Abstract
5-hydroxymethylcytosine (5hmC), an oxidized derivative of 5-methylcytosine (5mC), has been implicated as an important epigenetic regulator of mammalian development. Current procedures use DNA sequencing methods to discriminate 5hmC from 5mC, limiting their accessibility to the scientific community. Here we report a method that combines TET-assisted bisulfite conversion with Illumina 450 K DNA methylation arrays for a low-cost high-throughput approach that distinguishes 5hmC and 5mC signals at base resolution. Implementing this approach, termed “TAB-array”, we assessed DNA methylation dynamics in the differentiation of human pluripotent stem cells into cardiovascular progenitors and neural precursor cells. With the ability to discriminate 5mC and 5hmC, we identified a large number of novel dynamically methylated genomic regions that are implicated in the development of these lineages. The increased resolution and accuracy afforded by this approach provides a powerful means to investigate the distinct contributions of 5mC and 5hmC in human development and disease.
Keywords: 5-hydroxymethylcytosine, TAB-array, DNA methylation, Human pluripotent stem cells, Epigenetics, Differentiation, Neuronal cells, Cardiovascular cells
1. Introduction
Dramatic epigenetic changes occur during human embryonic development. Human pluripotent stem cells (hPSCs; embryonic stem cells and induced pluripotent stem cells) are a valuable model of epigenetic regulation in development because they can be directed in vitro to differentiate into many different cell types. DNA methylation has been shown to be an important epigenetic modification that regulates vital cellular processes including gene expression, retrotransposon silencing and imprinting. Most methylation occurs on cytosines in a CpG dinucleotide context, although non-CpG methylation (CpH), especially CpA methylation, is common in hPSCs and the brain [1–3]. DNA methylation has been shown to change dramatically as hPSCs differentiate [2]. In undifferentiated hPSCs, genes coding for lineage-specifying factors are uniformly methylated; as the cells differentiate, methylation increases in the promoters of pluripotency genes, while DNA methylation of regions associated with regulation of lineage-specific genes decreases in a cell type-specific manner [4].
Recently, an oxidized derivative of DNA methylation, 5-hydroxymethylcytosine (5hmC), has come into the spotlight and we are just now beginning to investigate its significance. 5hmC marks are found to be highest in specific types of neurons and in hPSCs, with 5hmC present on 0.7 and 0.4% of all cytosines in these cell types, respectively [5–7]. The Ten Eleven Translocation (TET) dioxygenases have been identified as the enzymes that convert 5-methylcytosine (5mC) to 5hmC [7,8]. Like most 5mC methylation, 5hmC typically occurs at CpG dinucleotides.
Several lines of evidence suggest that the TET proteins, and therefore 5hmC, play an important role in regulating the pluripotent state. It was recently shown that TET1 is dramatically induced during reprogramming of somatic cells to pluripotency and that knockdown of TET1 impedes the reprogramming process [9,10]. Additionally, TET1 can substitute for POU5F1 during reprogramming when MYC, KLF4 and SOX2 are also supplied [9]. Knockdown of the TET proteins in murine hPSCs induced precocious differentiation and increased 5mC in the promoters of pluripotency genes [8,11]. Collectively, these observations implicate the TET proteins, and 5hmC, as critical factors in the earliest stages of mammalian development.
While classical models of gene regulation highlight a clear repressive role for DNA methylation, with 5mC localized to heterochromatic stretches of the genome, early evidence suggests that 5hmC may have a functional association with euchromatin [11–13]. In stem cells, 5hmC has been shown to be highly enriched in bivalent promoters, which are defined by co-localization of H3K4me3 and H3K27me3 histone modifications [14–17]. Bivalency is considered to be a means to poise critical lineage-specifying genes for activation or silencing in response to specific developmental cues. On one hand, the presence of 5hmC in these regions could support the notion that 5hmC exists as an intermediate in a DNA demethylation cascade. On the other hand, the overlap of 5hmC with bivalent domains could support a role for 5hmC in establishing euchromatin and/or in the recruitment of transcription factors at critical stages of development. Additional evidence suggests a wider role for 5hmC beyond being an intermediary of DNA de-methylation. 5hmC has been detected in promoters and enhancers of actively transcribed genes in adult tissues [5,12,18,19] and it is abundant in terminally differentiated neurons in the brain [6]. Thus, existing evidence suggests that 5hmC may have a diverse, context-dependent set of functions. (See also reviews by Ficz and Pfeifer in this issue).
Resolving differences between 5mC and 5hmC will be critical in establishing a better understanding of how cytosine modifications contribute to the regulation of differentiation. The standard technique for studying DNA methylation is bisulfite treatment, which deaminates unmethylated cytosines to uracil (read as T following genome amplification) but does not affect 5mC or 5hmC, which are read as C. Subsequent sequencing or array-based approaches allow identification of methylated cytosines at single base resolution, but more complex technologies are required to distinguish 5hmC from 5mC. Several affinity-capture techniques have been developed for identification of 5hmC-enriched genomic regions [15,18]. A recent development, TET-Assisted Bisulfite sequencing (TAB-seq), has greatly improved mapping of 5hmC [14]; thereby permitting unbiased, genome-wide mapping of 5hmC at single-base resolution.
These technical advances have provided a powerful toolset for detailed investigations into 5hmC function. However, the considerable costs of sequencing and bioinformatic expertise required of its analysis limit the availability of protocols that discriminate 5hmC from 5mC. To make detailed mapping of 5hmC more accessible, we combined TAB conversion with DNA methylation profiling on the Illumina Infinium HumanMethylation450K BeadChip (450 K array). This method, called “TAB-array” provides a high-throughput method for analyzing both 5mC and 5hmC at less than 10% of the cost of sequence-based approaches [14,20,21]. The Illumina Infinium arrays contain internal quality control measures and can be easily analyzed with powerful user-friendly analysis packages in the R statistical environment [22–24].
We have detected dynamic methylation in the differentiation of human induced pluripotent stem cells (iPSCs) along two distinct lineages, cardiovascular progenitor cells (CVPs) and neural precursor cells (NPCs) using standard bisulfite conversion profiling techniques. However, assessment of these cell types using the TAB-array approach showed that we had initially underestimated the extent and complexity of dynamic methylation among these cell types. With the ability to distinguish 5hmC from5mC, we discovered, for example, that many regions of the genome identified as being 5mC in both lineages via standard methods, actually contained 5hmC in one lineage and 5mC in the other. Remarkably, we found that a majority of the genomic regions with developmentally dynamic 5mC among these cell types also displayed dynamic 5hmC. This suggests that 5hmC plays an active role in lineage choice during human development. Further analysis of a wider variety of lineages and stages of hPSC differentiation will allow us to better understand the epigenetic mechanisms underlying human cellular differentiation. With its lower cost and decrease in complexity of analysis, the TAB-array method will enable a greater number of investigations into 5hmC vs. 5mC function and improve our understanding of the role of differential DNA methylation in diverse cellular contexts.
