, β-glycan | Binds to TGF-β |
| TGIF2 | TGFβ-induced factor homeobox 2 | Transcriptional repressor. Probably represses transcription via the recruitment of histone deacetylase proteins | |
| THUMPD1 | THUMP domain containing 1 | ||
| TIAL1 | TIA1 cytotoxic granule-associated RNA binding protein-like 1 | TCBP, TIAR | RNA-binding protein. Possesses nucleolytic activity against cytotoxic lymphocyte target cells. May be involved in apoptosis |
| TM9SF3 | Transmembrane 9 superfamily member 3 | RP11-34E5.1, EP70-P-iso, SMBP | |
| TMED4 | Transmembrane emp24 protein transport domain containing 4 | ERS25, HNLF | Involved in endoplasmic reticulum stress response. May play a role in the regulation of heat-shock response and apoptosis |
| TMEM132B | Transmembrane protein 132B | ||
| TMEM14A | Transmembrane protein 14A | PTD011, C6orf73 | |
| TMEM192 | Transmembrane protein 192 | ||
| TMEM257 | Transmembrane protein 257 | CXorf1 | |
| TMEM45A | Transmembrane protein 45A | DERP7 | |
| TMEM64 | Transmembrane protein 64 | ||
| TMPO | Thymopoietin | CMD1T, LAP2, LEMD4, PRO0868, TP | May help direct the assembly of the nuclear lamina and thereby help maintain the structural organization of the nuclear envelope |
| TMPRSS11A | Transmembrane protease, serine 11A | ECRG1 | Probable serine protease, which may play a role in cellular senescence |
| TNIP1 | TNFAIP3 interacting protein 1 | ABIN-1, NAF1, VAN, nip40-1 | Interacts with zinc finger protein A20/TNFAIP3 and inhibits TNF-induced NF-κB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal |
| TNPO1 | Transportin 1 | IPO2, KPNB2, MIP, MIP1, TRN | Functions in nuclear protein import as nuclear transport receptor |
| TNRC6C | Trinucleotide repeat containing 6C | Plays a role in RNA-mediated gene silencing by miRNAs | |
| TRIM2 | Tripartite motif containing 2 | CMT2R, RNF86 | May contribute to the alteration of neural cellular mechanisms |
| TRUB1 | TruB psi synthase family member 1 | PUS4 | May be responsible for synthesis of psi from uracil in transfer RNAs |
| TSG101 | Tumor susceptibility 101 | TSG10, VPS23 | Component of the ESCRT-I complex, a regulator of vesicular trafficking process |
| TSHR | Thyroid stimulating hormone receptor | CHNG1, LGR3, hTSHR-I | Receptor for thyrothropin. Plays a central role in controlling thyroid cell metabolism |
| TUSC1 | Tumor suppressor candidate 1 | TSG-9, TSG9 | |
| TWF1 | Twinfilin actin-binding protein 1 | A6, PTK9 | Actin-binding protein involved in motile and morphological processes |
| UBA2 | Ubiquitin-like modifier activating enzyme 2 | HRIHFB2115, ARX, SAE2 | The dimeric enzyme acts as an E1 ligase for SUMO1, SUMO2, SUMO3, and probably SUMO4 |
| UCHL1 | Ubiquitin carboxyl-terminal esterase L1 | HEL-117, NDGOA, PARK5, PGP 9.5, PGP9.5, PGP95, Uch-L1 | Ubiquitin-protein hydrolase involved both in the processing of ubiquitin precursors and of ubiquitinated proteins |
| UGT3A1 | UDP glycosyltransferase 3 family, polypeptide A1 | UDP-glucuronosyltransferases catalyze phase II biotransformation reactions | |
| USP28 | Ubiquitin specific peptidase 28 | Deubiquitinase involved in DNA damage response checkpoint and MYC proto-oncogene stability | |
| VBP1 | Von Hippel-Lindau binding protein 1 | RP13-228J13.4, PFD3, PFDN3, VBP-1 | Binds specifically to c-CPN and transfers target proteins to it |
| WDR33 | WD repeat domain 33 | NET14, WDC146 | Essential for both cleavage and polyadenylation of pre-mRNA 3′ ends |
| WNT16 | Wingless-type MMTV integration site family, member 16 | Ligand for members of the Frizzled family of seven transmembrane receptors | |
| WNT2 | Wingless-type MMTV integration site family, member 2 | INT1L1, IRP | Ligand for members of the Frizzled family of seven transmembrane receptors |
| WNT3A | Wingless-type MMTV integration site family member 3A | Ligand for members of the Frizzled family of seven transmembrane receptors | |
| XIAP | X-linked inhibitor of apoptosis | RP1-315G1.5, API3, BIRC4, IAP-3, ILP1, MIHA, XLP2, hIAP-3, hIAP3 | Apoptotic suppressor |
| YOD1 | YOD1 deubiquitinase | RP11-164O23.1, DUBA8, OTUD2, PRO0907 | May play an important regulatory role at the level of protein turnover by preventing degradation |
| YY1 | YY1 transcription factor | DELTA, INO80S, NF-E1, UCRBP, YIN-YANG-1 | May play an important role in development and differentiation |
| ZEB2 | Zinc finger E-box binding homeobox 2 | HRIHFB2411, HSPC082, SIP-1, SIP1, SMADIP1, ZFHX1B | Transcriptional inhibitor that binds to DNA sequence 5′-CACCT-3′ in different promoters. Represses transcription of E-cadherin |
| ZFP36L2 | ZFP36 ring finger protein-like 2 | BRF2, ERF-2, ERF2, RNF162C, TIS11D | Probable regulatory protein involved in regulating the response to growth factors |
| ZIC2 | Zinc family member 2 | HPE5 | Involved in cerebellar development |
| ZNF12 | Zinc finger protein 12 | GIOT-3, HZF11, KOX3, ZNF325 | May be involved in transcriptional regulation |
| ZNF121 | Zinc finger protein 121 | D19S204, ZHC32, ZNF20 | May be involved in transcriptional regulation |
| ZNF132 | Zinc finger protein 132 | pHZ-12 | May be involved in transcriptional regulation |
| ZNF148 | Zinc finger protein 148 | BERF-1, BFCOL1, HT-β, ZBP-89, ZFP148, pHZ-52 | Involved in transcriptional regulation |
| ZNF180 | Zinc finger protein 180 | HHZ168 | May be involved in transcriptional regulation |
| ZNF238 | Zinc finger and BTB domain containing 18 | C2H2-171, MRD22, RP58, TAZ-1, ZNF18 | Sequence-specific DNA-binding protein with transcriptional repression activity |
| ZNF25 | Zinc finger protein 25 | KOX19, Zfp9 | May be involved in transcriptional regulation |
| ZNF30 | Zinc finger protein 30 | KOX28 | May be involved in transcriptional regulation |
| ZNF35 | Zinc finger protein 35 | HF.10, HF10, Zfp105 | May be involved in transcriptional regulation |
| ZNF350 | Zinc finger protein 350 | ZBRK1, ZFQR | Transcriptional repressor |
| ZNF426 | Zinc finger protein 426 | May be involved in transcriptional regulation | |
| ZNF445 | Zinc finger protein 445 | ZKSCAN15, ZNF168, ZSCAN47 | May be involved in transcriptional regulation |
| ZNF558 | Zinc finger protein 558 | May be involved in transcriptional regulation | |
| ZNF562 | Zinc finger protein 562 | May be involved in transcriptional regulation | |
| ZNF594 | Zinc finger protein 594 | hCG_1775942 | May be involved in transcriptional regulation |
| ZNF644 | Zinc finger protein 644 | BM-005, MYP21, NatF, ZEP-2 | May be involved in transcriptional regulation |
| ZNF652 | Zinc finger protein 652 | Functions as a transcriptional repressor | |
| ZNF700 | Zinc finger protein 700 | May be involved in transcriptional regulation | |
| ZNF703 | Zinc finger protein 703 | ZEPPO1, ZNF503L, ZPO1 | May function as a transcriptional repressor |
| ZNF711 | Zinc finger protein 711 | CMPX1, MRX97, ZNF4, ZNF5, ZNF6, Zfp711, dJ75N13.1 | May be involved in transcriptional regulation |
| ZNF763 | Zinc finger protein 763 | ZNF, ZNF440L | May be involved in transcriptional regulation |
| ZNF780A | Zinc finger protein 780A | ZNF780 | May be involved in transcriptional regulation |
Abbreviations: FGF, fibroblast growth factor; HLA, human leukocyte antigen; IL, interleukin; miRNA, microRNA; mRNA, messenger RNA; NK cells, natural killer cells; ssDNA, single-stranded DNA; TGF, Transforming growth factor; TNF, tumor necrosis factor; UV, ultraviolet; VEGF, vascular endothelial growth factor.
Table 12.
