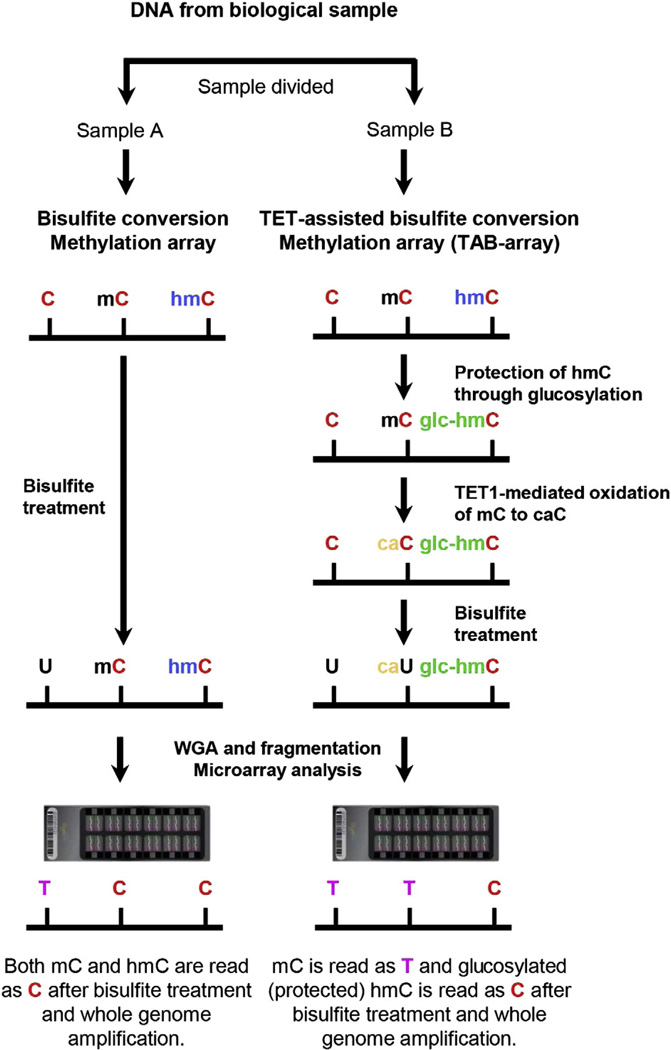
Figure 1.
Diagram of the TAB-array approach. Genomic DNA was prepared from each biological sample and split into two fractions. One fraction (Sample A) was bisulfite converted and analyzed by hybridization to the Illumina Infinium 450 K DNA Methylation BeadChip. Comparison to untreated DNA (not shown) allowed identification of methylated cytosines. The second fraction (Sample B) was glucosylated, protecting the 5hmC (hmC) but not the 5mC (mC) from oxidation by the TET1 enzyme. After bisulfite treatment, these samples are also hybridized to arrays. In the example, unmethylated cytosines were converted to thymidines by bisulfite treatment and amplification. In the absence of TAB treatment, both 5hmC and 5mC were protected and remain as cytosines. TAB treatment protected the 5hmC, while the 5mC was oxidized and converted by sodium bisulfite to thymidine. By comparing the hybridization results in the two samples, 5hmC could be distinguished from 5mC. WGA: whole genome amplification.