2. Results
In order to discriminate 5hmC from 5mC using methylation microarrays, we adapted a previously reported sequencing-based method [14]. As illustrated in Fig. 1, genomic DNA from each biological sample was split into two fractions for different downstream processing steps. One fraction was processed for standard bisulfite conversion, which deaminates both 5mC and 5hmC. This process identified cytosines as methylated or not methylated, without distinguishing between 5mC and 5hmC. The other fraction was used for TET-Assisted Bisulfite (TAB) conversion; DNA was first glucosylated to protect 5hmC, and then oxidized by TET1, followed by bisulfite treatment. Each replicate was then processed through the Illumina Infinium DNA methylation workflow and hybridized to 450 K arrays.
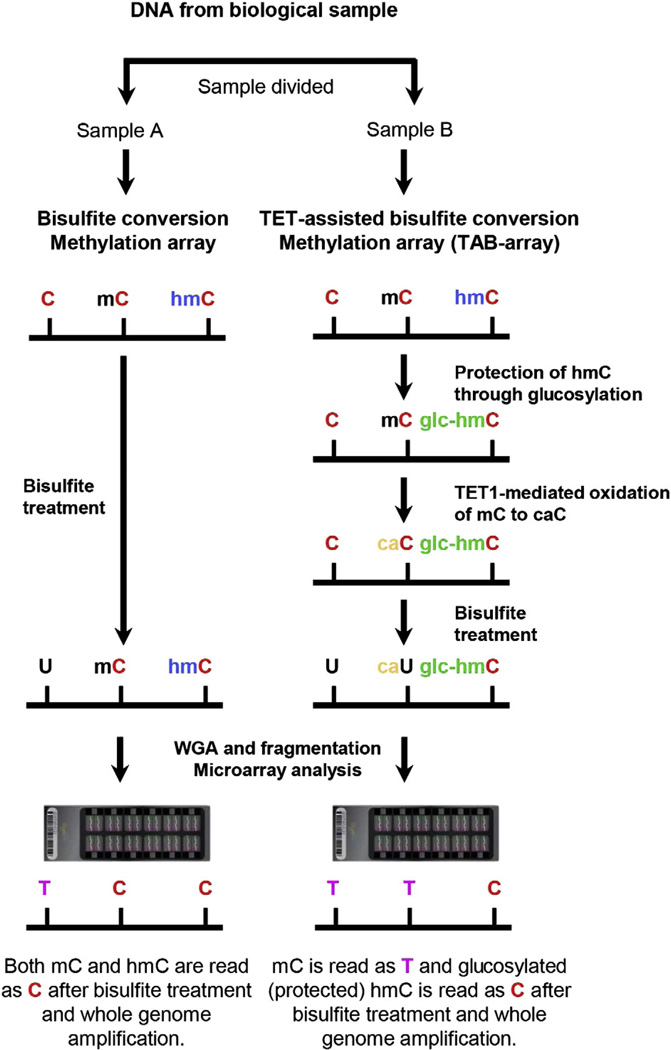
Figure 1.
Diagram of the TAB-array approach. Genomic DNA was prepared from each biological sample and split into two fractions. One fraction (Sample A) was bisulfite converted and analyzed by hybridization to the Illumina Infinium 450 K DNA Methylation BeadChip. Comparison to untreated DNA (not shown) allowed identification of methylated cytosines. The second fraction (Sample B) was glucosylated, protecting the 5hmC (hmC) but not the 5mC (mC) from oxidation by the TET1 enzyme. After bisulfite treatment, these samples are also hybridized to arrays. In the example, unmethylated cytosines were converted to thymidines by bisulfite treatment and amplification. In the absence of TAB treatment, both 5hmC and 5mC were protected and remain as cytosines. TAB treatment protected the 5hmC, while the 5mC was oxidized and converted by sodium bisulfite to thymidine. By comparing the hybridization results in the two samples, 5hmC could be distinguished from 5mC. WGA: whole genome amplification.
Glucosylation of 5hmC is a critical step in the TAB-array procedure. Therefore, we determined whether this treatment alone had any effects on DNA methylation. DNA was isolated from two biological replicates of human dermal fibroblasts (HDFs) and processed as described above, with the exception that the TAB treatment was terminated following glucosylation, prior to oxidation. Upon examination of the normalized Beta values in a scatterplot, we observed that the glucosylated samples were as similar to the untreated control samples as they were to each other (R ≥ 0.995, Fig. S1). Therefore, protection of 5hmC via glucosylation has no adverse effect on the stability of either 5hmC or 5mC.
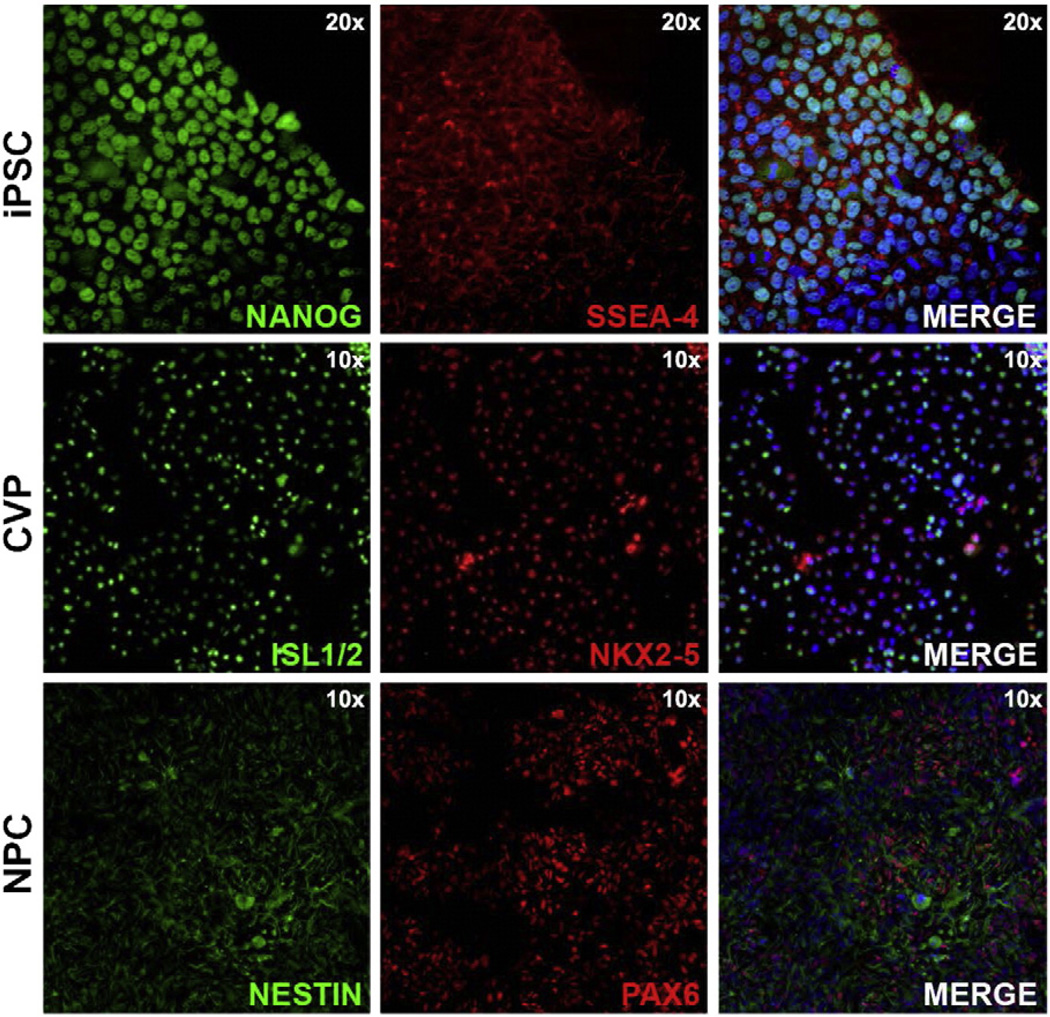