The top enriched clusters (enrich score >1) by DAVID for the combined targets of hsa-miR-181a-5p from both TarBase 6.0 and miRTarBase 4.0
| Category | Term | Gene count | P-value | FDR |
|---|---|---|---|---|
| Annotation cluster 1 | Enrichment score: 4.49 | |||
| GOTERM_BP_FAT | Response to hormone stimulus | 7 | 7.70E-06 | 1.10E-03 |
| GOTERM_BP_FAT | Response to endogenous stimulus | 7 | 1.40E-05 | 1.50E-03 |
| GOTERM_BP_FAT | Response to organic substance | 7 | 3.30E-04 | 8.80E-03 |
| Annotation cluster 2 | Enrichment score: 3.97 | |||
| GOTERM_BP_FAT | Negative regulation of apoptosis | 6 | 1.00E-04 | 5.60E-03 |
| GOTERM_BP_FAT | Negative regulation of programmed cell death | 6 | 1.10E-04 | 5.00E-03 |
| GOTERM_BP_FAT | Negative regulation of cell death | 6 | 1.10E-04 | 4.80E-03 |
| Annotation cluster 3 | Enrichment score: 3.57 | |||
| GOTERM_BP_FAT | DNA damage checkpoint | 4 | 3.90E-05 | 3.10E-03 |
| GOTERM_BP_FAT | DNA integrity checkpoint | 4 | 5.00E-05 | 3.60E-03 |
| GOTERM_BP_FAT | DNA damage response, signal transduction | 4 | 1.80E-04 | 6.50E-03 |
| GOTERM_BP_FAT | Cell cycle checkpoint | 4 | 2.60E-04 | 7.20E-03 |
| GOTERM_BP_FAT | Response to DNA damage stimulus | 4 | 1.50E-02 | 1.00E-01 |
| Annotation cluster 4 | Enrichment score: 3.38 | |||
| KEGG_PATHWAY | Bladder cancer | 4 | 1.80E-04 | 1.50E-03 |
| KEGG_PATHWAY | Endometrial cancer | 4 | 3.40E-04 | 2.30E-03 |
| KEGG_PATHWAY | Non-small-cell lung cancer | 4 | 3.80E-04 | 2.30E-03 |
| KEGG_PATHWAY | Acute myeloid leukemia | 4 | 4.70E-04 | 2.60E-03 |
| KEGG_PATHWAY | Glioma | 4 | 6.00E-04 | 3.00E-03 |
| KEGG_PATHWAY | Melanoma | 4 | 8.50E-04 | 3.60E-03 |
| Annotation cluster 5 | Enrichment score: 3.2 | |||
| GOTERM_BP_FAT | Regulation of apoptosis | 7 | 6.00E-04 | 1.40E-02 |
| GOTERM_BP_FAT | Regulation of programmed cell death | 7 | 6.30E-04 | 1.40E-02 |
| GOTERM_BP_FAT | Regulation of cell death | 7 | 6.50E-04 | 1.40E-02 |
| Annotation cluster 6 | Enrichment score: 3.13 | |||
| GOTERM_BP_FAT | Developmental growth | 4 | 2.30E-04 | 7.20E-03 |
| GOTERM_BP_FAT | Cell fate commitment | 4 | 9.10E-04 | 1.90E-02 |
| GOTERM_BP_FAT | Tissue morphogenesis | 4 | 1.90E-03 | 3.30E-02 |
| Annotation cluster 7 | Enrichment score: 2.74 | |||
| GOTERM_BP_FAT | Apoptosis | 6 | 1.20E-03 | 2.20E-02 |
| GOTERM_BP_FAT | Programmed cell death | 6 | 1.30E-03 | 2.30E-02 |
| GOTERM_BP_FAT | Cell death | 6 | 2.60E-03 | 3.90E-02 |
| GOTERM_BP_FAT | Death | 6 | 2.70E-03 | 3.80E-02 |
| Annotation cluster 8 | Enrichment score: 2.7 | |||
| GOTERM_BP_FAT | Positive regulation of macromolecule biosynthetic process | 6 | 1.70E-03 | 3.00E-02 |
| GOTERM_BP_FAT | Positive regulation of cellular biosynthetic process | 6 | 2.10E-03 | 3.50E-02 |
| GOTERM_BP_FAT | Positive regulation of biosynthetic process | 6 | 2.20E-03 | 3.70E-02 |
| Annotation cluster 9 | Enrichment score: 2.56 | |||
| GOTERM_BP_FAT | Positive regulation of apoptosis | 5 | 2.70E-03 | 3.90E-02 |
| GOTERM_BP_FAT | Positive regulation of programmed cell death | 5 | 2.70E-03 | 3.80E-02 |
| GOTERM_BP_FAT | Positive regulation of cell death | 5 | 2.80E-03 | 3.80E-02 |
| Annotation cluster 10 | Enrichment score: 2.41 | |||
| GOTERM_BP_FAT | Regulation of phosphorylation | 5 | 3.60E-03 | 4.40E-02 |
| GOTERM_BP_FAT | Regulation of phosphate metabolic process | 5 | 4.10E-03 | 4.50E-02 |
| GOTERM_BP_FAT | Regulation of phosphorus metabolic process | 5 | 4.10E-03 | 4.50E-02 |
| Annotation cluster 11 | Enrichment score: 2.22 | |||
| GOTERM_BP_FAT | Positive regulation of transcription, DNA-dependent | 5 | 3.90E-03 | 4.40E-02 |
| GOTERM_BP_FAT | Positive regulation of transcription | 5 | 7.00E-03 | 6.70E-02 |
| GOTERM_BP_FAT | Positive regulation of gene expression | 5 | 7.80E-03 | 7.20E-02 |
| Annotation cluster 12 | Enrichment score: 2.15 | |||
| GOTERM_BP_FAT | Positive regulation of protein kinase activity | 4 | 3.50E-03 | 4.40E-02 |
| GOTERM_BP_FAT | Positive regulation of kinase activity | 4 | 3.90E-03 | 4.40E-02 |
| GOTERM_BP_FAT | Positive regulation of transferase activity | 4 | 4.30E-03 | 4.60E-02 |
| GOTERM_BP_FAT | Regulation of protein kinase activity | 4 | 1.20E-02 | 9.20E-02 |
| GOTERM_BP_FAT | Regulation of kinase activity | 4 | 1.30E-02 | 9.80E-02 |
| GOTERM_BP_FAT | Regulation of transferase activity | 4 | 1.40E-02 | 1.00E-01 |
| Annotation cluster 13 | Enrichment score: 2.1 | |||
| GOTERM_BP_FAT | Positive regulation of cellular protein metabolic process | 4 | 4.00E-03 | 4.40E-02 |
| GOTERM_BP_FAT | Positive regulation of protein metabolic process | 4 | 4.50E-03 | 4.70E-02 |
| GOTERM_BP_FAT | Regulation of cellular protein metabolic process | 4 | 2.70E-02 | 1.80E-01 |
| Annotation cluster 14 | Enrichment score: 2.1 | |||
| GOTERM_BP_FAT | Branching morphogenesis of a tube | 3 | 3.70E-03 | 4.30E-02 |
| GOTERM_BP_FAT | Positive regulation of cell development | 3 | 4.10E-03 | 4.50E-02 |
| GOTERM_BP_FAT | Morphogenesis of a branching structure | 3 | 4.80E-03 | 4.90E-02 |
| GOTERM_BP_FAT | Tube morphogenesis | 3 | 1.30E-02 | 1.00E-01 |
| GOTERM_BP_FAT | Regulation of cell development | 3 | 3.30E-02 | 2.00E-01 |
| Annotation cluster 15 | Enrichment score: 2.06 | |||
| GOTERM_BP_FAT | Neuron projection morphogenesis | 4 | 3.10E-03 | 4.00E-02 |
| GOTERM_BP_FAT | Cell projection morphogenesis | 4 | 4.60E-03 | 4.80E-02 |
| GOTERM_BP_FAT | Neuron projection development | 4 | 5.20E-03 | 5.10E-02 |
| GOTERM_BP_FAT | Cell part morphogenesis | 4 | 5.20E-03 | 5.10E-02 |
| GOTERM_BP_FAT | Neuron development | 4 | 1.10E-02 | 9.00E-02 |
| GOTERM_BP_FAT | Cell morphogenesis | 4 | 1.30E-02 | 9.80E-02 |
| GOTERM_BP_FAT | Cell projection organization | 4 | 1.40E-02 | 1.00E-01 |
| GOTERM_BP_FAT | Cellular component morphogenesis | 4 | 1.70E-02 | 1.20E-01 |
| GOTERM_BP_FAT | Neuron differentiation | 4 | 2.20E-02 | 1.50E-01 |
| Annotation cluster 16 | Enrichment score: 2.03 | |||
| BIOCARTA | IGF-1 signaling pathway | 3 | 7.00E-03 | 3.00E-01 |
| BIOCARTA | IL-6 signaling pathway | 3 | 7.00E-03 | 3.00E-01 |
| BIOCARTA | Insulin signaling pathway | 3 | 7.00E-03 | 3.00E-01 |
| BIOCARTA | Signaling of hepatocyte growth factor receptor | 3 | 2.20E-02 | 3.10E-01 |
| Annotation cluster 17 | Enrichment score: 1.97 | |||
| GOTERM_BP_FAT | Embryonic appendage morphogenesis | 3 | 6.50E-03 | 6.30E-02 |
| GOTERM_BP_FAT | Embryonic limb morphogenesis | 3 | 6.50E-03 | 6.30E-02 |
| GOTERM_BP_FAT | Limb morphogenesis | 3 | 8.40E-03 | 7.50E-02 |
| GOTERM_BP_FAT | Appendage morphogenesis | 3 | 8.40E-03 | 7.50E-02 |
| GOTERM_BP_FAT | Limb development | 3 | 9.00E-03 | 7.70E-02 |
| GOTERM_BP_FAT | Appendage development | 3 | 9.00E-03 | 7.70E-02 |
| GOTERM_BP_FAT | Embryonic morphogenesis | 3 | 6.80E-02 | 3.40E-01 |
| Annotation cluster 18 | Enrichment score: 1.81 | |||
| GOTERM_BP_FAT | Response to ethanol | 3 | 3.60E-03 | 4.30E-02 |
| GOTERM_BP_FAT | Response to metal ion | 3 | 1.40E-02 | 1.00E-01 |
| GOTERM_BP_FAT | Response to inorganic substance | 3 | 3.30E-02 | 2.00E-01 |
| GOTERM_BP_FAT | Response to drug | 3 | 3.60E-02 | 2.10E-01 |
| Annotation cluster 19 | Enrichment score: 1.59 | |||
| GOTERM_BP_FAT | Response to estrogen stimulus | 3 | 9.40E-03 | 7.90E-02 |
| GOTERM_BP_FAT | Positive regulation of protein modification process | 3 | 2.80E-02 | 1.80E-01 |
| GOTERM_BP_FAT | Regulation of protein modification process | 3 | 6.40E-02 | 3.20E-01 |
| Annotation cluster 20 | Enrichment score: 1.57 | |||
| GOTERM_BP_FAT | Protein amino acid phosphorylation | 5 | 1.30E-02 | 9.70E-02 |
| GOTERM_BP_FAT | Phosphorylation | 5 | 2.30E-02 | 1.50E-01 |
| GOTERM_BP_FAT | Phosphate metabolic process | 5 | 4.30E-02 | 2.40E-01 |
| GOTERM_BP_FAT | Phosphorus metabolic process | 5 | 4.30E-02 | 2.40E-01 |