Considering that 5hmC levels have been reported to be very high in undifferentiated hPSCs and that hypomethylated genomic regions can be used to distinguish unrelated tissue types, we reasoned that 5hmC may play an important role in the activation of tissue-specific gene expression programs. To test this idea, we used an hPSC-based model system of lineage diversification. Specifically, two populations of human undifferentiated iPSCs were split three ways; a third of each population was harvested for profiling of the pluripotent state, while the two remaining fractions were kept in culture under conditions that promote differentiation into either cardiovascular (CVPs) or neural precursors (NPCs) for an additional 5 and 9 days, respectively. Undifferentiated cells were confirmed to be pluripotent as indicated by PluriTest™ [35] and the expression of the cell surface marker of SSEA-4 and NANOG (Fig. 2, top) [25,26]. In contrast, the CVPs expressed markers consistent with a CVP identity such as ISL-1 and NKX2.5 (Fig. 2, middle) [27,28]. The cells that differentiated along a neural lineage show expression of NESTIN and PAX6 (Fig. 2, bottom), as expected for NPCs. Undifferentiated iPSCs, CVPs, and NPCs were analyzed by the TAB-array approach outlined in Fig. 1.
Figure 2.
Characterization of undifferentiated pluripotent stem cells, cardiovascular progenitors and neural progenitors. Undifferentiated iPSCs were immunolabeled for pluripotency markers including NA NOG (top; green) and SSEA-4 (top; red). A merged image of NA NOG, SSEA-4 and a blue nuclear stain is shown at 20× magnification (top; right). CVPs were immunolabeled with ISL1/2 (middle; green) and NKX2.5 (middle; red). A merged image of ISL1/2, NKX2.5 and a blue nuclear stain is shown at 10× magnification (middle; right). NPCs were immunolabeled with NESTIN (bottom; green) and PAX6 (bottom; red). A merged image of NESTIN, PAX6 and a blue nuclear stain is shown at 10× magnification (bottom; right).
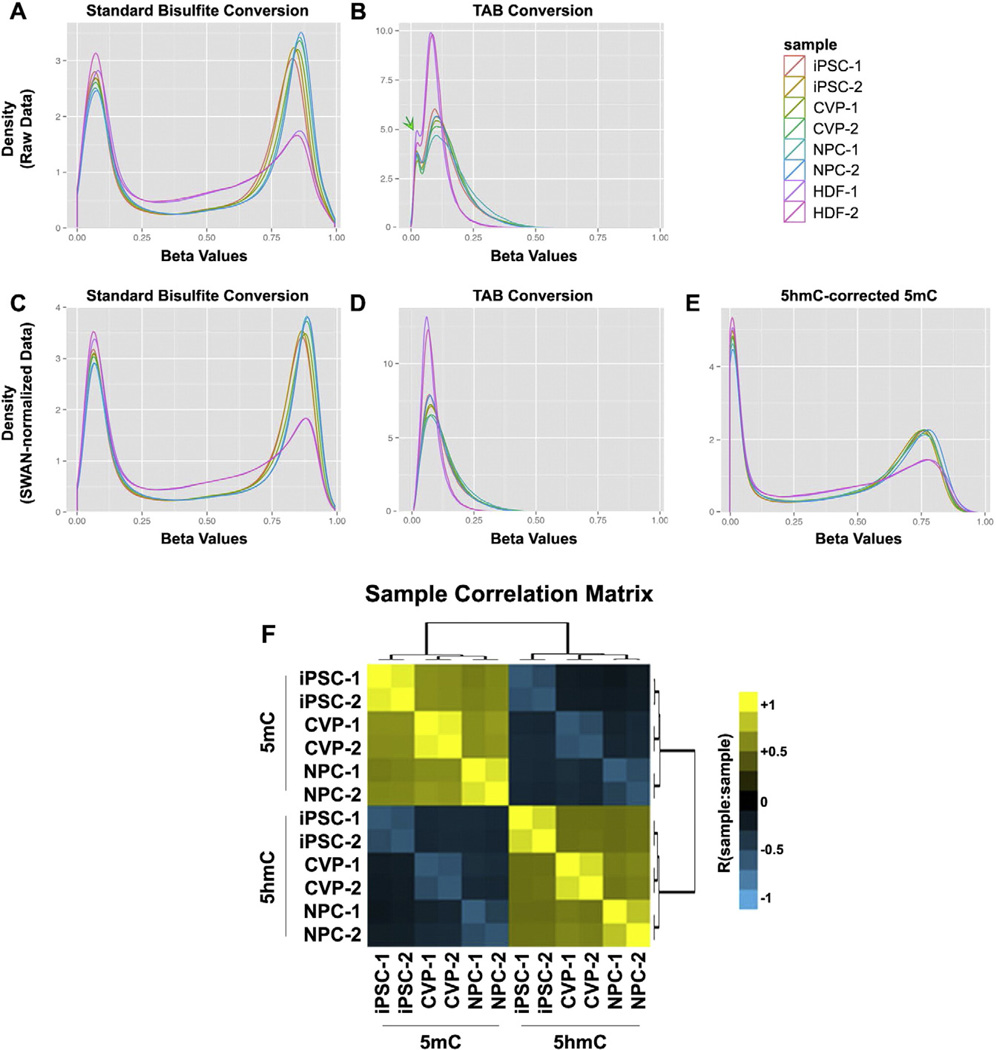
Prior to normalization and processing of the standard 450 K methylation and TAB-array data, we first examined the underlying distributions of the raw Beta values. As expected, the standard 450 K data fit a bimodal continuous distribution, with local maxima near the unmethylated (Beta = 0) and fully methylated (Beta = 1) extremes of the distribution (Fig. 3A). The TAB-array data appeared to more closely fit a unimodal distribution, with the exception of a small shoulder in the raw TAB-array samples density curve, near Beta = 0.05 (Fig. 3B, Fig. S2A). It was previously reported that approximately 80% of 5hmC was asymmetric across the genome, compared to only 8% for 5mC [14]. Therefore, we reasoned that the observed unimodal distribution was likely biological and not technical in nature. Given the striking differences in the distribution of these modifications, the standard 450 K and TAB-array data were independently normalized using the subsetquantile within array normalization (SWAN) method (Fig. 3C–D) [24].