| Annotation cluster 21 | Enrichment score: 1.55 | |||
| GOTERM_BP_FAT | Cell aging | 3 | 9.60E-04 | 1.90E-02 |
| GOTERM_BP_FAT | Negative regulation of neuron apoptosis | 3 | 2.30E-03 | 3.70E-02 |
| GOTERM_BP_FAT | Aging | 3 | 1.00E-02 | 8.50E-02 |
| GOTERM_BP_FAT | Actin cytoskeleton organization | 3 | 3.90E-02 | 2.20E-01 |
| GOTERM_BP_FAT | Actin filament-based process | 3 | 4.40E-02 | 2.40E-01 |
| GOTERM_BP_FAT | Membrane organization | 3 | 9.90E-02 | 4.40E-01 |
| GOTERM_CC_FAT | Membrane fraction | 3 | 3.20E-01 | 9.40E-01 |
| GOTERM_CC_FAT | Insoluble fraction | 3 | 3.30E-01 | 9.30E-01 |
| Annotation cluster 22 | Enrichment score: 1.47 | |||
| GOTERM_BP_FAT | Ras protein signal transduction | 3 | 9.40E-03 | 7.90E-02 |
| KEGG_PATHWAY | Long-term depression | 3 | 1.50E-02 | 4.40E-02 |
| KEGG_PATHWAY | B cell receptor signaling pathway | 3 | 1.70E-02 | 4.50E-02 |
| KEGG_PATHWAY | VEGF signaling pathway | 3 | 1.70E-02 | 4.50E-02 |
| KEGG_PATHWAY | Fc epsilon RI signaling pathway | 3 | 1.90E-02 | 4.60E-02 |
| KEGG_PATHWAY | ErbB signaling pathway | 3 | 2.30E-02 | 5.40E-02 |
| KEGG_PATHWAY | Gap junction | 3 | 2.40E-02 | 5.50E-02 |
| KEGG_PATHWAY | GnRH signaling pathway | 3 | 2.90E-02 | 6.30E-02 |
| KEGG_PATHWAY | T-cell receptor signaling pathway | 3 | 3.40E-02 | 7.20E-02 |
| KEGG_PATHWAY | Insulin signaling pathway | 3 | 5.20E-02 | 1.00E-01 |
| GOTERM_BP_FAT | Small GTPase-mediated signal transduction | 3 | 6.70E-02 | 3.40E-01 |
| KEGG_PATHWAY | Chemokine signaling pathway | 3 | 9.20E-02 | 1.70E-01 |
| KEGG_PATHWAY | Regulation of actin cytoskeleton | 3 | 1.20E-01 | 2.10E-01 |
| KEGG_PATHWAY | MAPK signaling pathway | 3 | 1.70E-01 | 2.80E-01 |
| Annotation cluster 23 | Enrichment score: 1.45 | |||
| GOTERM_BP_FAT | Axonogenesis | 3 | 3.00E-02 | 1.80E-01 |
| GOTERM_BP_FAT | Cell morphogenesis involved in neuron differentiation | 3 | 3.40E-02 | 2.00E-01 |
| GOTERM_BP_FAT | Cell morphogenesis involved in differentiation | 3 | 4.50E-02 | 2.50E-01 |
| Annotation cluster 24 | Enrichment score: 1.31 | |||
| GOTERM_CC_FAT | Nucleoplasm | 6 | 6.20E-03 | 4.90E-01 |
| GOTERM_CC_FAT | Nuclear lumen | 6 | 4.50E-02 | 5.70E-01 |
| GOTERM_CC_FAT | Intracellular organelle lumen | 6 | 9.40E-02 | 7.40E-01 |
| GOTERM_CC_FAT | Organelle lumen | 6 | 1.00E-01 | 7.20E-01 |
| GOTERM_CC_FAT | Membrane-enclosed lumen | 6 | 1.10E-01 | 7.10E-01 |
| Annotation cluster 25 | Enrichment score: 1.3 | |||
| GOTERM_BP_FAT | Hemopoiesis | 3 | 4.30E-02 | 2.40E-01 |
| GOTERM_BP_FAT | Hemopoietic or lymphoid organ development | 3 | 5.10E-02 | 2.70E-01 |
| GOTERM_BP_FAT | Immune system development | 3 | 5.60E-02 | 2.90E-01 |
| Annotation cluster 26 | Enrichment score: 1.06 | |||
| SP_PIR_KEYWORDS | Transcription regulation | 6 | 6.30E-02 | 3.80E-01 |
| SP_PIR_KEYWORDS | Transcription | 6 | 6.80E-02 | 3.90E-01 |
| GOTERM_BP_FAT | Transcription | 6 | 1.60E-01 | 6.10E-01 |
Abbreviations: DAVID, Database for Annotation, Visualization and Integrated Discovery; FDR, false discovery rate; GnRH, gonadotropin releasing hormone; IGF, insulin-like growth factor; IL, interleukin; VEGF, vascular endothelial growth factor.
Figure 9.
Endometrial carcinoma pathways in the combined target list of hsa-miR-181a-5p based on both TarBase and miRTarBase 4.0.
Notes: EC has two types. Type I EC, or estrogen-dependent endometrioid EC, represents the most common subtype. It is an estrogen-associated lesion often seen in conjunction with endometrial hyperplasia. The histological subtypes that correspond to endometrioid adenocarcinoma and its variants, as well as mucinous adenocarcinoma, are allocated to this group. Type II EC, or nonendometrioid EC, tends to affect older, postmenopausal women and is a non-estrogen-associated lesion. These cancers are not preceded by endometrial hyperplasia, though they can occasionally arise in endometrial polyps or from precancerous lesions, endometrial intraepithelial carcinoma, or in the vicinity of atrophic endometrium. The clinicopathological differences between the two types are paralleled by specific genetic alterations, with type I EC showing microsatellite instability and mutations in PTEN, PIK3CA, KRAS, and CTNNB1 (β-catenin), and type II exhibiting p53 mutations and chromosomal instability.17 hsa-miR-181a-5p has been found to regulate these genes and eventually promote EC initiation, development, growth, and metastasis.
Abbreviation: EC, endometrial cancer.
Furthermore, our DAVID analysis revealed that there were 33 KEGG pathways significantly enriched in the target list of hsa-miR-181a-5p, based on both TarBase and miR-TarBase (Table 13). These pathways included pathways in cancer, prostate cancer, thyroid cancer, renal cell carcinoma, chronic myeloid leukemia, neurotrophin signaling pathway, dorsoventral axis formation, bladder cancer, endometrial cancer, non-small-cell lung cancer, acute myeloid leukemia, glioma, long-term potentiation, melanoma, colorectal cancer, melanogenesis, cell cycle, natural killer cell–mediated cytotoxicity, focal adhesion, notch signaling pathway, long-term depression, pancreatic cancer, B cell receptor signaling pathway, VEGF signaling pathway, Fc epsilon RI signaling pathway, ErbB signaling pathway, gap junction, GnRH signaling pathway, T cell receptor signaling pathway, insulin signaling pathway, Jak-STAT signaling pathway, chemokine signaling pathway, and prion diseases.
Table 13.
KEGG pathways for the combined targets of hsa-miR-181a-5p based on TarBase 6.0 and miRTarBase 4.0
| Signaling pathway | Gene count | % | P-value | FDR |
|---|---|---|---|---|
| Pathways in cancer | 7 | 31.8 | 1.30E-04 | 1.30E-03 |
| Prostate cancer | 6 | 27.3 | 2.60E-06 | 7.80E-05 |
| Thyroid cancer | 5 | 22.7 | 8.20E-07 | 4.90E-05 |
| Renal cell carcinoma | 5 | 22.7 | 3.00E-05 | 5.90E-04 |
| Chronic myeloid leukemia | 5 | 22.7 | 3.90E-05 | 4.70E-04 |
| Neurotrophin signaling pathway | 5 | 22.7 | 2.80E-04 | 2.10E-03 |
| Dorsoventral axis formation | 4 | 18.2 | 3.70E-05 | 5.50E-04 |
| Bladder cancer | 4 | 18.2 | 1.80E-04 | 1.50E-03 |
| Endometrial cancer | 4 | 18.2 | 3.40E-04 | 2.30E-03 |
| Non-small-cell lung cancer | 4 | 18.2 | 3.80E-04 | 2.30E-03 |
| Acute myeloid leukemia | 4 | 18.2 | 4.70E-04 | 2.60E-03 |
| Glioma | 4 | 18.2 | 6.00E-04 | 3.00E-03 |
| Long-term potentiation | 4 | 18.2 | 7.50E-04 | 3.50E-03 |
| Melanoma | 4 | 18.2 | 8.50E-04 | 3.60E-03 |
| Colorectal cancer | 4 | 18.2 | 1.40E-03 | 5.50E-03 |
| Melanogenesis | 4 | 18.2 | 2.20E-03 | 8.30E-03 |
| Cell cycle | 4 | 18.2 | 4.30E-03 | 1.50E-02 |
| Natural killer cell–mediated cytotoxicity | 4 | 18.2 | 5.20E-03 | 1.70E-02 |
| Focal adhesion | 4 | 18.2 | 1.60E-02 | 4.50E-02 |
| Notch signaling pathway | 3 | 13.6 | 7.10E-03 | 2.20E-02 |
| Long-term depression | 3 | 13.6 | 1.50E-02 | 4.40E-02 |
| Pancreatic cancer | 3 | 13.6 | 1.60E-02 | 4.30E-02 |
| B-cell receptor signaling pathway | 3 | 13.6 | 1.70E-02 | 4.50E-02 |
| VEGF signaling pathway | 3 | 13.6 | 1.70E-02 | 4.50E-02 |
| Fc epsilon RI signaling pathway | 3 | 13.6 | 1.90E-02 | 4.60E-02 |
| ErbB signaling pathway | 3 | 13.6 | 2.30E-02 | 5.40E-02 |
| Gap junction | 3 | 13.6 | 2.40E-02 | 5.50E-02 |
| GnRH signaling pathway | 3 | 13.6 | 2.90E-02 | 6.30E-02 |
| T cell receptor signaling pathway | 3 | 13.6 | 3.40E-02 | 7.20E-02 |
| Insulin signaling pathway | 3 | 13.6 | 5.20E-02 | 1.00E-01 |
| Jak-STAT signaling pathway | 3 | 13.6 | 6.60E-02 | 1.30E-01 |
| Chemokine signaling pathway | 3 | 13.6 | 9.20E-02 | 1.70E-01 |
| Prion diseases | 2 | 9.1 | 9.20E-02 | 1.70E-01 |
Abbreviations: FDR, false discovery rate; GnRH, gonadotropin releasing hormone; KEGG, Kyoto Encyclopedia of Genes and Genomes; VEGF, vascular endothelial growth factor.