Figure 3.
Normalization and identification of differentially methylated cytosines in TAB-array data. 5hmC and standard 5mC data were initially independently normalized using the SWAN method in R to generate Beta Values. For each pair of fractions from the same biological sample, 5hmC-specific Beta values were subtracted from the corresponding 5mC replicate Beta values across all probes in order to remove 5hmC signal from the 5mC data. Differentially methylated cytosines were called independently for 5mC and 5hmC data using Limma. (A) Global density plot for all standard bisulfite conversion samples, prior to normalization. (B) Global density plot for all TAB-samples, prior to normalization. The green arrow identifies a unique feature of the Beta density distribution that is further analyzed in Fig. S2. (C) Global density plot for all standard bisulfite conversion samples, after normalization. (D) Global density plot for all TAB-samples, after normalization. (E) Global density plot for all standard bisulfite conversion samples, following correction for 5hmC signal contribution. (F) A correlation matrix highlighting the relationship between all 5mC and 5hmC samples according to the top 50% most variable probes in the 450 K array.
Comparison of the raw and normalized TAB-array data showed the small shoulder, near Beta = 0.05 in the raw data, resolved with normalization (Fig. 3B,D, Fig. S2A). As this feature was not observed in the standard 450 K array data, we took a closer look at the probes comprising this shoulder in the density curve. As CpG density has been shown to be inversely correlated with DNA methylation [29], we calculated CpG density across a 250 bp window centered on each probe and binned the probes into pentiles of increasing CpG density (Fig. S2B–C). Re-plotting of the Beta density curves for these data according to CpG density pentiles showed that this small shoulder in the TAB-array data was primarily from probes in the fifth CpG density pentile, although a small bump could also be seen in probes of the fourth pentile (Fig. S2D, green arrows). Interestingly, a less pronounced bump in the standard density curve was also apparent in the fifth CpG density pentile (Fig. S2D, red arrow), which was not apparent in the data as a whole. Therefore, we concluded that this feature in the distribution of 5hmC Beta density was not technical artifact, but instead reflected an inverse correlation between 5hmC and CpG density that was stronger than that of 5mC and CpG density. It is noteworthy that SWAN makes CpG density-dependent assumptions about variance in its normalization procedure, further supporting the independent normalization of TAB-array and standard 450 K array methylation data.
Because dynamic epigenetic changes accompany differentiation, we reasoned that some of the most biologically relevant changes in DNA methylation would be missed if 5mC and 5hmC could not be distinguished. For example, 5hmC is enriched near bivalent chromatin, which can arise at any point in development as progenitor cells set the stage for successive fate choice [15]. In such cases, the “poising” of these bivalent promoters would likely be associated with the conversion of 5mC to 5hmC. However, without the ability to discriminate these two methylated forms, any losses of 5mC would be masked in the data by concomitant gains in 5hmC. To obtain a more accurate representation of the distribution of 5mC prior to statistical analyses of these data, we subtracted the TAB-array signal from each biological sample's corresponding signal that was generated using standard bisulfite conversion. This significantly altered the distribution of Beta values in the 5hmC-corrected 5mC data (henceforth, 5mC) (Fig. 3E). Less than 1.0% of all probes were overcorrected to a Beta value < −0.05 (4482/478767 probes), and less than 0.17% to a Beta value < −0.1 (806/478767 probes). Over-corrected probes were reset to a Beta value of 0.001, flagged, and processed with the remainder of the data. The sample correlation matrix in Fig. 3F is based on the top 50% most variable probes (~225,000) on the array. Among replicate samples, the lowest observed correlation was R = 0.8. For 5hmC the correlation was 0.87, 0.88 and 0.80 for iPSCs, CVPs and NPCs, respectively. The corrected 5mC correlations were 0.94, 0.94, and 0.90.
To examine dynamically methylated cytosines accompanying the differentiation of iPSCs into CVPs and NPCs, the 5hmC and 5mC data were analyzed using Limma in R [22,23]. In Limma, each probe is fitted to a linear model and empirical Bayes methods are used to smooth the standard errors, allowing for more accurate detection of differentially methylated probes or expressed genes. As the 5mC and 5hmC data had drastically different distributions, these data were each analyzed separately to avoid over- and under-estimation of error in the 5mC and 5hmC data, respectively. Based on our experience in analyzing standard Illumina 450 K Methylation array data, we chose cutoffs at p < 0.01 for all comparisons and also required that ΔBeta ≥ 0.3 for 5mC, and ΔBeta ≥ 0.2 for 5hmC. Additionally, the uncorrected 5mC data were processed in the same manner as the corrected 5mC data for quality assurance.
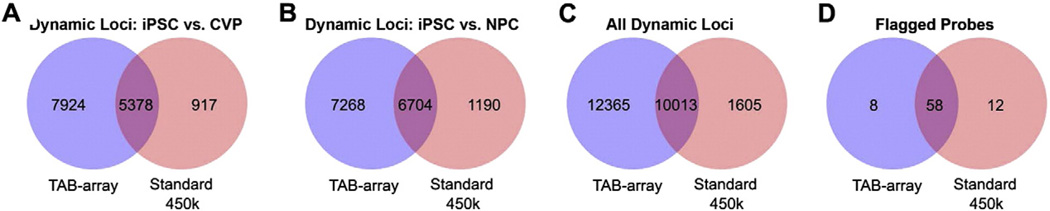
Fig. 4 compares the outcome of the TAB-array method to standard bisulfite conversion. Both approaches identified a common set of 5378 (A), 6704(B), and 10013(C) dynamically methylated loci, in CVPs, NPCs or in the union of the two, respectively (Table S1). The TAB-array method identified an additional 7924, 7268, 12365 dynamically methylated loci, in CVPs, NPCs or in the union of the two, compared to only 917, 1190, and 1605 unique to the standard method. In total, application of the TAB-array approach and correction of 5mC Beta values led to identification of twice as many differentially methylated loci compared to standard methods ((12365 + 10013) / (1605 + 10013) Fig. 4C). Of the probes that were uniquely significant using standard methods, 75% (1240/1650) would have been called significant in the 5mC TAB-array data at a slightly reduced ΔBeta threshold (≥0.2). However, only 33% (4092/12365) of the probes that were uniquely significant in the TAB-array data would have been called significant in the standard bisulfite data at this reduced threshold. We determined that the overcorrection of 5mC values during the normalization process had negligible impact on analytical outcome, as only 20/78 significant flagged probes were uniquely significant in either the TAB array or standard 450 K data (Fig. 4D).
Figure 4.
Comparison of statistical results from analysis of standard 450 K array and TAB-array data. Venn diagrams were used to plot the overlap in significantly dynamic loci identified in standard 450 K array data and TAB-array data for (A) iPSC vs. CVPs, (B) iPSC vs. NPCs, (C) the union of these two analyses, and (D) probes that were flagged as being overcorrected.
In order to determine the functional relevance of these dynamically regulated loci, we used a method designed for the identification of covariant molecular networks in gene expression data, weighted gene correlation network analysis (WGCNA) [30]. Although this method works best with linear scale gene expression data, running the current analysis at a high, soft thresholding power permitted approximation of the scale-free topology required of the technique (Fig. S3A–B, Supplementary Methods).We reasoned that through the identification of highly correlated patterns of dynamic methylation, the underlying genomic regions would be enriched for genes converging on specific pathways or biological processes critical in the establishment of the cardiovascular and neural lineages.
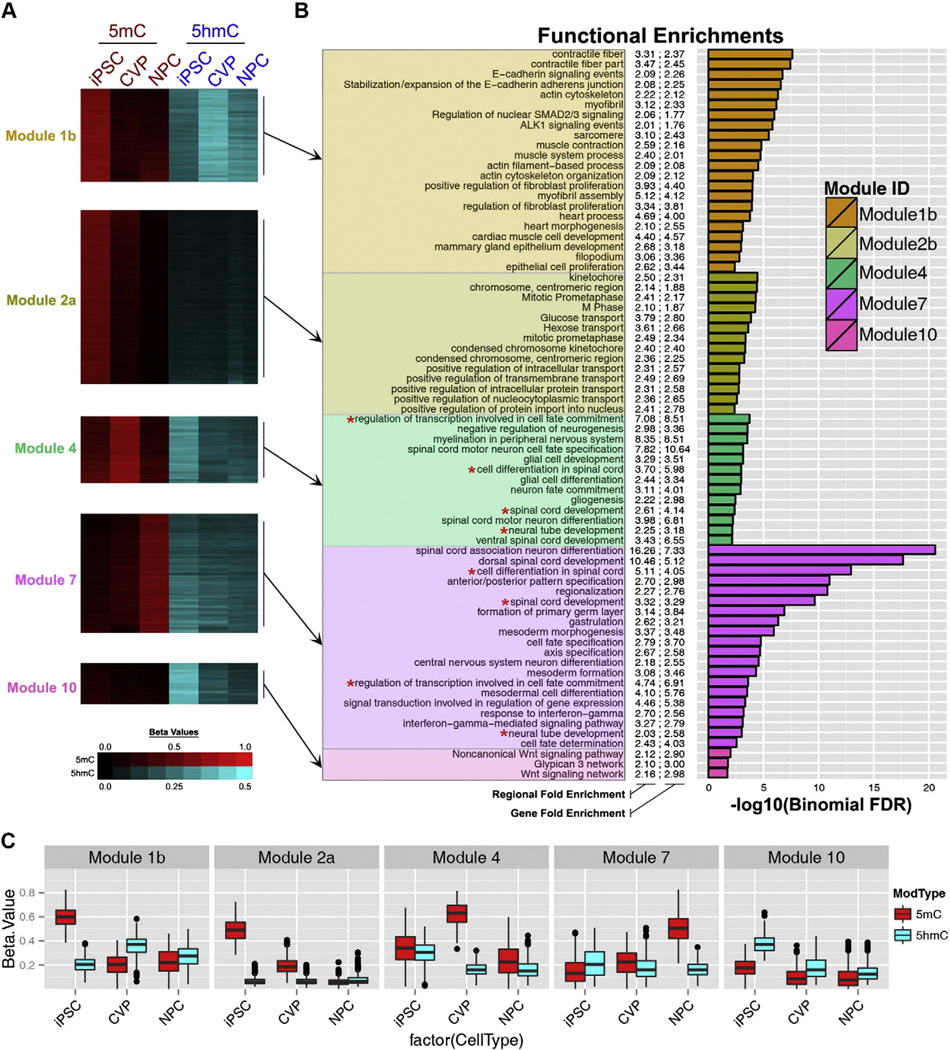
Initially, we identified 10 modules of covariant cytosines that were tested for functional enrichments using the genomic regions enrichment of annotations tool (GREAT) [31]. Analysis of each module's associated enrichments suggested that some modules could perhaps be resolved into multiple modules with distinct characteristics. Using targeted hierarchical clustering of cytosines within a given module, we generated two subclusters from each of modules 1, 2, and 6 that shared more functional enrichments compared to the complete module from which they were derived (Fig. S3C–E). In the end, we distinguished 13 modules with distinct patterns of 5mC and 5hmC (Table S2). Most of the 13 modules were dominated by processes relevant to pluripotent stem cells or to the development of the cardiovascular and neural lineages. Five of these modules were particularly noteworthy (Fig. 5), although many of the remaining modules were also interesting (Fig. S4).
Figure 5.
Functional enrichments for covariant DNA-methylation modules. Cytosines with significant differences in either 5hmC or 5mC levels among iPSCs, CVPs and NPCs were identified using Limma and subsequently binned into covariant modules using WGCNA. Beta values and functional enrichments for exemplary modules are shown. (A) Heatmap of Beta values for cytosines in Modules 1b, 2a, 4, 7, and 10. (B) Functional enrichments for each module as determined by GREAT. 5mc Beta values are plotted from black to red and 5hmC Beta values are plotted from black to cyan. For functional enrichments, both region and gene based fold-enrichment values are shown and the length of each bar represents the significance of the enrichment (-log10(binomial FDR)). (C) Box and whisker plots of Beta values of all cytosines in each of the modules in panels A–B. The corresponding data for modules not shown in this figure are provided in Fig. S4.
In iPSCs, the cytosines comprising Module 1b exhibited high levels of 5mC and low levels of 5hmC (Fig. 5A,C). Upon differentiation, 5mC levels were dramatically reduced in both lineages, with concomitant gains in 5hmC. Distribution of these data showed that CVP-5hmC levels were significantly higher than NPC-5hmC levels, with the median Beta for CVPs greater than the third quartile in NPCs (Fig. 5C). As expected, Module 1b was enriched for functions specific to derivatives of CVPs, such as muscle contraction and cardiac muscle cell development (Fig. 5B, Table S2). This pattern of dynamic methylation is consistent with the establishment of lineage-specific euchromatin in a progenitor cell type.