Among the 313 validated targets of hsa-miR-181a-5p, 22 were cancer genes (Table 14). These included ATM, BCL2, BRCA1, CCDC6, CCND1, CDX2, EP300, FBXO11, H3F3B, HOOK3, HOXA11, HRAS, KRAS, MAP2K1, NOTCH1, NOTCH2, PLAG1, PTPN11, RBM15, STAG2, TAF15, and TSHR. The KEGG pathway analysis also indicate that the targets regulated by hsa-miR-181a that are cancer genes are all involved in the tumorigenesis of bladder cancer, endometrial cancer, non-small-cell lung cancer, acute myeloid leukemia, glioma, melanoma, and colorectal cancer.
Table 14.
Combined targets of hsa-miR-181a-5p based on TarBase 6.0 and miRTarBase 4.0, that are cancer genes
| Gene symbol | Name | Tumor types (somatic) | Tumor types (germline) |
|---|---|---|---|
| ATM | Ataxia telangiectasia mutated | T-PLL | Leukemia; lymphoma; medulloblastoma; glioma |
| BCL2 | B-cell CLL/lymphoma 2 | NHL; CLL | |
| BRCA1 | Familial breast/ovarian cancer gene 1 | Ovarian | Breast; ovarian |
| CCDC6 | Coiled-coil domain containing 6 | NSCLC | |
| CCND1 | Cyclin D1 | CLL; B-ALL; breast | |
| CDX2 | Caudal type homeobox transcription factor 2 | AML | |
| EP300 | 300 kDa E1A-binding protein gene | Colorectal; breast; pancreatic; AML; ALL; DLBCL | |
| FBXO11 | F-box protein 11 | DLBCL | |
| H3F3B | H3 histone; family 3B (H3.3B) | Chondroblastoma | |
| HOOK3 | Hook homolog 3 | Papillary thyroid | |
| HOXA11 | Homeobox A11 | CML | |
| HRAS | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Infrequent sarcomas; rare other tumor types | Rhabdomyosarcoma; ganglioneuroblastoma; bladder |
| KRAS | v-Ki-ras2 Kirsten rat sarcoma 2 viral oncogene homolog | Pancreatic; colorectal; lung; thyroid; AML; other tumor types | |
| MAP2K1 | Mitogen-activated protein kinase kinase 1 | NSCLC; melanoma; colorectal | |
| NOTCH1 | Notch homolog 1; translocation-associated (Drosophila) (TAN1) | T-ALL | |
| NOTCH2 | Notch homolog 2 | Marginal zone lymphoma; DLBCL | |
| PLAG1 | Pleiomorphic adenoma gene 1 | Salivary adenoma | |
| PTPN11 | Protein tyrosine phosphatase; nonreceptor type 11 | JMML; AML; MDS | |
| RBM15 | RNA binding motif protein 15 | Acute megakaryocytic leukemia | |
| STAG2 | Stromal antigen 2 | Bladder carcinoma; glioblastoma; melanoma; Ewing’s sarcoma; myeloid neoplasms | |
| TAF15 | TAF15 RNA polymerase II; TBP-associated factor; 68 kDa | Extraskeletal myxoid chondrosarcoma; ALL | |
| TSHR | Thyroid stimulating hormone receptor | Toxic thyroid adenoma | Thyroid adenoma |
Abbreviations: ALL, acute lymphocytic leukemia; AML, adult acute myeloid leukemia; B-ALL, B-cell ALL; CLL, chronic lymphocytic leukemia; CML, chronic myelogenous leukemia; DLBCL, diffuse large B-cell lymphoma; JMML, juvenile myelomonocytic leukemia; MDS, myelodysplastic syndrome; NHL, non-Hodgkin lymphoma; NSCLC, non-small-cell lung cancer; T-ALL, T-cell ALL; T-PLL, T-cell subtype of prolymphocytic leukemia.
Now when we looked at the prediction accuracy of all the nine algorithms we used, we found that all the predicting programs behaved poorly. The reasons for this may include: a) the predicting criteria were not set up properly; b) the matching criteria for hsa-miR-181a with the complementary sites of the target mRNAs may have been too restrictive or too loose; c) the value for the threshold was not properly set prior to prediction; and d) the calculation of the P-values may have been too simple or too complicated. The poor predictive ability for all these algorithms also emphasizes the importance of experimental validation of the targets of any specific miRNAs.
Clinical validation of the role of hsa-miR-181a in EC tumorigenesis
Next, we aimed to validate the function of hsa-miR-181a in the pathogenesis of EC by measuring and comparing the expression levels of hsa-miR-181a in normal, benign, and malignant endometrial tissues. The association of disease progression of EC with the expression profile of hsa-miR-181a was also determined. There are two types of EC with distinct histological characteristics.15,51 Herein, a total of 78 tissue samples were classified by immunohistochemical staining. There were 47, 18, and 13 samples that were categorized as EC, endometrial hyperplasia, or normal endometrium, respectively. For the EC group, there were 37 samples, and 10 samples that belonged to type I and type II EC, respectively. The type I EC was ER-and PR-positive (Figure 10), and the type II EC was ER- and PR-negative (Figure 11).
Figure 10.
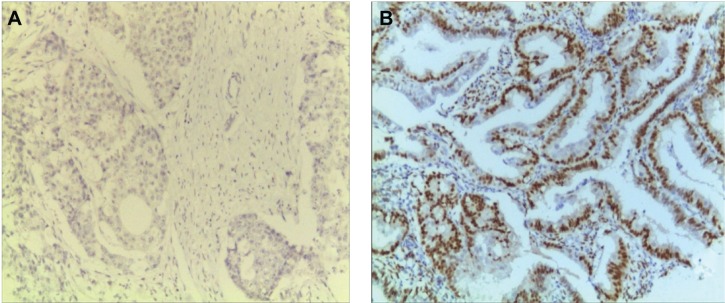
The expression of ER and PR in type I EC.
Notes: The expression of ER and PR was evaluated by immunohistochemistry. Dewaxed and dehydrated sections were washed with PBS and then, incubated with 3% peroxyl in methanol to terminate the activity of endogenous peroxidases. The sections were washed with PBS and immersed into boiled citrate-buffered solution for 10 minutes. The sections were blocked with 5% bovine serum albumin in PBS for 20 minutes at room temperature. Following that, the sections were probed with primary antibody against ER or PR, followed by biotinylated second anti-rabbit antibody. (A) Positive expression of ER; and (B) positive expression of PR.
Abbreviations: EC, endometrial cancer; ER, estrogen receptor; PBS, phosphate-buffered saline; PR, progesterone receptor.
Figure 11.
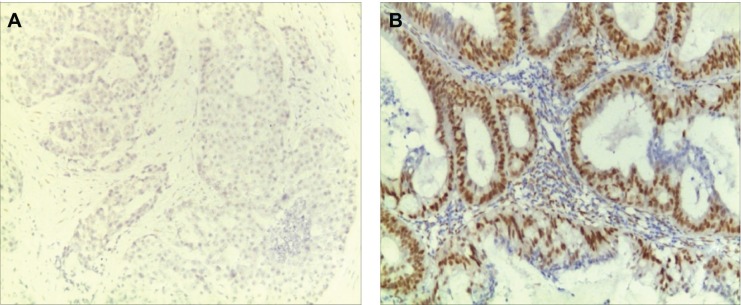
The expression of ER and PR in type II EC.
Notes: The expression of ER and PR was evaluated by immunohistochemistry. Dewaxed and dehydrated sections were washed with PBS and then, incubated with 3% peroxyl in methanol to terminate the activity of endogenous peroxidases. The sections were washed with PBS and immersed into boiled citrate-buffered solution for 10 minutes. The sections were blocked with 5% bovine serum albumin in PBS for 20 minutes at room temperature. Following that, the sections were probed with primary antibody against ER or PR, followed by biotinylated second anti-rabbit antibody. (A) Negative expression of ER; and (B) negative expression of PR.
Abbreviations: EC, endometrial cancer; ER, estrogen receptor; PBS, phosphate-buffered saline; PR, progesterone receptor.
To examine the potential role of hsa-miR-181a in the development and progression of EC, the expression profile of hsa-miR-181a was tested in all collected clinical samples, using RT-PCR (Figure S1). In comparison with normal endometrium, the expression level of hsa-miR-181a was increased 8.5-, 31.2-, and 4.1-fold in type I EC, type II EC, and endometrial hyperplasia, respectively (P<0.05, by one-way ANOVA) (Tables 15 and 16). The expression level of hsa-miR-181a in type II EC was higher (3.7-fold) than that in type I EC (P<0.05). In addition, hsa-miR-181a had a higher expression level in EC than that in endometrial hyperplasia.
Table 15.
Expression level of hsa-miR-181a in normal endometrium, endometrial hyperplasia, and EC
| Tissue | N | ΔCt | 2−ΔΔCt |
|---|---|---|---|
| Endometrial cancer | 47 | −3.356±2.401 | 10.240 |
| Endometrial hyperplasia | 18 | −1.893±2.568 | 4.073 |
| Normal endometrium | 13 | 0.133±2.527 | 1.000 |
Note: ΔCt, normalized threshold cycle; 2−ΔΔCt, a method used to calculate relative changes in the gene expression determined from real-time quantitative polymerase chain reaction experiments and it means the fold change.
Abbreviations: ΔCt, normalized threshold cycle; EC, endometrial cancer.
Table 16.