Module 2a was completely devoid of 5hmC in all cell types. However, these cytosines exhibited very high levels of 5mC in iPSCs that were reduced to very low levels in CVPs and completely abolished in NPCs (Fig. 5A, Fig. 5C). This module was almost completely devoid of CpGs (Fig. 6A) and contained a large number of genes categorized as cell-cycle associated chromosome condensation and nucleocytoplasmic protein transport (Fig. 5B, Table S2). Module 10 was also specific to undifferentiated iPSCs, but was characterized by high levels of 5hmC and low levels of 5mC (Fig. 5A,C). Upon differentiation, 5hmC levels were diminished to low levels in CVPs and to slightly lower levels in NPCs. Intriguingly, these cytosines were often found in genomic regions associated with Wnt signaling, which is critical to the exit from pluripotency in the early stages of both CVP and NPC differentiation (Fig. 5B). Therefore, we reasoned that this pattern of change is consistent with the proposed role for 5hmC in developmental poising of lineage-specifying factors.
Figure 6.
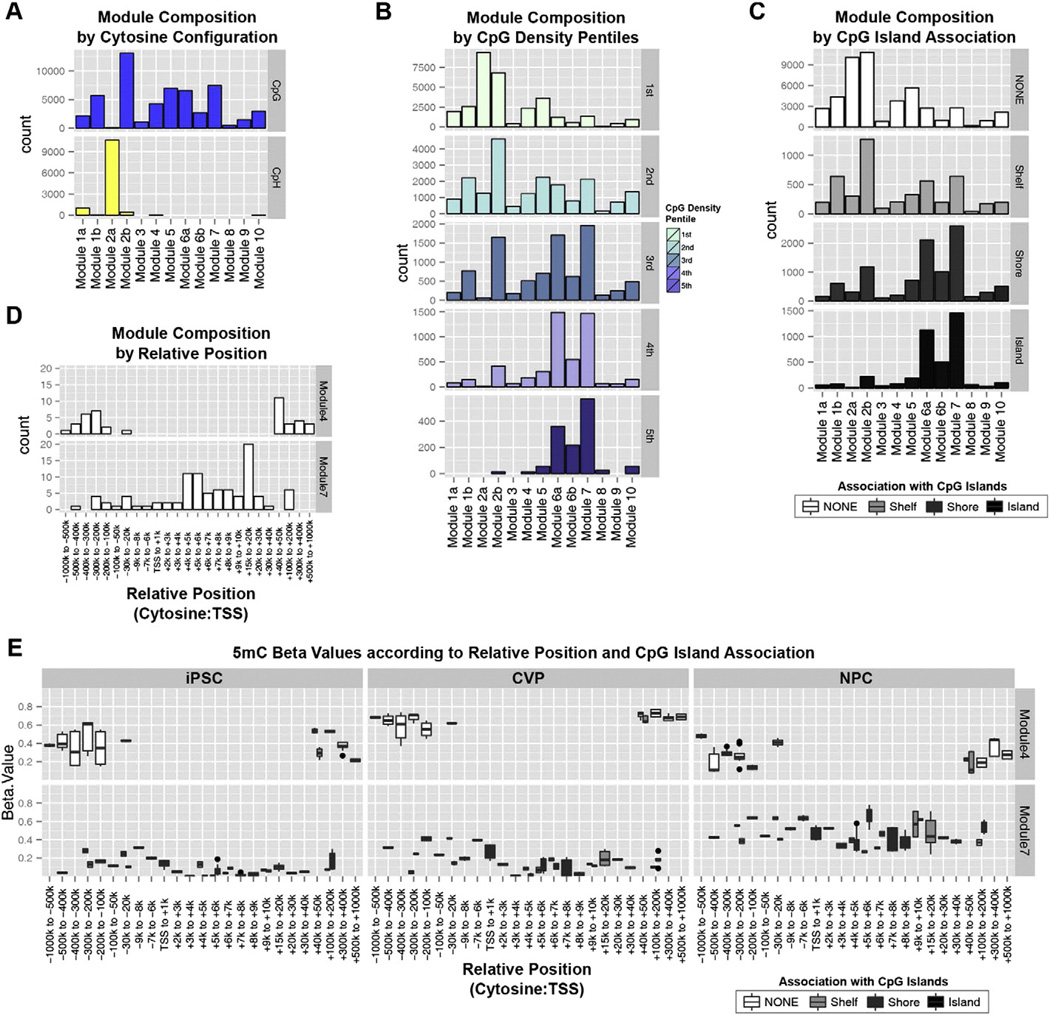
Definition of DNA methylation modules according to multiple parameters. Each module composition is defined according to (A) cytosine configuration, (B) CpG density pentiles, and (C) association with CpG islands. (D) Genomic position relative to the TSS of each cytosine's associated transcripts. (E) 5mC Beta values for the collection of cytosines associated with functional enrichment terms that were significant in both Modules 4 and 7. These data are plotted according to each cytosine's position relative to the transcriptional start site of its associated transcript, and its association with CpG islands.
Similar to Module 10, Module 4 cytosines had high levels of 5hmC in iPSCs, but also had nearly equivalent levels of 5mC (Fig. 5A,C). Upon differentiation, CVPs experienced highly significant gains of 5mC and losses of 5hmC, while both of these marks were significantly reduced in NPCs (Fig. 5C). These NPC-hypomethylated genomic regions were dominated by neurodevelopmental processes, especially those associated with motor neuron differentiation and spinal cord development (Fig. 5B, Table S2). Interestingly, the NPC differentiation protocol used here has recently been shown to correct the paralytic effects in a demyelinated mouse model of multiple sclerosis following NPC intraspinal transplantation [32]. Therefore, our epigenetic analyses are consistent with experimental evidence that these NPCs are particularly suited for stimulating spinal cord repair and gliogenesis.
The pattern of methylation in Module 7 was most clearly characterized by high levels of 5mC in NPCs (Fig. 5A). Additionally, we observed intermediate levels of 5hmC in iPSCs that were slightly reduced in both CVPs and NPCs upon differentiation (Fig. 5C). Surprisingly, the most significant enrichments for this module paralleled those of Module 4, with several of the enrichment terms exactly matched between these modules (Fig. 5B, red asterisks). Further, 13/59 genes within these matched terms were associated with cytosines from both Module 4 and Module 7. Of these genes, 11/13 were potent lineage-specifying transcription factors (DLX2, GLI3, HES1, LHX1, LHX5, NKX6-2, NR2F2, PAX3, PAX6, SKI, SOX4) and two were critical players in neuronal development and migration (COBL, RGMA). Given the disparate patterns of methylation characteristic of Modules 4 and 7, we reasoned that the cytosines within these modules must be distinguished by underlying sequence determinants and/or genomic positioning relative to each associated gene's transcriptional start site (TSS). Examining the composition of these modules according to CpG density, we found that Module 4 cytosines were primarily found in CpG-poor regions. Conversely, Module 7 contained more cytosines in CpG dense regions and CpG islands than any other module (Fig. 6B–C). Therefore, Module 4 and 7 cytosines can be distinguished according to CpG density.
Next we focused our analysis on 68 cytosines from Modules 4 and 7 that were associated with the overlapping set of 13 genes from the functional enrichment results. Module 4 cytosines were nearly exclusively found in regions 100–1000 kb upstream or 40–1000 kb downstream of each genes TSS, with a single cytosine 20–30 kb upstream (Fig. 6D, top). On the contrary, the vast majority of Module 7 cytosines were localized to regions immediately downstream of the TSS (Fig. 6D, bottom). The distinctions between Modules 4 and 7 are summarized in a plot of 5mC Beta values for these 68 cytosines in iPSCs, CVPs and NPCs according to the annotations from Fig. 6D (x-axis) and Fig. 6C (split boxplots to color scale). Fig. 6E shows Module 4 cytosines within CpG-poor gene distal regions, and Module 7 cytosines within CpG-rich gene proximal regions downstream of the TSS. The pattern of dynamic methylation in the distal and CpG-poor regions of Module 4 is consistent with developmental poising in iPSCs (Fig. 5C, 6E). Upon differentiation, these poised regions undergo heterochromatic resolution in CVPs and euchromatic resolution in NPCs with concomitant increases in 5mC methylation across gene body elements (Module 7).
3. Discussion
DNA methylation is critical to the regulation of both developmental and homeostatic processes. Well-established technologies have shed light on how the interplay between DNA methylation, histone modifications and non-coding RNA expression maintain the fidelity required of these processes. Recently, an oxidized derivative of DNA methylation, 5hmC, was found to be widespread throughout the genomes of hPSCs and differentiated cell types, introducing a new major player in the regulation of epigenetics. To date, targeted studies of 5hmC have been limited, in part because these studies have relied on cost-prohibitive DNA sequencing. Furthermore, discrimination of 5mC and 5hmC in sequencing data is inherently complex, requiring deconvolution of 4 distinct signals from standard bisulfite (C, 5mC + 5hmC) and TAB-seq data (C + 5mC, and 5hmC). The cost required to generate these data and bioinformatics expertise required to analyze them limit the accessibility of 5hmC-targeted studies for many researchers. The TAB-array method described here will allow for the widespread incorporation of 5hmC-targeted studies by the research community, providing a cost effective means to generate single base resolution 5mC and 5hmC specific data that can be easily analyzed within the R statistical environment.
Our analysis of 5mC and 5hmC dynamics in the in vitro differentiation of iPSCs into CVPs or NPCs highlights the utility of the TAB-array method. At the most basic level, we were able to identify nearly twice as many dynamically methylated cytosines in our differentiation model compared to those identified by standard measures. More specifically, the discrimination of 5mC and 5hmC allowed for the identification of complex patterns of these two modifications that were suggestive of their functional interplay. An example is the convergence of methylation Modules 4 and 7 on genes associated with spinal cord development. Within Module 4, CpG regions distal to spinal cord genes appear poised, due to the presence of both 5mC and 5hmC at these locations. Module 7, however, lacked appreciable levels of 5hmC in iPSCs, but contained significant gene body methylation in CpG dense regions of the same spinal cord genes. An in vivo study in mouse embryos showed 5hmC enrichment in gene bodies of genes associated with dorsal forebrain neuronal fates [33]. In contrast, we did not observe 5hmC enrichment in gene bodies in our human NPCs in any of the modules; however, this is not surprising, since the NPCs we studied were not forebrain precursors, but rather a unique in vitro population that was effective in restoring function in a model of multiple sclerosis [32].
The implication of function for non-CpG methylation in pluripotent stem cells is another example of the analytical strength afforded by the TAB-array approach. Specifically, the sub-clustering of Module 2 to generate Modules 2a and b was based on the absence of 5hmC at cytosines from Module 2a. This distinction, which indicated a highly significant and uniform enhancement of genes regulating cell-cycle associated chromosome condensation and nucleocytoplasmic protein transport, would not have been possible if the analysis were restricted to standard 5mC data from the 450 K array. In conclusion, our study supports diverse roles for 5hmC, with evidence of both developmental poising and establishment of euchromatin. Additionally, sequence determinants such as CpG density, gene-relative genomic location and cytosine configuration (CpG vs CpH) underlie the localization of 5mC vs 5hmC modifications in differentiating human cells.
4. Materials and methods
4.1. Cell culture and characterization
The iPSC line used for these studies was derived from dermal fibroblasts from donor 68 [34], using Sendai virus (Cytotune®, Life Technologies) according to the manufacturer's instructions. Cells were tested for pluripotency using embryoid body formation, teratoma formation and PluriTest [35] (data not shown). Cells were adapted to feeder-free conditions on Geltrex® (Life Technologies), expanded in StemPro® culture medium (Life Technologies), and passaged with Accutase (Life Technologies).
For differentiation into CVPs, iPSCs were seeded at 125,000 cells per well of a 6 well plate coated with bovine collagen type 1 (BD Bioscience) at 8 µg/cm2. Cells were cultured for 5 days in cardiac differentiation medium (Genea Biocells), changing the medium every other day.
Differentiation of NPCs from iPSCs was carried out using a specific protocol that produced cells that restored function after transplant to a mouse model of multiple sclerosis [32]. Briefly, 1 × 104 cells per cm2 were seeded on Matrigel (BD Biosciences)-coated 6 well plates in StemPro® medium (Life Technologies) for 24 hours. The medium was then changed to NPC differentiation medium which contains the following: DMEM/F-12 (Life Technologies), 20% Knockout Serum Replacement (Life Technologies), 1 × Glutamax™ (Life Technologies), 0.1 mM 2-mercaptoethanol (Life Technologies), 20 ng/mL midkine (EMD-Millipore), 2 µM dorsomorphin (Sigma-Aldrich), 2 µM A 83-01 (Tocris), and 2 µM PNU-74654 (Sigma-Aldrich). Cells were Accutase® passaged (1:3) on days 3 and 6 and harvested on day 9.
4.2. Immunocytochemistry
Cells were fixed in 4% paraformaldehyde for 15 minutes at room temperature. After blocking in 5% FBS, cells were incubated with primary antibodies against Nanog (Cell Signaling, 1:400), SSEA4 (Cell Signaling, 1:400), Pax6 (Developmental Studies Hybridoma Bank, 1:100) and Nestin (Millipore, 1:15000). Cells were incubated with diluted secondary antibodies conjugated to AlexaFluor 488 and 555 (Life Technologies, 1:1000). Cells were washed twice and mounted using 4',6-diamidino-2-phenylindole (DAPI)-containing Vectashield® mounting medium (Vector Laboratories). For CVP immunolabeling, cells were fixed in formalin for 20 minutes and blocked in 1% BSA. Cells were incubated with primary antibodies including Isl1/2 (Developmental Studies Hybridoma Bank, 1:200) or Nkx2.5 (Santa Cruz Biotechnology, sc-14033, 1:200) at room temperature for 1 hour. Cells were then incubated with secondary antibodies conjugated with AlexaFluor 488 and 594 (Life technologies, 1:1000) for 1 hour at room temperature.
4.3. DNA isolation
Two wells of a 6well plate were harvested to generate two biological replicates for each sample type. Cells were removed from the dish using Accutase and DNA was isolated using a DNeasy kit (Qiagen) according to the manufacturer's recommendations. DNA was quantified using a Qubit and dsDNA BR Reagent (Life Technologies).