Differential expression level of hsa-miR181a in endometrium tissues
| Tissue | 2−ΔΔCt | P |
|---|---|---|
| Endometrial hyperplasia vs normal endometrial tissue | 4.073 | 0.027 |
| Type II endometrial cancer vs type I endometrial cancer | 3.668 | 0.032 |
| Type II endometrial cancer vs endometrial hyperplasia | 7.669 | 0.003 |
| Type II endometrial cancer vs normal endometrial tissue | 31.233 | 0.000 |
| Type I endometrial cancer vs endometrial hyperplasia | 2.091 | 0.127 |
| Type I endometrial cancer vs normal endometrial tissue | 8.515 | 0.000 |
Note: 2−ΔΔCt: a method used to calculate relative changes in the gene expression determined from real-time quantitative polymerase chain reaction experiments and it means the fold change.
Since we have observed the differential expression profile of hsa-miR-181a in all examined clinical samples, we further assessed the association between the expression of hsa-miR-181a and the development of EC with regard to histological type, stage, grade, migration, and invasion. As shown in Table 17, compared with the stage I and II EC, there was a remarkable increase in the expression level of hsa-miR-181a in stage III and VI EC (P=0.01, by one-way ANOVA). The expression level of hsa-miR-181a was increased in EC with lymph migration and myometrial invasion. In addition, compared with the grade 1 EC, there was an increase in the expression level of hsa-miR-181a in grade 2 and grade 3 EC (P>0.05; Table 17). Taken together, these results indicate that there is an association between the expression level of hsa-miR-181a and the clinical development and progression of EC.
Table 17.
Association between hsa-miR-181a expression and pathological characteristics of EC
| Pathological characteristics | N | ΔCt | F | P |
|---|---|---|---|---|
| FIGO stage | ||||
| I+II | 38 | −2.929±2.306 | 7.120 | 0.011 |
| III+VI | 9 | −5.160±2.002 | ||
| Tumor grade | ||||
| G1 | 32 | −3.152±2.528 | 0.723 | 0.400 |
| G2 + G3 | 15 | −3.792±2.118 | ||
| Lymph migration | ||||
| No | 42 | −3.183±2.370 | 2.095 | 0.155 |
| Yes | 5 | −4.808±2.401 | ||
| Myometrial invasion | ||||
| ≤1/2 | 39 | −3.086±2.406 | 3.033 | 0.088 |
| >1/2 | 8 | −4.674±2.015 | ||
Abbreviations: ΔCt, normalized threshold cycle; EC, endometrial cancer; FIGO, International Federation of Gynecology and Obstetrics.
Discussion
miRNAs play critical roles in regulating proliferation, differentiation, apoptosis, development, metabolism, and immunity.2 miRNAs may act as oncogenes or tumor suppressors, and they could play a potential role as diagnostic and prognostic biomarkers of cancers.7,52 Specific miRNAs are expressed in various tissues, and changes in regulation of gene expression are thought to cause carcinogenesis. Thus, tissue-specific miRNAs may be used as effective biomarkers for cancer diagnosis, treatment, and prognosis.8 Hsa-miR-181a has been proposed to play a role in the pathogenesis, development, progression, metastasis, prognosis, and therapeutic response to chemo- and radiotherapy in EC,4,53 ovarian cancer,54 glioma,30,55 liver cancer,56 colorectal cancer,57–59 gastric cancer,60,61 lung cancer,62 breast cancer,63–67 cervical carcinoma,68,69 pancreatic cancer,46 osteosarcoma,70 oral squamous cell carcinoma,71,72 B-cell lymphoma,73 thyroid cancer,74 salivary adenoid cystic carcinoma,75 and acute and chronic leukemias.76–81 Ciafrè et al,82 firstly reported that the expression of hsa-miR-181a was significantly downregulated in primary glioblastomas and human glioblastoma cell lines compared with normal brain tissue. As in glioblastoma, significant downregulation of hsa-miR-181a was also observed in squamous lung cell carcinoma,83 oral squamous cell carcinoma,71 luminal A-like breast cancer,67 and non-small-cell lung cancer.84 However, hsa-miR-181a was significantly overexpressed in MCF-7 breast cancer cells,85 colorectal cancer,28 and hepatocellular carcinoma cells.56,86,87 Hsa-miR-181a was upregulated in acute myeloid leukemia,76 especially in the M1 and M2 subtypes, and in myelodysplastic syndromes88 but downregulated in multiple myeloma89 and chronic lymphocyte leukemia.79,90 hsa-miR-181a can serve as an oncogene46,54,56–58,60,61,65,66,68–70,87,91 or tumor suppressor,55,67,71,75,78,79,92 implicating its multifaceted and complex roles in the regulation of its target genes and signaling pathways associated with cancer initiation, growth, development, progression, and metastasis.
In the present study, our bioinformatic study predicted that hsa-miR-181a could regulate a large number of targets, including proteins that participate in regulation of cell proliferation, cell cycle, apoptosis, autophagy, metabolism, signaling transduction, and transport. A further search in TarBase and miRTarBase identified 313 targets of hsa-miR-181a-5p, and 22 of these genes are cancer genes that play critical roles in the tumorigenesis of various cancers.
During the prediction process, we employed ten different predicting programs that are based on different matching criteria and calculating algorithms. All the algorithms displayed disappointing predictive accuracy and ability when compared with the validated targets of hsa-miR-181a. It appears that there is a need to refine or combine these algorithms to improve their predictive accuracy and ability. Indeed, most of these prediction algorithms, including RNAhybrid, miRanda, TargetScan, DIANA microT, and PicTar exhaustively analyze all the possible miRNA: mRNA pairs, searching for structural evidence that could suggest the existence of an interaction. Although these approaches are significantly cheaper than those based on experimental validation, results of these methods are in many cases uncorrelated to each other, and their degree of overlap is low as shown in this study. The weakness of these algorithms depends on many factors, especially on the impossibility of incorporating in a single model all the possible interplaying variants/factors that can affect miRNA targeting and the prediction outcomes, especially in mammals. Different results can also depend on the approach used and on the rules considered for the miRNA targeting, as well as on the type of resource of sequences they use as a reference dataset.93 Shirdel et al94 found that the precision and recall values computed against validated interactions of a specific algorithm were generally poor, but a combination of these algorithms can improve the prediction precision. Recently, some machine learning approaches have been incorporated, to learn to combine the outputs of distinct prediction algorithms and improve their accuracy.95–97 Zhang and Verbeek97 proposed the application of a supervised learning algorithm, ie, a Bayesian network learner, to distinct sets of features considered in three prediction algorithms, including RNAhybrid, miRanda, and TargetScan. Pio et al96 proposed a semisupervised ensemble learning approach using miRTarBase as the set of labeled (positive) interactions and microRNA Data Integration Portal (mirDIP) as the set of unlabeled interactions, and the predictive accuracy was improved.
We next compared the expression levels of hsa-miR-181a in normal endometrium, endometrial hyperplasia, and type I and type II EC. We found that the expression level of hsa-miR-181a was significantly higher in EC than that in normal endometrium and that advanced EC exhibited a higher expression level of hsa-miR-181a. These observations demonstrate that there was an association between the expression level of hsa-miR-181a and the progression of EC and that hsa-miR-181a might serve as an oncogene in the development and progression of EC.
Many miRNAs are aberrantly expressed in cancer, resulting in functional alterations in cell differentiation, proliferation, migration, invasion, programmed cell death, and survival.5,6,8,53 A number of oncogenes and tumor-suppressor genes could be potentially regulated by miRNAs. miRNAs are presumed to be a class of genes involved in human tumorigenesis, and miRNA-mediated gene regulation is an important cellular biologic process in cancer development.5,6,8,53 For example, let-7 acts as tumor suppressor gene, which was found to be downregulated in lung tumors and associated with a poor postoperative prognosis.98 It has been showed that the RAS oncogene was regulated by let-7 and that a decreased expression level of let-7 in lung cancer resulted in an increase in the expression level of the RAS oncogene.99
Many studies have showed that upregulation of hsa-miR-181a promotes carcinogenesis, cancer cell growth, and metastasis in a variety of cancers, via regulation of a number of molecular targets and signaling pathways related to cell proliferation, invasion, migration, survival, and cell death.46,54,56–58,60,61,65,66,68–70,87,91 Zou et al87 observed an increase in the expression level of hsa-miR-181a, which may contribute to the development and progression of hepatocellular carcinoma via targeting of E2F transcription factor 5, p130-binding (E2F5). hsa-miR-181a was also upregulated in hepatocellular cancer stem cells.86,100 Silencing hsa-miR-181 led to a decreased motility and invasion of hepatocellular cancer stem cells, via targeting of the putative tumor suppressor Ras association domain family 1 isoform A (RASSF1), metalloproteinase inhibitor 3 (ie, TIMP3), and nemo-like kinase (NLK).100 has-miR-181 could directly target hepatic transcriptional regulators of differentiation, including caudal type homeobox 2 (CDX2), GATA binding protein 6 (GATA6), and NLK, an inhibitor of Wnt/β-catenin signaling.86 hsa-miR-181a promoted tumor growth and liver metastasis in colorectal cancer patients by targeting the tumor suppressor WNT inhibitory factor 1 (WIF1).28 hsa-miR-181a was most elevated in these colorectal cancer patients with liver metastases and could serve as an independent prognostic factor of poor overall survival.28 hsa-miR-181a showed a potent tumor-promoting effect through inhibition of the expression of WIF1 and promotion of epithelial–mesenchymal transition.28 Moreover, upregulation of hsa-miR-181a plays a potential role in the development of gastric cancer by targeting the tumor suppressor ATM serine/threonine kinase (ATM). Consequently, it leads to promotion of gastric cancer cell proliferation and inhibition of apoptosis. Wei et al101 showed that the PTEN/Akt signaling pathway was involved in the regulatory effect of hsa-miR-181a in the development of colon cancer, by promoting cell growth. hsa-miR-181a also played an important role in ovarian cancer progression, by promoting epithelial–mesenchymal transition.54 These data indicate that hsa-miR-181a may function as oncogenic miRNA in cancer development and progression. In agreement with previous studies, our findings showed that there was a significant increase in the expression level of hsa-miR-181a in EC compared with that in normal endometrium. Moreover, our results showed that advanced EC had a significant higher expression level of hsa-miR-181a than that in early stage of EC, suggesting that hsa-miR-181a may have a critical role in tumor metastasis of advanced EC.
On the other hand, hsa-miR-181 may function as a tumor suppressor. In glioma, hsa-miR-181a was shown to be downregulated.30 Both hsa-miR-181a and hsa-miR-181b triggered growth inhibition, induced apoptosis, and inhibited invasion in glioma cells. Transiently overexpressed hsa-miR-181a significantly sensitized malignant glioma U87MG cells to radiation with downregulated BCL2.55 In chronic lymphocytic leukemia, hsa-miR-181a together with hsa-miR-15a, hsa-miR-16-1, hsa-miR-29b, and hsa-miR-181b were all downregulated.90 These miRNAs may play a role in the pathogenesis of chronic lymphocytic leukemia and serve as new biomarkers for the prediction of prognosis in chronic lymphocytic leukemia. hsa-miR-181a expression level was found to be significantly lower in poor prognosis patients, and a low expression of hsa-miR-181a and hsa-miR-181b was associated with shorter overall survival and treatment-free survival in patients with chronic lymphocytic leukemia.79 Furthermore, hsa-miR-181a inhibited the migration, invasion, and proliferation of salivary adenoid cystic carcinoma cells, and suppressed tumor growth and lung metastasis in nude mice, via targeting of MAP2K1, MAPK1, and SNAI2.75
Based on our DAVID and KEGG pathway analysis, PI3K/Akt, MAPK, and Wnt signaling pathways played important roles in the development of type I EC. CCND1, HRAS, and KRAS are all key components in these pathways, which are all validated targets of hsa-miR-181a.
The expression level of hsa-miR-181a has been proposed as a potential biomarker for assessing prognosis and therapeutic response in cancer. Ouyang et al102 suggested that hsa-miR-181a may be a potential biomarker for predicting chemoresistance in the treatment of triple negative breast cancer. It also has been showed that hsa-miRNA-181a enhanced the chemoresistance of human cervical squamous cell carcinoma to cisplatin by targeting protein kinase Cδ69 and that hsa-miR-181a may serve an oncologic miRNA biomarker for luminal A-like breast cancer.67 Interestingly, Pichler et al59 showed a reverse correlation between hsa-miR-181a expression level and survival rate in patients with colorectal cancer. In our study, we observed a significant difference in the expression level of hsa-miR-181a among normal endometrium, endometrial hyperplasia, and EC, and a higher expression level of hsa-miR-181a in advanced EC. Our findings suggest that the expression level of hsa-miR-181a might serve as a useful biomarker for the prediction of prognosis of EC in clinic.
In summary, our bioinformatics studies have showed that hsa-miR-181a might regulate a large number of target genes that are important in the regulation of critical cell processes. To date, 313 targets of hsa-miR-181a have been validated, and 22 of these targets are cancer genes. Many of these genes are involved in tumorigenesis of various cancers, including EC. Our data demonstrate that hsa-miR-181a is upregulated in EC, with a possible role in the development and progression of EC (Figure 12). It might serve as a new biomarker for prognosis prediction in EC in clinical practice and has important implication in the treatment of EC. More mechanistic and functional studies are needed to validate the role of hsa-miR-181a in the pathogenesis of EC and to establish the association between the expression level of hsa-miR-181a and the clinical phenotypes, including disease status and therapeutic response of EC to chemo- and radiotherapy.
Figure 12.
Proposed mechanisms for how hsa-miR-181a regulates cancer genes and may serve as an oncogene for the development of EC.
Notes: hsa-miR-181a may play a role in the tumorigenesis of EC via multiple pathways that interplay with many important oncogenes and tumor suppressors. hsa-miR-181a may interact with H3F3B, ATM, CCDC6, TAF15, RAS, and PLAG1 to promote cell proliferation. It may suppress cell apoptosis via interaction with NOTCH1, NOTCH2, MAPK1, and BCL2. It may affect ubiquitin-mediated proteolysis via regulation of FBXO11, STAG2, BRCA2, HOXA11, and RBM15. All these actions may transform the normal endometrial cells into tumor cells.
Abbreviation: EC, endometrial cancer; ATM, ATM serine/threonine kinase; BCL2, B-cell lymphoma 2; CCDC6, coiled-coil domain containing 6; PLAG1, pleiomorphic adenoma gene 1; MAPK1, mitogen activated kinase-like protein 1; RAS, rat sarcoma viral oncogene homolog; BRCA2, breast cancer 2, early onset; STAG2, stromal antigen 2; HOXA11, homeobox A11; RBM15, RNA binding motif protein 15; NOTCH1, notch 1; H3F3B, H3 histone, family 3B; FBXO11, F-box protein 11; TAF15, TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68 kDa; HOOK3, hook microtubule-tethering protein 3; PTPN11, protein tyrosine phosphatase, non-receptor type 11; TSHR, thyroid stimulating hormone receptor; CDX2, caudal type homeobox 2; EP300, E1A binding protein p300.
Supplementary materials
Table S1 A full list of cancer genes, based on Futreal et al1. Reprinted by permission from Macmillan Publishers Ltd: Nature Reviews Cancer. Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. Copyright © 2004.1
Table S2 Predicted targets of hsa-miR-181a-5p, by DIANA microT v5.0
Table S3 Predicted targets of hsa-miR-181a-5p, by miRanda-mirSVR
Table S4 Predicted targets of hsa-miR-181a-5p, by miRanda-mirSVR, that are cancer genes
Table S5 Predicted targets of hsa-miR-181a-5p, by miRDB
Table S6 Predicted targets of hsa-miR-181a-5p, by RNA22 v2
Table S7 Predicted targets of hsa-miR-181a-5p, by RNA22 v2, that are cancer genes
Table S8 Predicted targets of hsa-miR-181a-5p, by TargetMiner
Table S9 Predicted targets of hsa-miR-181a-5p, by TargetScan 6.2
Table S10 Predicted targets of hsa-miR-181a-5p, by PicTar
Table S11 Predicted targets of hsa-miR-181a-5p, by MicroCosm Targets v5
Table S12 Predicted targets of hsa-miR-181a-3p, by DIANA microT v5.0
Table S13 Predicted targets of hsa-miR-181a-3p, by miRanda-mirSVR
Table S14 Predicted targets of hsa-miR-181a-3p, by miRanda-mirSVR, that are cancer genes
Table S15 Predicted targets of hsa-miR-181a-3p, by miRDB
Table S16 Predicted targets of hsa-miR-181a-3p, by RNA22 v2
Table S17 Predicted targets of hsa-miR-181a-3p, by TargetMiner
Table S18 Predicted targets of hsa-miR-181a-5p, by MicroCosm Targets v5
Table S19 Predicted targets of hsa-miR-181a, by miRWALK
Table S20 Predicted genes of hsa-miR-181a, by miRWALK
Table S21 A summarized table that includes all the targets predicted to be regulated by hsa-miR-181a by the eight algorithms
Analysis of hsa-miR-181a expression in a human endometrial specimen by real-time PCR.
Notes: (A) Melting curve showing the single melt peak for hsa-miR-181a and U6, respectively; and (B) the amplification plot of the target gene.
Abbreviation: PCR, polymerase chain reaction.
Reference
- 1.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Acknowledgments
This work was supported by Guangdong Natural Science Foundation (grant number S2012010010024), Guangzhou, People’s Republic of China. Dr Zhi-Wei Zhou is a postdoctoral fellow supported by the College of Pharmacy, University of South Florida, Tampa, FL, USA.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122(1):6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 2.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153(3):516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 4.Banno K, Yanokura M, Kisu I, Yamagami W, Susumu N, Aoki D. MicroRNAs in endometrial cancer. Int J Clin Oncol. 2013;18(2):186–192. doi: 10.1007/s10147-013-0526-9. [DOI] [PubMed] [Google Scholar]
- 5.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482(7385):347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nana-Sinkam SP, Croce CM. Clinical applications for microRNAs in cancer. Clin Pharmacol Ther. 2013;93(1):98–104. doi: 10.1038/clpt.2012.192. [DOI] [PubMed] [Google Scholar]
- 9.Ritchie W, Rasko JE. Refining microRNA target predictions: sorting the wheat from the chaff. Biochem Biophys Res Commun. 2014;445(4):780–784. doi: 10.1016/j.bbrc.2014.01.181. [DOI] [PubMed] [Google Scholar]
- 10.Peterson SM, Thompson JA, Ufkin ML, Sathyanarayana P, Liaw L, Congdon CB. Common features of microRNA target prediction tools. Front Genet. 2014;5:23. doi: 10.3389/fgene.2014.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferlay J, Soerjomataram I, Ervik M, et al. GLOBOCAN 2012 v.10, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet] Lyon: International Agency for Research on Cancer; 2013. [Accessed November 10, 2014]. Available from: http://globocan.iarc.fr. [Google Scholar]
- 12.Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2011. Bethesda, MD: National Cancer Institute; 2014. [Google Scholar]
- 13.Balch C, Matei DE, Huang TH, Nephew KP. Role of epigenomics in ovarian and endometrial cancers. Epigenomics. 2010;2(3):419–447. doi: 10.2217/epi.10.19. [DOI] [PubMed] [Google Scholar]
- 14.Amant F, Moerman P, Neven P, Timmerman D, Van Limbergen E, Vergote I. Endometrial cancer. Lancet. 2005;366(9484):491–505. doi: 10.1016/S0140-6736(05)67063-8. [DOI] [PubMed] [Google Scholar]
- 15.Colombo N, Preti E, Landoni F, et al. ESMO Guidelines Working Group Endometrial cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013;24(Suppl 6):vi33–vi38. doi: 10.1093/annonc/mdt353. [DOI] [PubMed] [Google Scholar]
- 16.Hecht JL, Mutter GL. Molecular and pathologic aspects of endometrial carcinogenesis. J Clin Oncol. 2006;24(29):4783–4791. doi: 10.1200/JCO.2006.06.7173. [DOI] [PubMed] [Google Scholar]
- 17.Yeramian A, Moreno-Bueno G, Dolcet X, et al. Endometrial carcinoma: molecular alterations involved in tumor development and progression. Oncogene. 2013;32(4):403–413. doi: 10.1038/onc.2012.76. [DOI] [PubMed] [Google Scholar]
- 18.National Cancer Institute . Endometrial Cancer: 2014. [Accessed December 15, 2014]. Available from http://www.cancer.gov/cancertopics/types/endometrial. [Google Scholar]