4.4. TAB conversion
For Tet-Assisted bisulfite conversion, 1 µg of genomic DNA with C/5mC/5hmC spike-in controls was first sheared, then glucosylated and oxidized as reported [14] before subsequent bisulfite treatment. The 1 µg of genomic DNA containing 0.5% M. SssI methylated lambda DNA and 0.25% hydroxymethylated pUC19 control was sheared to an average of 2,000 bp by sonication (Covaris). The glucosylation reaction was performed in a 20 µl volume containing 50 mM HEPES (pH 8.0), 25 mM MgCl2, 50 ng/µl DNA, 200 mM UDP-Glc, and 2 µM βGT. The reaction was incubated at 37 °C for 1 hr. The glucosylated DNA was purified with QIAquick PCR Purification Kit (Qiagen). The oxidation reaction was performed in two 50 µl aliquots containing 50 mM HEPES (pH 8.0), 100 mM ammonium iron (II) sulfate, 1 mM α-ketoglutarate, 2 mM ascorbic acid, 2.5 mM DTT, 100 mM NaCl, 1.2 mM ATP, 6 ng/µl glucosylated DNA, and 5 µM recombinant Tet1. The reaction was incubated at 37 °C for 75 min. After proteinase K treatment, the oxidized DNA was purified with Micro Bio-Spin P-30 Gel Column (Bio-Rad) and QIAquick PCR Purification Kit (Qiagen).
4.5. Bisulfite conversion and DNA processing
The Bisulfite conversion was performed using the EZ DNA Methylation Kit (Zymo Research) according to the manufacturer's instructions. Whole genome amplification, fragmentation and preparation of the DNA for hybridization were performed using the Infinium HumanMethylation450 BeadChip kit (Illumina) as described in the manufacturer's protocol.
4.6. Data normalization
Illumina IDATS were read directly into R and normalized using Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Details about the normalization procedure and correction of 5mC Beta values are provided in the supplementary methods.
4.7. Identification of dynamic methylation
All analyses were performed using Limma in R. Details of these procedures are provided in the supplementary methods.
4.8. Identification of DNA methylationmodules and functional enrichments
DNA methylation modules were identified using WGCNA in R and functional enrichments were identified using GREAT. Details of these procedures are provided in the supplementary methods.
4.9. Data accession
Raw and normalized data have been deposited in the GEO repository under the accession GSE60225.
Supplementary Material
Acknowledgments
We thank the members of the Loring lab for their many insightful discussions, especially Yu-Chieh Wang and Eyitayo S. Fakunle. We acknowledge the contributions of Leslie Caron at Genea BioCells. JFL, KLN, SEP, MJB, VLGP, JPS, and RLC are supported by grants from the California Institute for Regenerative Medicine [CIRM; CL1-00502, TR01250, RM1-01717 (Loring); TR3-05603 (Loring and T. Lane); and RB3-05022 (J. Gottesfeld)], and the NIH [5R33MH087925-04 (Loring) and 1R21 DA032975-01 (P. Sanna and Loring), R01 HG006827 (C. He)]. KLN is supported by an Autism Speaks Fellowship. RLC is supported by a Ken and Karen Craven Multiple Sclerosis Fellowship. MY is a HHMI International Predoctoral Fellow. We wish to also acknowledge Y. Sasai's inspiring work on in vitro embryonic development.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ygeno.2014.08.014.
Contributor Information
Jeanne F. Loring, Email: jloring@scripps.edu.
Jian-Bing Fan, Email: JFan@illumina.com.
References
- 1.Altun G, Loring JF, Laurent LC. DNA methylation in embryonic stem cells. J. Cell. Biochem. 2010;109:1–6. doi: 10.1002/jcb.22374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, Wei CL. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, Garitaonandia I, Muller FJ, Wang YC, Boscolo FS, Fakunle E, Dumevska B, Lee S, Park HS, Olee T, D'Lima DD, Semechkin R, Parast MM, Galat V, Laslett AL, Schmidt U, Keirstead HS, Loring JF, Laurent LC. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Y, Chen J, Li K, Wu T, Huang B, Liu W, Kou X, Zhang Y, Huang H, Jiang Y, Yao C, Liu X, Lu Z, Xu Z, Kang L, Wang H, Cai T, Gao S. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell. 2013;12:453–469. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Wang T, Wu H, Li Y, Szulwach KE, Lin L, Li X, Chen IP, Goldlust IS, Chamberlain SJ, Dodd A, Gong H, Ananiev G, Han JW, Yoon YS, Rudd MK, Yu M, Song CX, He C, Chang Q, Warren ST, Jin P. Subtelomeric hotspots of aberrant 5-hydroxymethylcytosine-mediated epigenetic modifications during reprogramming to pluripotency. Nat. Cell Biol. 2013;15:700–711. doi: 10.1038/ncb2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 12.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, Eberl HC, Mensinga A, Brinkman AB, Lephikov K, Muller U, Walter J, Boelens R, van Ingen H, Leonhardt H, Carell T, Vermeulen M. Dynamic readers for 5-(hydroxy) methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, Yoon YS, Ren B, He C, Jin P. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B, He C. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S, Rao A. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, Fan JB, Shen R. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; [Google Scholar]
- 23.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 24.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct 4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 26.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 27.Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119:969. doi: 10.1242/dev.119.3.969. [DOI] [PubMed] [Google Scholar]
- 28.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, Evans SM, Laugwitz KL, Chien KR. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 29.Pelizzola M, Koga Y, Urban AE, Krauthammer M, Weissman S, Halaban R, Molinaro AM. MEDME: an experimental and analytical methodology for the estimation of DNA methylation levels based on microarray derived MeDIP-enrichment. Genome Res. 2008;18:1652–1659. doi: 10.1101/gr.080721.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Coleman R, Leang R, Tran H, Kopf A, Walsh CM, Sears-Kraxberger I, Steward O, Macklin WB, Loring JF, Lane TE. Human neural precursor cells promote neurologic recovery in a viral model of multiple sclerosis. Stem Cell Rep. 2014;2:825–837. doi: 10.1016/j.stemcr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, Lu Q. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laurent LC, Nievergelt CM, Lynch C, Fakunle E, Harness JV, Schmidt U, Galat V, Laslett AL, Otonkoski T, Keirstead HS, Schork A, Park HS, Loring JF. Restricted ethnic diversity in human embryonic stem cell lines. Nat. Methods. 2010;7:6–7. doi: 10.1038/nmeth0110-06. [DOI] [PubMed] [Google Scholar]
- 35.Muller FJ, Schuldt BM, Williams R, Mason D, Altun G, Papapetrou EP, Danner S, Goldmann JE, Herbst A, Schmidt NO, Aldenhoff JB, Laurent LC, Loring JF. A bioinformatic assay for pluripotency in human cells. Nat. Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.