- 19.Dong P, Kaneuchi M, Konno Y, Watari H, Sudo S, Sakuragi N. Emerging therapeutic biomarkers in endometrial cancer. Biomed Res Int. 2013;2013:130362. doi: 10.1155/2013/130362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ow TJ, Sandulache VC, Skinner HD, Myers JN. Integration of cancer genomics with treatment selection: from the genome to predictive biomarkers. Cancer. 2013;119(22):3914–3928. doi: 10.1002/cncr.28304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitra S, Das S, Chakrabarti J. Systems biology of cancer biomarker detection. Cancer Biomark. 2013;13(4):201–213. doi: 10.3233/CBM-130363. [DOI] [PubMed] [Google Scholar]
- 22.Widschwendter M, Jones A, Teschendorff AE. Epigenetics makes its mark on women-specific cancers – an opportunity to redefine oncological approaches? Gynecol Oncol. 2013;128(1):134–143. doi: 10.1016/j.ygyno.2012.09.027. [DOI] [PubMed] [Google Scholar]
- 23.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 24.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11(12):849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 26.Lin Y, Nie Y, Zhao J, et al. Genetic polymorphism at miR-181a binding site contributes to gastric cancer susceptibility. Carcinogenesis. 2012;33(12):2377–2383. doi: 10.1093/carcin/bgs292. [DOI] [PubMed] [Google Scholar]
- 27.Havelange V, Stauffer N, Heaphy CC, et al. Functional implications of microRNAs in acute myeloid leukemia by integrating microRNA and messenger RNA expression profiling. Cancer. 2011;117(20):4696–4706. doi: 10.1002/cncr.26096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji D, Chen Z, Li M, et al. MicroRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1. Mol Cancer. 2014;13:86. doi: 10.1186/1476-4598-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fei J, Li Y, Zhu X, Luo X. miR-181a post-transcriptionally downregulates oncogenic RalA and contributes to growth inhibition and apoptosis in chronic myelogenous leukemia (CML) PLoS One. 2012;7(3):e32834. doi: 10.1371/journal.pone.0032834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi L, Cheng Z, Zhang J, et al. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185–193. doi: 10.1016/j.brainres.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 31.Panda H, Chuang TD, Luo X, Chegini N. Endometrial miR-181a and miR-98 expression is altered during transition from normal into cancerous state and target PGR, PGRMC1, CYP19A1, DDX3X, and TIMP3. J Clin Endocrinol Metab. 2012;97(7):E1316–E1326. doi: 10.1210/jc.2012-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11(8):R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maragkakis M, Alexiou P, Papadopoulos GL, et al. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics. 2009;10:295. doi: 10.1186/1471-2105-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41(Web Server issue):W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14(6):1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 40.Bandyopadhyay S, Mitra R. TargetMiner: microRNA target prediction with systematic identification of tissue-specific negative examples. Bioinformatics. 2009;25(20):2625–2631. doi: 10.1093/bioinformatics/btp503. [DOI] [PubMed] [Google Scholar]
- 41.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 42.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18(10):1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 44.Dweep H, Sticht C, Pandey P, Gretz N. miRWALK – database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44(5):839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 45.Vergoulis T, Vlachos IS, Alexiou P, et al. TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2012;40(Database issue):D222–D229. doi: 10.1093/nar/gkr1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsu SD, Tseng YT, Shrestha S, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42(Database issue):D78–D85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 50.Huang da W, Sherman BT, Tan Q, et al. DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pecorelli S. Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol Obstet. 2009;105(2):103–104. doi: 10.1016/j.ijgo.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 52.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gilabert-Estelles J, Braza-Boils A, Ramon LA, et al. Role of microRNAs in gynecological pathology. Curr Med Chem. 2012;19(15):2406–2413. doi: 10.2174/092986712800269362. [DOI] [PubMed] [Google Scholar]
- 54.Parikh A, Lee C, Peronne J, et al. microRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial-mesenchymal transition. Nat Commun. 2014;5:2977. doi: 10.1038/ncomms3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen G, Zhu W, Shi D, et al. MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol Rep. 2010;23(4):997–1003. doi: 10.3892/or_00000725. [DOI] [PubMed] [Google Scholar]
- 56.Brockhausen J, Tay SS, Grzelak CA, et al. miR-181a mediates TGF-β-induced hepatocyte EMT and is dysregulated in cirrhosis and hepatocellular cancer. Liver Int. 2015;35(1):240–253. doi: 10.1111/liv.12517. [DOI] [PubMed] [Google Scholar]
- 57.Nishimura J, Handa R, Yamamoto H, et al. microRNA-181a is associated with poor prognosis of colorectal cancer. Oncol Rep. 2012;28(6):2221–2226. doi: 10.3892/or.2012.2059. [DOI] [PubMed] [Google Scholar]
- 58.Zou C, Chen J, Chen K, et al. Functional analysis of miR-181a and Fas involved in hepatitis B virus-related hepatocellular carcinoma pathogenesis. Exp Cell Res. 2015 doi: 10.1016/j.yexcr.2014.11.007. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 59.Pichler M, Winter E, Ress AL, et al. miR-181a is associated with poor clinical outcome in patients with colorectal cancer treated with EGFR inhibitor. J Clin Pathol. 2014;67(3):198–203. doi: 10.1136/jclinpath-2013-201904. [DOI] [PubMed] [Google Scholar]
- 60.Zhang X, Nie Y, Du Y, Cao J, Shen B, Li Y. MicroRNA-181a promotes gastric cancer by negatively regulating tumor suppressor KLF6. Tumour Biol. 2012;33(5):1589–1597. doi: 10.1007/s13277-012-0414-3. [DOI] [PubMed] [Google Scholar]
- 61.Zhang X, Nie Y, Li X, et al. MicroRNA-181a functions as an oncomir in gastric cancer by targeting the tumour suppressor gene ATM. Pathol Oncol Res. 2014;20(2):381–389. doi: 10.1007/s12253-013-9707-0. [DOI] [PubMed] [Google Scholar]
- 62.Galluzzi L, Morselli E, Vitale I, et al. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 2010;70(5):1793–1803. doi: 10.1158/0008-5472.CAN-09-3112. [DOI] [PubMed] [Google Scholar]
- 63.Farazi TA, Horlings HM, Ten Hoeve JJ, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71(13):4443–4453. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo LJ, Zhang QY. Decreased serum miR-181a is a potential new tool for breast cancer screening. Int J Mol Med. 2012;30(3):680–686. doi: 10.3892/ijmm.2012.1021. [DOI] [PubMed] [Google Scholar]
- 65.Taylor MA, Sossey-Alaoui K, Thompson CL, Danielpour D, Schiemann WP. TGF-β upregulates miR-181a expression to promote breast cancer metastasis. J Clin Invest. 2013;123(1):150–163. doi: 10.1172/JCI64946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bisso A, Faleschini M, Zampa F, et al. Oncogenic miR-181a/b affect the DNA damage response in aggressive breast cancer. Cell Cycle. 2013;12(11):1679–1687. doi: 10.4161/cc.24757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McDermott AM, Miller N, Wall D, et al. Identification and validation of oncologic miRNA biomarkers for luminal A-like breast cancer. PLoS One. 2014;9(1):e87032. doi: 10.1371/journal.pone.0087032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ke G, Liang L, Yang JM, et al. MiR-181a confers resistance of cervical cancer to radiation therapy through targeting the pro-apoptotic PRKCD gene. Oncogene. 2013;32(25):3019–3027. doi: 10.1038/onc.2012.323. [DOI] [PubMed] [Google Scholar]
- 69.Chen Y, Ke G, Han D, Liang S, Yang G, Wu X. MicroRNA-181a enhances the chemoresistance of human cervical squamous cell carcinoma to cisplatin by targeting PRKCD. Exp Cell Res. 2014;320(1):12–20. doi: 10.1016/j.yexcr.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 70.Jianwei Z, Fan L, Xiancheng L, Enzhong B, Shuai L, Can L. MicroRNA 181a improves proliferation and invasion, suppresses apoptosis of osteosarcoma cell. Tumour Biol. 2013;34(6):3331–3337. doi: 10.1007/s13277-013-0902-0. [DOI] [PubMed] [Google Scholar]
- 71.Shin KH, Bae SD, Hong HS, Kim RH, Kang MK, Park NH. miR-181a shows tumor suppressive effect against oral squamous cell carcinoma cells by downregulating K-ras. Biochem Biophys Res Commun. 2011;404(4):896–902. doi: 10.1016/j.bbrc.2010.12.055. [DOI] [PubMed] [Google Scholar]
- 72.Liu M, Wang J, Huang H, Hou J, Zhang B, Wang A. miR-181a-Twist1 pathway in the chemoresistance of tongue squamous cell carcinoma. Biochem Biophys Res Commun. 2013;441(2):364–370. doi: 10.1016/j.bbrc.2013.10.051. [DOI] [PubMed] [Google Scholar]
- 73.Alencar AJ, Malumbres R, Kozloski GA, et al. MicroRNAs are independent predictors of outcome in diffuse large B-cell lymphoma patients treated with R-CHOP. Clin Cancer Res. 2011;17(12):4125–4135. doi: 10.1158/1078-0432.CCR-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keutgen XM, Filicori F, Crowley MJ, et al. A panel of four miRNAs accurately differentiates malignant from benign indeterminate thyroid lesions on fine needle aspiration. Clin Cancer Res. 2012;18(7):2032–2038. doi: 10.1158/1078-0432.CCR-11-2487. [DOI] [PubMed] [Google Scholar]
- 75.He Q, Zhou X, Li S, et al. MicroRNA-181a suppresses salivary adenoid cystic carcinoma metastasis by targeting MAPK-Snai2 pathway. Biochim Biophys Acta. 2013;1830(11):5258–5266. doi: 10.1016/j.bbagen.2013.07.028. [DOI] [PubMed] [Google Scholar]
- 76.Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-McIver A, Young BD. MicroRNA miR-181a correlates with morphological subclass of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21(5):912–916. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- 77.Cuesta R, Martínez-Sánchez A, Gebauer F. miR-181a regulates cap-dependent translation of p27(kip1) mRNA in myeloid cells. Mol Cell Biol. 2009;29(10):2841–2851. doi: 10.1128/MCB.01971-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bai H, Cao Z, Deng C, Zhou L, Wang C. miR-181a sensitizes resistant leukaemia HL-60/Ara-C cells to Ara-C by inducing apoptosis. J Cancer Res Clin Oncol. 2012;138(4):595–602. doi: 10.1007/s00432-011-1137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu DX, Zhu W, Fang C, et al. miR-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis. 2012;33(7):1294–1301. doi: 10.1093/carcin/bgs179. [DOI] [PubMed] [Google Scholar]
- 80.Dahlhaus M, Schult C, Lange S, Freund M, Junghanss C. MicroRNA 181a influences the expression of HMGB1 and CD4 in acute leukemias. Anticancer Res. 2013;33(2):445–452. [PubMed] [Google Scholar]
- 81.Lin S, Pan L, Guo S, et al. Prognostic role of microRNA-181a/b in hematological malignancies: a meta-analysis. PLoS One. 2013;8(3):e59532. doi: 10.1371/journal.pone.0059532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ciafrè SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334(4):1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 83.Gao W, Shen H, Liu L, Xu J, Xu J, Shu Y. MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J Cancer Res Clin Oncol. 2011;137(4):557–566. doi: 10.1007/s00432-010-0918-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao W, Yu Y, Cao H, et al. Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed Pharmacother. 2010;64(6):399–408. doi: 10.1016/j.biopha.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 85.Miller TE, Ghoshal K, Ramaswamy B, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283(44):29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ji J, Yamashita T, Budhu A, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50(2):472–480. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zou C, Li Y, Cao Y, et al. Up-regulated MicroRNA-181a induces carcinogenesis in hepatitis B virus-related hepatocellular carcinoma by targeting E2F5. BMC Cancer. 2014;14:97. doi: 10.1186/1471-2407-14-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pons A, Nomdedeu B, Navarro A, et al. Hematopoiesis-related microRNA expression in myelodysplastic syndromes. Leuk Lymphoma. 2009;50(11):1854–1859. doi: 10.3109/10428190903147645. [DOI] [PubMed] [Google Scholar]
- 89.Pichiorri F, Suh SS, Ladetto M, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci U S A. 2008;105(35):12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu DX, Miao KR, Fang C, et al. Aberrant microRNA expression in Chinese patients with chronic lymphocytic leukemia. Leuk Res. 2011;35(6):730–734. doi: 10.1016/j.leukres.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 91.Albulescu R. TGF-β upregulates miR-181a expression to promote breast cancer metastasis. Biomark Med. 2013;7(2):204. [PubMed] [Google Scholar]
- 92.Jiao X, Zhao L, Ma M, et al. MiR-181a enhances drug sensitivity in mitoxantone-resistant breast cancer cells by targeting breast cancer resistance protein (BCRP/ABCG2) Breast Cancer Res Treat. 2013;139(3):717–730. doi: 10.1007/s10549-013-2607-x. [DOI] [PubMed] [Google Scholar]
- 93.Ritchie W, Flamant S, Rasko JE. Predicting microRNA targets and functions: traps for the unwary. Nat Methods. 2009;6(6):397–398. doi: 10.1038/nmeth0609-397. [DOI] [PubMed] [Google Scholar]
- 94.Shirdel EA, Xie W, Mak TW, Jurisica I. NAViGaTing the micronome – using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS One. 2011;6(2):e17429. doi: 10.1371/journal.pone.0017429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Godsey B. Discovery of miR-mRNA interactions via simultaneous Bayesian inference of gene networks and clusters using sequence-based predictions and expression data. J Integr Bioinform. 2013;10(1):227. doi: 10.2390/biecoll-jib-2013-227. [DOI] [PubMed] [Google Scholar]
- 96.Pio G, Malerba D, D’Elia D, Ceci M. Integrating microRNA target predictions for the discovery of gene regulatory networks: a semi-supervised ensemble learning approach. BMC Bioinformatics. 2014;15(Suppl 1):S4. doi: 10.1186/1471-2105-15-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang Y, Verbeek FJ. Comparison and integration of target prediction algorithms for microRNA studies. J Integr Bioinform. 2010;7(3) doi: 10.2390/biecoll-jib-2010-127. [DOI] [PubMed] [Google Scholar]
- 98.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 99.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 100.Meng F, Glaser SS, Francis H, et al. Functional analysis of microRNAs in human hepatocellular cancer stem cells. J Cell Mol Med. 2012;16(1):160–173. doi: 10.1111/j.1582-4934.2011.01282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wei Z, Cui L, Mei Z, Liu M, Zhang D. miR-181a mediates metabolic shift in colon cancer cells via the PTEN/AKT pathway. FEBS Lett. 2014;588(9):1773–1779. doi: 10.1016/j.febslet.2014.03.037. [DOI] [PubMed] [Google Scholar]
- 102.Ouyang M, Li Y, Ye S, et al. MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS One. 2014;9(5):e96228. doi: 10.1371/journal.pone.0096228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 104.MacDonald BT, Tamai K, He X. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pijnenborg JM, Kisters N, van Engeland M, et al. APC, β-catenin, and E-cadherin and the development of recurrent endometrial carcinoma. Int J Gynecol Cancer. 2004;14(5):947–956. doi: 10.1111/j.1048-891X.2004.014534.x. [DOI] [PubMed] [Google Scholar]
- 106.van der Zee M, Jia Y, Wang Y, et al. Alterations in Wnt-β-catenin and Pten signalling play distinct roles in endometrial cancer initiation and progression. J Pathol. 2013;230(1):48–58. doi: 10.1002/path.4160. [DOI] [PubMed] [Google Scholar]
- 107.Liu Y, Meng F, Xu Y, et al. Overexpression of Wnt7a is associated with tumor progression and unfavorable prognosis in endometrial cancer. Int J Gynecol Cancer. 2013;23(2):304–311. doi: 10.1097/IGC.0b013e31827c7708. [DOI] [PubMed] [Google Scholar]
- 108.Zhao Y, Yang Y, Trovik J, et al. A novel wnt regulatory axis in endometrioid endometrial cancer. Cancer Res. 2014;74(18):5103–5117. doi: 10.1158/0008-5472.CAN-14-0427. [DOI] [PubMed] [Google Scholar]
- 109.Liu Y, Patel L, Mills GB, et al. Clinical significance of CTNNB1mutation and Wnt pathway activation in endometrioid endometrial carcinoma. J Natl Cancer Inst. 2014;106(9):dju245. doi: 10.1093/jnci/dju245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 111.Plotnikov A, Zehorai E, Procaccia S, Seger R. The MAPK cascades: signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim Biophys Acta. 2011;1813(9):1619–1633. doi: 10.1016/j.bbamcr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 112.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11(5):338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 113.Suzuki T, Aoki D, Susumu N, Udagawa Y, Nozawa S. Imbalanced expression of TAN-1 and human Notch4 in endometrial cancers. Int J Oncol. 2000;17(6):1131–1139. doi: 10.3892/ijo.17.6.1131. [DOI] [PubMed] [Google Scholar]
- 114.Cobellis L, Caprio F, Trabucco E, et al. The pattern of expression of Notch protein members in normal and pathological endometrium. J Anat. 2008;213(4):464–472. doi: 10.1111/j.1469-7580.2008.00963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mitsuhashi Y, Horiuchi A, Miyamoto T, Kashima H, Suzuki A, Shiozawa T. Prognostic significance of Notch signalling molecules and their involvement in the invasiveness of endometrial carcinoma cells. Histopathology. 2012;60(5):826–837. doi: 10.1111/j.1365-2559.2011.04158.x. [DOI] [PubMed] [Google Scholar]
- 116.Jonusiene V, Sasnauskiene A, Lachej N, et al. Down-regulated expression of Notch signaling molecules in human endometrial cancer. Med Oncol. 2013;30(1):438. doi: 10.1007/s12032-012-0438-y. [DOI] [PubMed] [Google Scholar]
- 117.Nickkho-Amiry M, McVey R, Holland C. Peroxisome proliferator-activated receptors modulate proliferation and angiogenesis in human endometrial carcinoma. Mol Cancer Res. 2012;10(3):441–453. doi: 10.1158/1541-7786.MCR-11-0233. [DOI] [PubMed] [Google Scholar]
- 118.Pengchong H, Tao H. Expression of IGF-1R, VEGF-C and D2-40 and their correlation with lymph node metastasis in endometrial adenocarcinoma. Eur J Gynaecol Oncol. 2011;32(6):660–664. [PubMed] [Google Scholar]
- 119.Dobrzycka B, Mackowiak-Matejczyk B, Kinalski M, Terlikowski SJ. Pretreatment serum levels of bFGF and VEGF and its clinical significance in endometrial carcinoma. Gynecol Oncol. 2013;128(3):454–460. doi: 10.1016/j.ygyno.2012.11.035. [DOI] [PubMed] [Google Scholar]
- 120.Saarelainen SK, Staff S, Peltonen N, et al. Endoglin, VEGF, and its receptors in predicting metastases in endometrial carcinoma. Tumour Biol. 2014;35(5):4651–4657. doi: 10.1007/s13277-014-1609-6. [DOI] [PubMed] [Google Scholar]
- 121.Wang J, Taylor A, Showeil R, et al. Expression profiling and significance of VEGF-A, VEGFR2, VEGFR3 and related proteins in endometrial carcinoma. Cytokine. 2014;68(2):94–100. doi: 10.1016/j.cyto.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 122.Saharinen P, Eklund L, Pulkki K, Bono P, Alitalo K. VEGF and angiopoietin signaling in tumor angiogenesis and metastasis. Trends Mol Med. 2011;17(7):347–362. doi: 10.1016/j.molmed.2011.01.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of hsa-miR-181a expression in a human endometrial specimen by real-time PCR.
Notes: (A) Melting curve showing the single melt peak for hsa-miR-181a and U6, respectively; and (B) the amplification plot of the target gene.
Abbreviation: PCR, polymerase chain reaction.